Abstract
While advances in network and pathway analysis have flourished in the era of genome-wide association analysis, understanding the genetic mechanism of individual loci on phenotypes is still readily accomplished using genetic modeling approaches. Here, we demonstrate two novel genotype-phenotype models implemented in a flexible genetic modeling platform. The examples come from analysis of families with specific language impairment (SLI), a failure to develop normal language without explanatory factors such as low IQ or inadequate environment. In previous genome-wide studies, we observed strong evidence for linkage to 13q21 with a reading phenotype in language-impaired families. First, we elucidate the genetic architecture of reading impairment and quantitative language variation in our samples using a bivariate analysis of reading impairment in affected individuals jointly with language quantitative phenotypes in unaffected individuals. This analysis largely recapitulates the baseline analysis using the categorical trait data (posterior probability of linkage (PPL) = 80%), indicating that our reading impairment phenotype captured poor readers who also have low language ability. Second, we performed epistasis analysis using a functional coding variant in the brain-derived neurotrophic factor (BDNF) gene previously associated with reduced performance on working memory tasks. Modeling epistasis doubled the evidence on 13q21 and raised the PPL to 99.9%, indicating that BDNF and 13q21 susceptibility alleles are jointly part of the genetic architecture of SLI. These analyses provide possible mechanistic insights for further cognitive neuroscience studies based on the models developed herein.
Key Words: Genetic epistasis, Genetic association study, Genetic linkage, Penetrance, Bayesian analysis
Introduction
The hallmark of complex disease genetics is a lack of determinism between phenotype and the underlying susceptibility genotype(s). Unlike Mendelian diseases with underlying genes of major effect, where the relationship between phenotype and genotype may be complex but is largely deterministic and readily modeled by the concept of penetrances and disease allele frequencies, complex diseases lack such clear major effects. Unsurprisingly, statistical models used to map complex disease susceptibility loci have largely abandoned the concept of uniquely determined effects parameterized in terms of penetrance and disease allele frequencies for models that emphasize the overall strength of the genetic effect. Such models therefore ignore the specific allelic effects that underlie the relative risk defined by Risch [1], as one very common example, since any given relative risk value may have infinite combinations of multi-locus penetrances and susceptibility allele frequencies. However, the concept of modeling deterministic relationships is not without merit in complex disease under certain circumstances. Rather than focusing on mapability, where perhaps the strength of the effect is the main predictor of success, employing specific models of the genotype-phenotype relationship after evidence for a locus has been determined remains an underutilized tool in complex disease genetics.
Along these lines, we demonstrate a flexible genetic modeling platform (described in detail below) that allows interrogation of specific genotype-phenotype relationships in a collection of large extended pedigrees ascertained for specific language impairment (SLI). SLI is a neurodevelopmental disorder clinically defined as a significant delay in the acquisition and/or use of language despite adequate education. Classifying a subject as having SLI excludes a history of mental retardation, neurological or psychiatric impairments and speech-motor or gross sensory deficits that could have affected language acquisition. Affecting approximately 7% of children entering into kindergarten [2], SLI is associated with generally poor academic outcomes if unresolved [3, 4, 5]. Phenotypic presentation is heterogeneous. A large proportion of individuals with SLI will exhibit difficulty with some aspects of higher level phonological processing necessary for the development of both language and reading, while grammatical deficits are also commonly reported [for review see [6]]. Some subjects demonstrate concomitant difficulties in processing dynamic (rapidly changing) sensory information within a very brief time range [7, 8, 9, 10, 11, 12, 13]. Although some individuals with SLI appear to largely compensate as adults, many do not [4, 14, 15, 16, 17, 18, 19].
Familial aggregation studies, twin studies and prospective studies collectively indicate that SLI is heritable. Numerous case-control familial aggregation studies of SLI have previously been reported, and despite SLI incidence rates varying due to differences in the applied definition of SLI, the frequency of impairment is significantly increased in first-degree relatives in families containing a proband (18–42%) versus control families (3–26%) [20, 21, 22, 23, 24]. When using the categorical diagnosis of SLI, twin studies demonstrate near 100% concordance for monozygotic twins and ∼50–70% concordance for dizygotic twins [25, 26], which shows that SLI as defined by categorical affection status does indeed have a genetic component. DeThorne et al. [27] determined heritability of language difficulty to be 0.54, which was congruent with previously reported heritability rates [28, 29]. An additional Twin Early Development Study determined the heritability of normal language in a set of 4-year-old twins to be 0.39 [30].
Prior linkage studies have found LOD scores greater than 3 at loci on chromosomes 13, 16, and 19 [31, 32, 33, 34]. The SLI Consortium found the locus on chromosome 16 with a non-word repetition measure (non-words are pseudowords that are consistent with standard English pronunciation) and the locus on chromosome 19 using an expressive language score trait [33]. Two years later, in 2004, an independent sample replicated the locus on chromosome 16, increasing the total LOD score from 3.6 to 7.5 [34]. However, the picture on chromosome 19 was more difficult to interpret since the replication sample only showed linkage with non-word repetition as opposed to the discovery sample that used the expressive language score. Pooling all samples for analysis with either univariate trait reduced the LOD to background noise. However, later investigations using multivariate analysis [35] demonstrated that the best-fitting model for the chromosome-19 locus gave greater weight to the expressive language score trait over the non-word repetition trait and indicated that differences seen in the 2004 linkage analysis were due primarily to the differences in ascertainment between the two samples sets. Evidence suggests that variations in both c-maf-inducing protein (CMIP) and calcium-transporting ATPase, type2C, member2 (ATP2C2) genes have independent effects that are responsible for the observed linkage to chromosome 16 [36]. In a separate line of inquiry using these samples, a marginally significant association was seen between the contactin-associated protein-like 2 gene, CNTNAP2, and SLI [37]. This gene is of interest due to its regulation by FOXP2, a protein known to influence human speech development, though it is not commonly mutated in developmental dyspraxias, autism or language impairments [38, 39, 40, 41, 42, 43].
Our group implicated 13q21 as a region containing an SLI susceptibility allele with an initial HLOD of 3.92 (posterior probability of linkage (PPL) of 53%) using a reading impairment phenotype [32]; however, a common language impairment phenotype did not provide evidence for linkage. Addition of a second group of families selected for SLI increased the PPL to 92% using the same reading impairment phenotype, thus supporting a susceptibility allele at 13q21 [31], though again, no linkage was observed with a language impairment phenotype. Despite very strong evidence for linkage to the region in a sample ascertained for language deficits, it has remained unclear why a reading impairment phenotype would provide stronger evidence for linkage in an SLI sample, i.e., a sample selected for language deficits, than a language impairment phenotype. While these findings are consistent with prior behavioral studies that showed a link between impaired speech and language abilities with lower reading scores [44], it remains to be explained if the 13q21 locus is related primarily to language, primarily to reading, or some combination thereof.
As the relationship between reading, language and the susceptibility allele on 13q21 clearly requires further elucidation, we sought ways to model this complex genotype-(multiple) phenotype relationship directly by constructing models that have strong assumptions and by assessing if the data are consistent with those assumptions. The PPL statistical framework has been extended to allow for joint analysis of clinical diagnosis (or other categorical phenotype) and a putatively related quantitative trait [45]. This analysis has been successfully applied to datasets where (1) the relationship between the diagnosis and the quantitative trait was unknown (alcoholism diagnosis and EEG [45]); (2) quantitative assessment could not be completed for some individuals (autism diagnosis and parent's broader autism phenotype score), or (3) treatment effects render the quantitative trait uninformative in affected subjects, but the trait remains informative in the untreated/unaffected persons (autoimmune thyroid disease and serum auto-antibodies [46]). This method models the categorical phenotype as being related to the quantitative trait through an unspecified threshold, thus dichotomizing the quantitative trait into a categorical trait. In the present case, we are seeking to understand if reading impairment in our sample represents the low scores from the distribution of quantitative language variation. If true, then it would clearly demonstrate that the reading impairment phenotype is consistent with identification of language-impaired subjects. If the language trait is not linked in a univariate analysis this would additionally imply that the reading impairment phenotype preferentially selects a subset of language-impaired individuals from the sample. The converse relationship need not be present. Language- impaired individuals may not be a subset of reading-impaired individuals. Therefore, as part of our ongoing efforts to map the locus on 13q21, we sought to elucidate the relationship between reading and language as it relates to the susceptibility allele on 13q21.
We were also interested in examining the relationship between a known functional allele at a separate locus and our findings on 13q21. The advantage of incorporating gene × gene interaction, also known as epistasis, into a family study is theoretically very strong when such gene × gene interactions exist. While there are difficulties in assessing epistasis in the analysis of some study designs, our collection of large extended pedigrees contains suitable information about epistasis to make the analysis reasonable [47, 48]. Based on evidence for a marginal effect with reading impairment in our sample [49] (see also suppl. materials, www.karger.com/doi/10.1159/000320367), we decided to test epistasis by examining a coding variant in brain-derived neurotrophic factor gene (BDNF), a member of the nerve growth factor family known to regulate synaptic growth, proliferation, differentiation and the survival of neurons in the developing nervous system [50, 51]. A naturally occurring polymorphism in BDNF, rs6265, results in an amino acid substitution from valine to a methionine (p.Val66Met) within the prodomain of the precursor peptide affecting trafficking of the protein within the cell [52]. Found only in humans, this variant has a population frequency of 20–30% in Caucasian populations and accounts for significant behavioral variance during N-back memory tasks [52] and word recall [53, 54]. The met allele has been linked to decreased neuronal integrity and synaptic abundance as well as reduced levels of episodic memory [50]. Based on this body of compelling evidence that (1) the BDNF val/met alleles are functional and (2) these alleles produce a measurable effect on human cognition, we genotyped the underlying SNP, rs6265, in our SLI families with the hypothesis that working memory would have a detectable influence on language development, though we did not postulate as to the nature or the strength of the effect.
Using the PPL framework as a platform for examining evidence for and against specific genetic models, we examined the genotype-phenotype relationship of language impairment in a collection of large extended families. We first demonstrate the relationship between reading and language measures at our locus of interest to provide context for further work on 13q21. We then extended our previous work by examining the evidence for 13q21 when incorporating epistasis with a functional BDNF SNP. These complementary modeling approaches will be discussed in the context of the complex SLI phenotype.
Subjects, Materials and Methods
Subjects and Phenotyping
The data set is derived from Bartlett et al. [32] with minor changes to the original samples as noted below. The study sample was comprised of 1 nuclear and 3 extended Canadian families of Celtic ancestry (n = 158). Of these, 71 subjects had both phenotypic and genotypic information, 14 only DNA for genotypes and 1 only phenotypic information. The remaining subjects were required to fully reconstruct the pedigrees but do not have phenotypic or genotypic information. We have added 55 new subjects to the existing 4 families and removed the 1 nuclear family (n = 4 subjects) from analysis due to limited DNA availability. These numbers represent a change of 59 samples from the 2002 publication. These pedigrees were originally identified while recruiting for a linkage study of schizophrenia [102] and were noted to have a history of language or reading impairments.
For inclusion in the study, the families were screened by a speech language pathologist via telephone interview for a history of language impairment segregating in the family. Those families with a strong family history of language impairment were scheduled for assessment. The largest family identified this way is only related to participants in the schizophrenia study by marriage (i.e., unrelated) and for the remaining families, no phenotypes from subjects with schizophrenia are used in any analysis. Prior to enrollment and testing, informed consent that conformed to the guidelines for treatment of human subjects approved by Rutgers University was obtained. All subjects received a comprehensive battery administered by an experienced tester in their own homes [see [32] for more details]. Assessment tools for the present study included:
-
(1)
Test of Language Development (TOLD; age appropriate version [55, 56, 57]), which is a comprehensive test of language functioning including specific subtypes of language processes such as comprehension, expression, syntax, grammar and phonology. Subtests may be combined for composite scores in listening, speaking, grammar, vocabulary, receptive language and expressive language as well as an overall language score known as the spoken language quotient or SLQ.
-
(2)
Wechsler Intelligence Test (performance portions, age appropriate version [58, 59, 60, 61]).
-
(3)
Woodcock Reading Mastery [62] subtests Word Attack (single non-word reading), Word Identification (Word ID; single word reading) and Passage Comprehension (Pass Comp; reading comprehension of short passages).
For our quantitative analysis of language we used the TOLD Speaking (expressive language measures), Grammar (syntax and morphology measures) and SLQ composite scores. For our quantitative analysis of reading we used Word ID, Word Attack and Pass Comp. A table with the correlations between the quantitative measures used in this study is presented in online supplementary table 1.
We used two diagnostic classifications of impairment from Bartlett et al. [32]. Due to the classifications not being mutually exclusive, it was possible for an individual subject to meet the criteria for more than one of the following classifications. To be language impaired, a subject's SLQ on the TOLD [56, 57] was ≤85. Further, a subject was classified as reading impaired if their single non-word reading score (Word Attack [62]) was one standard deviation below their performance IQ. There were 10 subjects in the language-impaired-only group, 4 in the reading-impaired-only group and 15 in the language-impaired + reading-impaired group. In our sample, it was not necessary to exclude any subject from analysis because of mental retardation, abnormal hearing, oral motor or structural defects.
Single Nucleotide Polymorphism Genotyping
To further assess linkage to the 13q21 region, we genotyped 30 SNPs at roughly 0.3 cM interval across the 10 cM region. These SNPs were selected from HapMap [63] (Phase 3) requiring a minor allele frequency >0.4 and a linkage disequilibrium (LD) (R2) <0.2 as calculated from the CEPH (Utah residents with ancestry from northern and western Europe) HapMap samples, and at least 200 base pairs away from any (1) dbSNP (build 129) variant [64]; (2) repetitive element as identified by RepeatMasker (http://www.repeatmasker.org/); (3) repetitive elements identified by Tandem Repeats Finder (http://tandem.bu.edu/trf/trf.html), or (4) ‘structural variations’ listed in the UCSC genome browser [65]. From the candidates that met these criteria, final SNPs were chosen by walking down the chromosome in 0.3-cM intervals as determined by the Rutgers combined genetic map (for the list of chosen SNPs, see online suppl. table 2) [66]. Genotyping was performed by multiplex PCR of amplicons containing the chosen SNPs followed by the ligase detection reaction (LDR) method of Bruse et al. [67] on a Luminex 200 Multiplex Bio-Assay Analyzer. All samples were stored and handled as described previously [32]. Genotypes were called based on the same metric as in Bruse et al. [67], but initial clustering analysis was conducted by in-house Python 2.5.1 scripts using the scipy 0.6 library clustering routines (http://www.scipy.org/). This script automatically produces allele intensity plots color coded by called genotype. Markers with less than 85% completion were not considered further (n = 3). All plots were visually inspected (by T.R.S. and C.W.B.); SNPs that did not cluster appropriately were either dropped (n = 3) or called manually (n = 5).
Previously, a panel of 28 functional SNPs was also genotyped using the same genotyping procedures listed above and tested for association with SLI in a pilot study to examine genetic variation related to (possibly non-language) cognitive traits and their potential role in development of SLI [49]. These 28 SNPs were chosen based on three criteria. The SNP was required to: (1) be associated with a cognitive trait in the literature; (2) have evidence of biological function from cell lines, mouse models or postmortem human brain tissue, and (3) affect a gene expressed in brain. Based on these results (for a summary, see online suppl. table 3), only the BDNF SNP, rs6265, showed evidence of a marginal effect and was chosen for analysis as a possible gene ×gene interaction partner to the 13q21 locus in this study. We re-genotyped rs6265 in single-plex PCR and LDR reactions to confirm the quality of the initial multiplex PCR/LDR genotypes. Since genotyping error detection in an isolated SNP is inefficient, even in our large extended pedigrees, we also validated the assay by direct-sequencing 2 samples in the center mass of each of the 3 genotyping clusters and 2 samples on each edge. Results from direct sequencing confirmed all genotype calls from the LDR assay.
Genotype Cleaning
All genotype data were automatically processed as they were generated via in-house Python scripts for consistent error checking to minimize human errors. Files containing the called genotypes were output into a pre-MAKEPED linkage format file. A linkage format locus data file was generated to form the two required inputs to PEDCHECK v1.1 [68] which detects all Mendelian inconsistencies. PEDCHECK output was tabulated and merged with raw allele intensity data for manual evaluation of the genotype calls in the flagged nuclear family containing the error. Ambiguous genotypes were repeated and irreconcilable genotypes were handled by setting all of the flagged nuclear family's genotypes for that SNP to missing. SIMWALK2 v2.91 [69, 70] was used to detect Mendelian-consistent errors through its implementation of the Sobel et al. [71] pedigree likelihood that allows for errors. This likelihood is a multipoint method for detecting unlikely genotypes (input files were created with MEGA2 v4.0.r1 [72]). Files were analyzed 3 times using slightly different parameters and random number seeds to ensure convergence on a stable solution. Any genotype flagged in 2–3 of the runs was set to missing. Genotypes flagged in a single run were noted; repeating of analyses with these other potential genotyping errors yielded no or trivial changes in the results reported here (data not shown). Lastly, PEDSTATS v0.6.3 was used to check markers for Hardy-Weinberg equilibrium using both the founders-only function as well as the unrelated samples functions [73]. Markers flagged at p < 0.01 were dropped (n = 4). After cleaning the data, a total of 20 SNP markers within the 10 cM 13q21 region remained for further analysis. Information content was estimated using Merlin v1.1.2 [74] on 3 of the 4 families. The family that was too large to be run through Merlin did not differ in genotyping completion rates or heterozygosity.
Statistical Analyses
We chose a unified analytical strategy for the analysis of categorical and quantitative traits in a framework that incorporates both linkage and LD with the causal disease variant on (essentially) the same scale. Use of the same scale across multiple analyses allows us to readily interpret results from one analysis in the context of another. The PPL framework is therefore used in this paper. It additionally has the advantage that it is essentially model-free, since the nuisance trait parameters have been integrated out of the likelihood and its implementation, the software program Kelvin [75, 76] has already been adapted for use with the types of analyses presented here. For additional information on how the models are calculated we refer the reader to the following papers [46, 77, 78, 79, 80, 81, 82, 83], cited in context below. Kelvin [75, 76] implements the PPL class of models for measurement of the strength of genetic evidence. The PPL is parameterized in terms of a general approximating likelihood, and all parameters of the trait model are then integrated out numerically, permitting the use of Bayes’ theorem for computation of the posterior probability of the hypothesis of interest [84]. All results were based on multipoint analysis; Hardy-Weinberg equilibrium has been assumed throughout. The Rutgers combined genetic map was used for genetic locations [66]. Both linkage and LD analysis can be performed in this framework; the difference between the two involves adding a single parameter for the trait-locus – marker-locus phase probability (in LD analysis) [81].
Our previous SLI studies have shown linkage to a dichotomous trait, reading impairment, as described above. Therefore, our baseline linkage analysis for interpretation of reading and language measures uses the dichotomous trait PPL, which is the standard admixture linkage likelihood with the trait parameters integrated out (penetrances, disease gene frequency, admixture). It is of interest to understand the genetic relationship between reading impairment or language impairment and various quantitative measures of language and reading because of the strong co-occurrence between the two disorders. To characterize these relationships we apply PPL analysis with a quantitative trait threshold. This model, as described previously [45, 46, 82], allows for the mixture of quantitative and dichotomous phenotypes in the same pedigree within the same analysis model. In this case, we classified individuals who were ‘affected’ with reading impairment or ‘affected’ with language impairment as having a phenotype beyond an unspecified threshold (this threshold is treated as a nuisance parameter and is integrated out of the likelihood to retain its model-free approach) while all ‘unaffected’ individuals were assigned their quantitative values. This analysis can be used to explore the relationship between two, in the most informative case, distinctly derived phenotypes that are not both quantitative or both dichotomous.
To model epistasis, an additional layer can be added to the above models in that multiple sets of penetrances (dichotomous traits) or multiple sets of genotypic means and variances may be modeled to have dependence on a separate, unlinked, genomic location, i.e., epistasis [83]. Our method is an extension of the liability class model from linkage analysis which includes additional penetrances for the second (epistatic) locus and is therefore different from conditioning on a second locus in a regression framework which does not have the same genetic interpretation [85]. The models are calculated the same way except for the inclusion of genotype-dependent parameters, where the genotype comes not from the locus under study, but a different locus of interest at an unlinked location. While there is a multiplicity of other statistical models for epistasis available in the literature [see [85]for general discussion], we have chosen this particular model for its transparent interpretation in terms of the fundamental biology (see Discussion). Specifically, the model allows separate sets of 13q21 penetrances to differ based on genotypes from a functional SNP within BDNF. One limitation of the method we employ is that liability classes may not be unknown if a phenotype is specified due to practical limitations in computing. Therefore, 3 subjects who did not have DNA available but did have phenotypic information were set to unknown for both the phenotype and the liability class. This creates a different baseline with which the epistasis analysis must be compared since the non-epistasis baseline must also have those same 3 subjects set to phenotype unknown.
Of final note, Bayesian methods that quantify the probability of the linkage/association evidence do not generate p values [86]. However, it is still true that a large PPL is more intellectually palatable if the specific value in question is not commonly seen across the genome. As a loose guide, we have performed a simulation where we generated 10,000 datasets. These datasets were identical to the families analyzed here in both phenotypes and configuration of available phenotypic and genotypic information. SNPs in an identical configuration of allele frequencies and map distances were simulated, but no genetic effect was simulated. In this simulation, we would characterize p values to be related to PPLs as follows: p < 0.05 related to PPL >1.5%; p < 0.01 related to PPL >4%; p < 0.001 related to PPL >23%. These numbers are virtually the same as suggested in another large-scale null simulation with the PPL [87].
Results
Baseline Linkage Analysis
As described in the Methods section, the pedigrees used differ slightly from our previous reports [31, 32]. We therefore did not make direct comparisons with those reported results. We first calculated a new baseline linkage analysis across the 13q21 region. Using the previously described reading impairment phenotype [31, 32], the maximum PPL in the region was 80% at rs1413119 (solid black line, fig. 1). This was larger than the previously reported 53% in Bartlett et al. [31, 32].
Fig. 1.
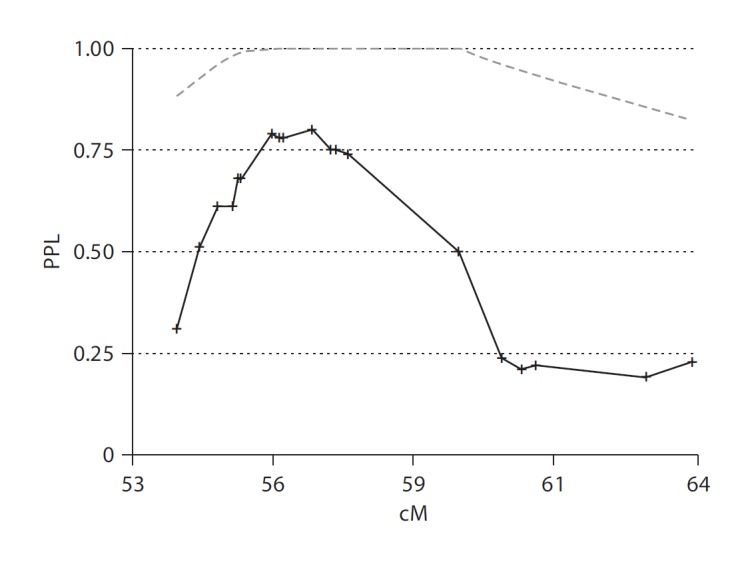
Baseline PPL linkage analysis of 13q21 in the Canadian sample. Using the four SLI families as described in the Methods section and a dense map of SNP genotypes, this figure shows the baseline linkage results used for further analyses in the current study (solid black line). Information content is also displayed (dashed gray line).
Relationship between Quantitative Language Variation and Reading Impairment in Our Sample
In order to determine if the observed linkage to the categorical reading impairment phenotype was indirectly selecting for language ability we assessed the relationship between quantitative language variation and the reading impairment in our sample. We first characterized the relationship between language and reading measures on a purely phenotypic level and then characterized the relationship between these measures in the context of the 13q21 linkage analysis. Figure 2 shows a boxplot analysis of how the dichotomous reading impairment trait separated out low versus high language ability. Based on these data, we may predict a clear dissociation between reading and language impairment in the linkage analysis just as was originally observed [31, 32].
Fig. 2.
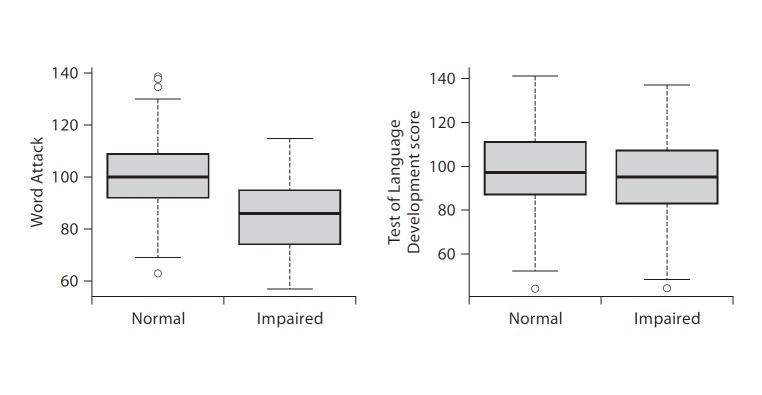
Language impairment predicts reading ability (left), but reading impairment does not predict language ability (right). Using language impairment status as a category, the boxplot shows the Word Attack (single non-word reading from the WRM) scores of all family members available for phenotyping. Note the clear dissociation of readers based on language ability in this sample. On the right is the converse where reading impairment status is used as a category and the boxplot shows the expressive language scores from the TOLD. The thick black line is the mean and circles are data points outside the quartiles.
We then investigated whether the 13q21 locus was related to poor reading ability due to poor language ability, using the quantitative trait threshold PPL. This analysis model explicitly assumes that reading-impaired persons are below an unspecified threshold on the quantitative language ability scale, which necessarily implies the two traits are related by way of the reading impairment, implying a co-occurring language impairment. We applied this method to 3 combinations of quantitative language measures from the TOLD family of tests (Speaking, Grammar and SLQ) and reading impairment (table 1; see also online suppl. table 4). All 3 were consistent with linkage to 13q21. Both Speaking and SLQ showed strong evidence for linkage but were attenuated compared to reading impairment alone. Grammar was attenuated more where the evidence for linkage dropped below standards for declaring genome-wide linkage, but was still very suggestive (PPL = 18% where PPL = 21% is needed to declare a genome-wide linkage). These data are consistent with language processes being involved in the 13q21 linkage, though none of the three language traits selected for analysis are in fact the ‘optimal’ language trait to couple with the reading impairment phenotype highlighting the limitations of mapping complex phenotypes to the available constructs.
Table 1.
PPL (location on chromosome 13) for univariate quantitative language and reading traits and threshold models including dichotomous language impairment (LI) or reading impairment (RI)
| Univariate | + LI threshold | + RI threshold | |
|---|---|---|---|
| LI (dichotomous) | 0.03 (55 cM) | ||
| SLQ (quantitative trait) | 0.014 (59 cM) | ||
| Word identification | 0.021 (56 cM) | 0.017 (56 cM) | |
| Word attack | 0.017 (56 cM) | 0.021 (56 cM) | |
| Pass comp | 0.021 (60 cM) | 0.018 (59 cM) | |
| RI (dichotomous) | 0.80 (56 cM) | ||
| Speaking | 0.018 (59 cM) | 0.63 (55 cM) | |
| SLQ | 0.014 (59 cM) | 0.54 (55 cM) | |
| Grammar | 0.022 (59 cM) | 0.18 (56 cM) | |
The reverse analogous scenario produced the opposite result, as expected. Using our language impairment trait and the quantitative reading measures Word ID, Word Attack and Passage Comprehension we examined a model that assumed language impairment was a consequence of being on the low end of quantitative reading variation. PPLs from these models were all inconsistent with linkage to 13q21. As our sample contains many language-impaired individuals who are not poor readers (see table 1 and fig. 2), analysis that mixed the two trait domains assuming a one-to-one relationship did not show linkage to 13q21. We also performed univariate quantitative trait analysis using 3 language measures and 3 reading measures, of which none showed evidence for linkage (table 2; see also online suppl. materials). Thus, out of all the reading and language analyses presented, only the reading impairment phenotype with or without additional language phenotypic information showed evidence for linkage. Taken together, our reading and language analysis results are consistent with the reading impairment phenotype being related to impaired language processes.
Table 2.
Maximum likelihood estimates of penetrances for single-locus and two-locus models of 13q21 risk alíele
| wt/wt | wt/risk | risk/risk | |
|---|---|---|---|
| No BDNF information | 0.0 | 0.2 | 0.999 |
| BDNF Val/Val or Val/Met | 0.0 | 0.0 | 0.999 |
| BDNF Met/Met | 0.0 | 0.999 | 0.999 |
Epistasis Analysis with a Genetic Variant Associated with Working Memory
Epistasis analysis by conditioning on alleles known to be functionally related to cognitive tasks relevant for language development was considered the most reasonable step to improve our mapping signal with this relatively small sample using the reading impairment phenotype. We previously indicated that rs6265 within BDNF showed evidence for association with the reading impairment phenotype (LD-PPL = 6%, see online suppl. materials). BDNF has a well-documented role in working memory which, although not a language measure itself, is related to language processing [88]. We examined the effect of incorporating the BDNF rs6265 risk allele into our linkage analysis on 13q21, assuming either a dominant or recessive epistatic effect. The dominant epistatic model did not change the PPL appreciably (<2%) but the recessive epistatic model increased the PPL to 1 (fig. 3). We would interpret this result to mean a posterior probability that is close to 1, but not 1 (which would mean certainty). The difficulty of such a PPL is that the scale has reached its ceiling, where one can no longer distinguish any changes in the strength of the evidence. In order to overcome this issue we also examined the LOD maximized over all parameters (MOD) for the two conditions since the LOD scale is unbounded. The MOD score is available as part of the standard Kelvin output as it is the maximum LOD encountered during the numerical integration procedure (and the associated model parameters are therefore the MLEs for the trait model). The increase in evidence was 4.19 LOD units to a MOD of 8.44, indicating a very clear effect of BDNF SNP rs6265 at this locus.
Fig. 3.
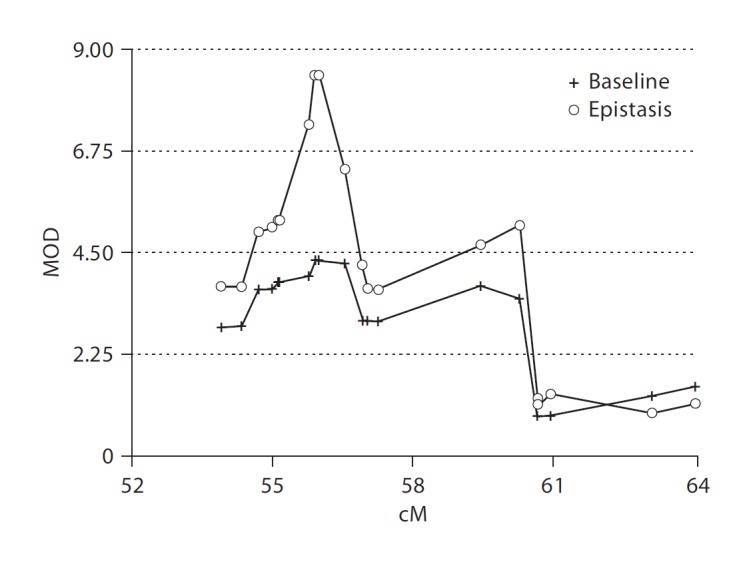
Epistasis analysis showing that BDNF SNP rs6265 increases MOD at the chromosome 13q21 locus over baseline using the reading impairment phenotype. The MOD essentially doubles in the vicinity of 56 cM, which represents a large change from the baseline linkage analysis as shown by the open circles.
We sought to further improve interpretation of the difference in MOD scores, given that the allowing for epistasis adds more parameters and therefore introduces a different scale than that for the baseline (non-epistasis) MOD. Therefore, we also examined the integrated likelihood, which retains the same scale regardless of the number of parameters in the model since it is not maximized. This metric showed an increase of 2.26 log10 units to a final value of 4.86 that is nearly double the strength in evidence for linkage to the region. This measure therefore also supports a strong increase in the evidence for linkage to 13q21 and indicates that the large increase in the MOD score was not due to inflation from additional parameters in the epistasis likelihood. In 1,000 null simulations we did not find any replicates that were more extreme than this, suggesting p < 0.001. Notably, this p value would meet criteria for genome-wide significance even though the analysis was focused on 13q21, thus there was no need for genome-wide correction. Taken together, the results suggest that evidence for a susceptibility allele in 13q21 is substantially greater when an interaction with BDNF is included in the analysis.
The effect of the BDNF functional SNP can be quantified by changes in the maximum likelihood estimated penetrances from the single-locus model to the two-locus model. Table 2 displays these results. In all cases, the penetrance for non-carriers (phenocopies) was estimated to be 0. Heterozygotes for the 13q21 risk allele show a clear dichotomy between BDNF SNP genotypes, where those with 0 or 1 less efficient met alleles are not at risk, but those with two met alleles have as much risk as persons who are homozygous for the 13q21 risk allele. The single-locus model shows some risk for 13q21 risk allele heterozygotes but substantially less than the two-locus model indicates for persons who are homozygous for the BDNF met allele.
Discussion
In 2002 our group initially hypothesized that the reading impairment phenotype ‘in a population selected for SLI is most likely measuring the resultant reading outcome of an underlying language deficit as opposed to a reading deficit in isolation’ [32]. However, this conjecture remained without evidence until two developments took place. First, we required a deeper understanding of the relationship between reading and language deficits as well as normal reading and language. In the past 8 years, numerous studies have been published that suggest reading impairments in persons with SLI may be related to their underlying language deficits [16, 89, 90, 91, 92]. Taken together, these studies indicate that abnormal language development may start the child on an abnormal developmental trajectory for the acquisition of reading skills, which increases the likelihood of a co-occurring reading deficit. Second, we required the correct analytical tools to be developed. Bartlett and Vieland [82] clean up presented a detailed series of simulations examining the characteristics of the PPL reparameterized for quantitative traits. That method was later adapted to allow for the unspecified threshold analysis as used in this paper and successfully applied to elucidate the genetic architecture of autoimmune thyroid disease in Vieland et al. [46]. In this publication, we capitalized on advances in these two areas to perhaps explain why a sample selected for language impairment gives stronger linkage with a reading impairment phenotype. While the data do not suggest a reading impairment phenotype is universally better for finding SLI susceptibility alleles, as we have only examined one locus in depth in this paper, the reading impairment phenotype does appear to be the most relevant trait for the locus on 13q21.
To explore if the categorical reading impairment phenotype is indirectly selecting for a language trait we examined the data using models that assume reading impairment is related to quantitative language variation, specifically that reading-impaired persons will be on the low end of observed language variation. Our results are consistent with this assumption. The reverse was not true. Language-impaired subjects when assumed to be on the low end of reading variation did not show linkage to the 13q21 region. As one explanation for this pattern we suggest that the reading impairment trait in our sample is selecting subjects that have both reading and language impairments. This pattern is also largely seen in the phenotypic overlap between our definitions of language and reading impairments and is supported by the fact that language ability predicts reading impairment in our sample (fig. 2). The individual quantitative traits did not show linkage to the region, indicating that no univariate analysis contains the most relevant information for linkage to 13q21. Taken together, these data support the notion that joint language and reading deficits are the most relevant phenotype for the 13q21 SLI locus.
Our analysis of reading and language phenotypes involved two levels: (1) the phenotypic analysis of how reading impairment separated out (essentially) non-overlapping groups on language scores but language impairment did not separate out groups on reading scores, and (2) the linkage analysis using the same principles. Both analyses displayed the same trends. However, we note that these two analyses were conducted on the same subjects from the same families and No. 1 is neither a replication study of the observed effects of No. 2, nor vice versa. The two analyses did not have to agree with one another by virtue of the fact that even though our families were selected based on phenotypic information, they were not selected on the basis of genotypic information. Therefore, the genotypes at 13q21 were not required to be correlated with our phenotypes (i.e., not selected for) as observed. In essence, the linkage information at this locus was a ‘free parameter’ in the second analysis that was not a simple recapitulation of the first phenotypic analysis. While this approach is intuitively not as revealing as a replication, it still represents convergence of evidence. Nonetheless, although our phenotypic analysis is concordant with the specific genetic analysis of 13q21, this finding alone does not imply that such a relationship will be observed generally in SLI. Ascertaining any subjects on language may distort the distribution of correlated reading scores, and here we have the additional complexity that these pedigrees were ascertained for apparent genetic transmission of language impairment. The relationship between language and reading may not generalize to other populations.
Further studies will be required to identify what mediates the reading-language relationship presented here. Our measure of reading involves only the recognition of isolated pseudowords (which relies strongly on phonological processing) and hence does not model the multidimensional components of reading that include vocabulary, grammar, and other language processes required for reading comprehension. In fact, it is possible that a measure of pseudoword reading actually measures a cognitive process that is fundamental to both reading and language – a process that presumably is more influenced by genetic variation on 13q21. If true, then selection for families based on language ability is a de facto selection of families based on phonological processing skills. Additionally, it is possible that aspects of language apart from phonological skills are more sensitive to effects of remediation or natural normalization of performance with age (compensation) and are therefore less suited as behavioral biomarkers.
This paper also applies a method for analysis of epistasis in pedigrees. We believe this is the first epistasis analysis to so dramatically increase the strength of the evidence for linkage to a cognitive trait in humans. The results provide much greater signal-to-noise compared to a typical genome-wide screen (either linkage or association), which is one of the theoretical benefits of exploiting epistasis. It is hoped that this will translate into greater signal for association fine mapping which is ongoing. However, even in the absence of the specific allele in 13q21 that increases susceptibility, the known functional allele in BDNF informs us about SLI in a limited but important fashion. The penetrance estimates from the two-locus modeling indicate that persons who are homozygous for the met allele at the BDNF SNP rs6265 are at a dramatically higher risk for developing SLI in conjunction with the as yet unidentified SLI risk allele for within the 13q21 locus. It remains to be seen if this effect translates to other loci as well.
Several studies have been done to try to establish the effects that the BDNF genotype has on hippocampal-dependent memory. As seen in mouse studies, human studies also give evidence to support a gene-dosage affect. It has been shown that Met carriers (Val/Met and Met/Met) consistently perform worse than their Val/Val counterparts on tasks that have been shown to be hippocampal dependent. Hariri et al. [50] have shown through functional magnetic resonance imaging (fMRI) that when asked to perform a test of place recognition, Met carriers exhibited significantly lower levels of activation in the hippocampus. This same result was seen during both the encoding and retrieval phases of the task. In further support, another set of fMRI studies was performed using a task (N-back task) that tested an individual's working memory and typically would result in the disengagement of the hippocampus. It was found that the Met carriers failed to disengage the hippocampus to the extent that the Val/Val individuals did [52]. Another robust finding is a Met-associated decrease in hippocampal volume of up to 15% as measured by structural MRI. Transgenic mouse studies have reinforced the findings of human studies by showing a significant decrease in hippocampal volume of the Val/Met allele as compared to the Val/Val allele. Also, studies of the transgenic mouse have shown that the p.Val66Met is involved in activity-dependent secretion of Bdnf from neurons and results in a decrease in dendrite arbor complexity. Interestingly, there seems to be an additive gene dosage affect; Val/Met heterozygotes have less activity-dependent secretion than the Val/Val homozygote counterparts, but are still more active than the Met/Met homozygotes [93].
Although molecular neuroscience explanations for the interaction of BDNF with the SLI susceptibility allele on 13q21 cannot be confirmed until we have compelling evidence for a specific allele in 13q21, there are cognitive neuroscience theories that make predictions regarding possible interactions at the level of neural systems. One theory of SLI posits that language deficits either stem from, or are reinforced by, a deficient form of short-term memory for speech sounds known as phonological short-term memory [94]. This buffer is required for the processing of speech sounds in order to string the sounds into more meaningful units for analysis, presumably at the linguistic level. Given the role of BDNF in hippocampal-related processes, if the phonological short-term memory buffer involves the hippocampus, then the interaction between BDNF and SLI alleles on 13q21 follows directly from the 13q21 SLI deficit being exacerbated by poor phonological short-term memory via hippocampal processes. Another theory of SLI suggests that an auditory processing deficit limits the ability of children with SLI to appropriately detect subtle but critical distinctions in the speech sound stream such as the subtle ‘-ed’ ending that is critical for learning the past tense [95]. If a sensory processing deficit is present, such that phonemes are not processed correctly to begin with, an additional deficit in the ability to store those phonemes for additional processing, which may have included possible compensatory processing, could result in the kind of interaction suggested by our data. At present this is speculation, but it is possible to test these alternatives in a cognitive neuroscience framework. It is unfortunate that our Canadian pedigrees do not have an adequate measure of phonological short-term memory to test these alternatives in this sample. However, using suitable measures in other populations with rs6265 genotyping should be sufficient, given a large enough sample to test such a hypothesis.
Overall, these analyses demonstrate that thoughtful modeling of the genotype-phenotype relationship can elucidate important relationships in specific datasets that inform notions of which genetic mechanisms appear to mediate effects on phenotypic manifestations as well relationships between phenotypes. In the present case, the locus on 13q21 appears to have an interesting effect on non-word reading ability through effects on global language, presumably mediated developmentally through phonological processing. This last point is best tested at the phenotypic level and indeed evidence is provided in the literature [19, 96, 97]. Additionally, the strong genetic interaction between a BDNF variant associated with reduced working memory performance and the locus on 13q21 adds another level of phenotypic complexity in that it provides a potential genetic basis for the well-documented relationship between working memory and SLI [98, 99, 100, 101]. While it would be interesting to further test our interaction with phonological working memory phenotypes so prominently featured in the SLI literature, we do not have such a measure in these families. However, since the behavioral literature is quite extensive on the relationship of working memory as an important part of the SLI phenotype, it would appear reasonable to posit that the BDNF-13q21 interaction belies phonological working memory impairment. This possibility awaits testing in other samples.
Supplementary Material
Supplementary data
Acknowledgements
We would like to thank the participating families, who contributed their time and patience to make this study possible, and Veronica J. Vieland for insightful comments on a previous version of the manuscript. This research was supported by Research Grant 12-FY02-107 (L.M.B.) from the March of Dimes, R01DC009453 (C.W.B.) and also supported in part by an allocation of computing time from the Ohio Supercomputer Center Grant PCCR0001-2 (C.W.B.).
References
- 1.Risch N. Linkage strategies for genetically complex traits. Ii. The power of affected relative pairs. Am J Hum Genet. 1990;46:229–241. [PMC free article] [PubMed] [Google Scholar]
- 2.Tomblin JB, Records NL, Buckwalter P, Zhang X, Smith E, O'Brien M. Prevalence of specific language impairment in kindergarten children. J Speech Lang Hear Res. 1997;40:1245–1260. doi: 10.1044/jslhr.4006.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dale PS, Price TS, Bishop DV, Plomin R. Outcomes of early language delay: I. Predicting persistent and transient language difficulties at 3 and 4 years. J Speech Lang Hear Res. 2003;46:544–560. doi: 10.1044/1092-4388(2003/044). [DOI] [PubMed] [Google Scholar]
- 4.Snowling M, Bishop DV, Stothard SE. Is preschool language impairment a risk factor for dyslexia in adolescence? J Child Psychol Psychiatry. 2000;41:587–600. doi: 10.1111/1469-7610.00651. [DOI] [PubMed] [Google Scholar]
- 5.Whitehouse AJ, Line EA, Watt HJ, Bishop DV. Qualitative aspects of developmental language impairment relate to language and literacy outcome in adulthood. Int J Lang Commun Disord. 2009:1–22. doi: 10.1080/13682820802708080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leonard LB. Children with specific language impairment. Cambridge, MA: MIT Press; 1998. [Google Scholar]
- 7.Tallal P, Piercy M. Defects of non-verbal auditory perception in children with developmental aphasia. Nature. 1973;241:468–469. doi: 10.1038/241468a0. [DOI] [PubMed] [Google Scholar]
- 8.Tallal P, Piercy M. Developmental aphasia: impaired rate of non-verbal processing as a function of sensory modality. Neuropsychologia. 1973;11:389–398. doi: 10.1016/0028-3932(73)90025-0. [DOI] [PubMed] [Google Scholar]
- 9.Wright BA, Lombardino LJ, King WM, Puranik CS, Leonard CM, Merzenich MM. Deficits in auditory temporal and spectral resolution in language-impaired children. Nature. 1997;387:176–178. doi: 10.1038/387176a0. [DOI] [PubMed] [Google Scholar]
- 10.Wright BA, Bowen RW, Zecker SG. Nonlinguistic perceptual deficits associated with reading and language disorders. Curr Opin Neurobiol. 2000;10:482–486. doi: 10.1016/s0959-4388(00)00119-7. [DOI] [PubMed] [Google Scholar]
- 11.Witton C, Stein JF, Stoodley CJ, Rosner BS, Talcott JB. Separate influences of acoustic am and fm sensitivity on the phonological decoding skills of impaired and normal readers. J Cogn Neurosci. 2002;14:866–874. doi: 10.1162/089892902760191090. [DOI] [PubMed] [Google Scholar]
- 12.Talcott JB, Witton C, McClean M, Hansen PC, Rees A, Green GG, Stein JF. Can sensitivity to auditory frequency modulation predict children's phonological and reading skills? Neuroreport. 1999;10:2045–2050. doi: 10.1097/00001756-199907130-00010. [DOI] [PubMed] [Google Scholar]
- 13.Witton C, Talcott JB, Hansen PC, Richardson AJ, Griffiths TD, Rees A, Stein JF, Green GG. Sensitivity to dynamic auditory and visual stimuli predicts nonword reading ability in both dyslexic and normal readers. Curr Biol. 1998;8:791–797. doi: 10.1016/s0960-9822(98)70320-3. [DOI] [PubMed] [Google Scholar]
- 14.Snowling MJ, Bishop DV, Stothard SE, Chipchase B, Kaplan C. Psychosocial outcomes at 15 years of children with a preschool history of speech-language impairment. J Child Psychol Psychiatry. 2006;47:759–765. doi: 10.1111/j.1469-7610.2006.01631.x. [DOI] [PubMed] [Google Scholar]
- 15.Reed MA. Speech perception and the discrimination of brief auditory cues in reading disabled children. J Exp Child Psychol. 1989;48:270–292. doi: 10.1016/0022-0965(89)90006-4. [DOI] [PubMed] [Google Scholar]
- 16.Catts HW, Bridges MS, Little TD, Tomblin JB. Reading achievement growth in children with language impairments. J Speech Lang Hear Res. 2008;51:1569–1579. doi: 10.1044/1092-4388(2008/07-0259). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catts HW, Fey ME, Tomblin JB, Zhang X. A longitudinal investigation of reading outcomes in children with language impairments. J Speech Lang Hear Res. 2002;45:1142–1157. doi: 10.1044/1092-4388(2002/093). [DOI] [PubMed] [Google Scholar]
- 18.Catts HW, Fey ME, Proctor-Williams K. The relationship between language and reading. Preliminary results from a longitudinal investigation. Logoped Phoniatr Vocol. 2000;25:3–11. doi: 10.1080/140154300750045858. [DOI] [PubMed] [Google Scholar]
- 19.Catts HW. The relationship between speech-language impairments and reading disabilities. J Speech Hear Res. 1993;36:948–958. doi: 10.1044/jshr.3605.948. [DOI] [PubMed] [Google Scholar]
- 20.Rice ML, Haney KR, Wexler K. Family histories of children with sli who show extended optional infinitives. J Speech Lang Hear Res. 1998;41:419–432. doi: 10.1044/jslhr.4102.419. [DOI] [PubMed] [Google Scholar]
- 21.Tallal P, Hirsch LS, Realpe-Bonilla T, Miller S, Brzustowicz LM, Bartlett C, Flax JF. Familial aggregation in specific language impairment. J Speech Lang Hear Res. 2001;44:1172–1182. doi: 10.1044/1092-4388(2001/091). [DOI] [PubMed] [Google Scholar]
- 22.Tallal P, Ross R, Curtiss S. Familial aggregation in specific language impairment. J Speech Hear Disord. 1989;54:167–173. doi: 10.1044/jshd.5402.167. [DOI] [PubMed] [Google Scholar]
- 23.Tomblin JB. Familial concentration of developmental language impairment. J Speech Hear Disord. 1989;54:287–295. doi: 10.1044/jshd.5402.287. [DOI] [PubMed] [Google Scholar]
- 24.Van der Lely HK, Stollwerck L. A grammatical specific language impairment in children: an autosomal dominant inheritance? Brain Lang. 1996;52:484–504. doi: 10.1006/brln.1996.0026. [DOI] [PubMed] [Google Scholar]
- 25.Bishop DV, North T, Donlan C. Genetic basis of specific language impairment: evidence from a twin study. Dev Med Child Neurol. 1995;37:56–71. doi: 10.1111/j.1469-8749.1995.tb11932.x. [DOI] [PubMed] [Google Scholar]
- 26.Tomblin JB, Buckwalter PR. Heritability of poor language achievement among twins. J Speech Lang Hear Res. 1998;41:188–199. doi: 10.1044/jslhr.4101.188. [DOI] [PubMed] [Google Scholar]
- 27.DeThorne LS, Petrill SA, Hayiou-Thomas ME, Plomin R. Low expressive vocabulary: higher heritability as a function of more severe cases. J Speech Lang Hear Res. 2005;48:792–804. doi: 10.1044/1092-4388(2005/055). [DOI] [PubMed] [Google Scholar]
- 28.Bishop DV. The role of genes in the etiology of specific language impairment. J Commun Disord. 2002;35:311–328. doi: 10.1016/s0021-9924(02)00087-4. [DOI] [PubMed] [Google Scholar]
- 29.Kovas Y, Hayiou-Thomas ME, Oliver B, Dale PS, Bishop DV, Plomin R. Genetic influences in different aspects of language development: the etiology of language skills in 4.5-year-old twins. Child Dev. 2005;76:632–651. doi: 10.1111/j.1467-8624.2005.00868.x. [DOI] [PubMed] [Google Scholar]
- 30.Colledge E, Bishop DV, Koeppen-Schomerus G, Price TS, Happe FG, Eley TC, Dale PS, Plomin R. The structure of language abilities at 4 years: a twin study. Dev Psychol. 2002;38:749–757. doi: 10.1037//0012-1649.38.5.749. [DOI] [PubMed] [Google Scholar]
- 31.Bartlett CW, Flax JF, Logue MW, Smith BJ, Vieland VJ, Tallal P, Brzustowicz LM. Examination of potential overlap in autism and language loci on chromosomes 2, 7, and 13 in two independent samples ascertained for specific language impairment. Hum Hered. 2004;57:10–20. doi: 10.1159/000077385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartlett CW, Flax JF, Logue MW, Vieland VJ, Bassett AS, Tallal P, Brzustowicz LM. A major susceptibility locus for specific language impairment is located on 13q21. Am J Hum Genet. 2002;71:45–55. doi: 10.1086/341095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The SLI Consortium A genomewide scan identifies two novel loci involved in specific language impairment. Am J Hum Genet. 2002;70:384–398. doi: 10.1086/338649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The SLI Consortium Highly significant linkage to the sli1 locus in an expanded sample of individuals affected by specific language impairment. Am J Hum Genet. 2004;74:1225–1238. doi: 10.1086/421529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monaco AP. Multivariate linkage analysis of specific language impairment (sli) Ann Hum Genet. 2007;71:660–673. doi: 10.1111/j.1469-1809.2007.00361.x. [DOI] [PubMed] [Google Scholar]
- 36.Newbury DF, Winchester L, Addis L, Paracchini S, Buckingham LL, Clark A, Cohen W, Cowie H, Dworzynski K, Everitt A, Goodyer IM, Hennessy E, Kindley AD, Miller LL, Nasir J, O'Hare A, Shaw D, Simkin Z, Simonoff E, Slonims V, Watson J, Ragoussis J, Fisher SE, Seckl JR, Helms PJ, Bolton PF, Pickles A, Conti-Ramsden G, Baird G, Bishop DV, Monaco AP. Cmip and atp2c2 modulate phonological short-term memory in language impairment. Am J Hum Genet. 2009;85:264–272. doi: 10.1016/j.ajhg.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcon M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 39.Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. Foxp2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:1120–1127. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
- 40.MacDermot KD, Bonora E, Sykes N, Coupe AM, Lai CS, Vernes SC, Vargha-Khadem F, McKenzie F, Smith RL, Monaco AP, Fisher SE. Identification of foxp2 truncation as a novel cause of developmental speech and language deficits. Am J Hum Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meaburn E, Dale PS, Craig IW, Plomin R. Language-impaired children: no sign of the foxp2 mutation. Neuroreport. 2002;13:1075–1077. doi: 10.1097/00001756-200206120-00020. [DOI] [PubMed] [Google Scholar]
- 42.Newbury DF, Bonora E, Lamb JA, Fisher SE, Lai CS, Baird G, Jannoun L, Slonims V, Stott CM, Merricks MJ, Bolton PF, Bailey AJ, Monaco AP. Foxp2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70:1318–1327. doi: 10.1086/339931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernes SC, Macdermot KD, Monaco AP, Fisher SE. Assessing the impact of FOXP1 mutations on developmental verbal dyspraxia. Eur J Hum Genet. 2009;17:1354–1358. doi: 10.1038/ejhg.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Thorne LS, Hart SA, Petrill SA, Deater-Deckard K, Thompson LA, Schatschneider C, Davison MD. Children's history of speech-language difficulties: Genetic influences and associations with reading-related measures. J Speech Lang Hear Res. 2006;49:1280–1293. doi: 10.1044/1092-4388(2006/092). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartlett CW, Vieland VJ. Two novel quantitative trait linkage analysis statistics based on the posterior probability of linkage: application to the coga families. BMC Genet. 2005;6(suppl 1):S121. doi: 10.1186/1471-2156-6-S1-S121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vieland VJ, Huang Y, Bartlett C, Davies TF, Tomer Y. A multilocus model of the genetic architecture of autoimmune thyroid disorder, with clinical implications. Am J Hum Genet. 2008;82:1349–1356. doi: 10.1016/j.ajhg.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vieland VJ, Huang J. Reply to Cordell and Farrall. Am J Hum Genet. 2003;73:1471–1473. [Google Scholar]
- 48.Vieland VJ, Huang J. Two-locus heterogeneity cannot be distinguished from two-locus epistasis on the basis of affected-sib-pair data. Am J Hum Genet. 2003;73:223–232. doi: 10.1086/376563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simmons TR, Weaver PJ, Andrews-Bryant T, Flax JF, Tallal P, Bartlett CW, Brzustowicz LM. Multivariate genetic analysis of specific language impairment (sli) in extended pedigrees: the relationship between reading and language measures on 13q21: American Society of Human Genetics. Philadelphia: Pennsylvania; 2008. Abstract program number 1830. [Google Scholar]
- 50.Hariri AR, Goldberg TE, Mattay VS, Kolachana BS, Callicott JH, Egan MF, Weinberger DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690–6694. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tyler WJ, Alonso M, Bramham CR, Pozzo-Miller LD. From acquisition to consolidation: on the role of brain-derived neurotrophic factor signaling in hippocampal- dependent learning. Learn Mem. 2002;9:224–237. doi: 10.1101/lm.51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The bdnf val66met polymorphism affects activity-dependent secretion of bdnf and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 53.Komulainen P, Pedersen M, Hanninen T, Bruunsgaard H, Lakka TA, Kivipelto M, Hassinen M, Rauramaa TH, Pedersen BK, Rauramaa R. BDNF is a novel marker of cognitive function in ageing women: the DR's EXTRA study. Neurobiol Learn Mem. 2008;90:596–603. doi: 10.1016/j.nlm.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Miyajima F, Ollier W, Mayes A, Jackson A, Thacker N, Rabbitt P, Pendleton N, Horan M, Payton A. Brain-derived neurotrophic factor polymorphism val66met influences cognitive abilities in the elderly. Genes Brain Behav. 2008;7:411–417. doi: 10.1111/j.1601-183X.2007.00363.x. [DOI] [PubMed] [Google Scholar]
- 55.Hammil DD, Brown VL, Larsen SC, Wiederhold JL: Test of adolescent language-2. Austin, TX, Pro-Ed, 1987.
- 56.Hammil DD, Newcomer PL: Test of language development-2, intermediate. Austin, TX, Pro-Ed, 1988.
- 57.Newcomer PL, Hammil DD: Test of language development-2, primary. Austin, TX, Pro-Ed, 1988.
- 58.Wechsler D. Wechsler intelligence scale for children-revised. San Antonio, TX: The Psychological Corporation; 1974. [Google Scholar]
- 59.Wechsler D. Wechsler adult intelligence scale-revised. San Antonio, TX: The Psychological Corporation; 1981. [Google Scholar]
- 60.Wechsler D. Wechsler pre-school and primary scale of intelligence -revised. San Antonio, TX: The Psychological Corporation; 1989. [Google Scholar]
- 61.Wechsler D. Wechsler abrreviated scale of intelligence. San Antonio, TX: The Psychological Corporation; 1999. [Google Scholar]
- 62.Woodcock RW. Woodcock reading mastery tests-revised. Minnesota: American Guidance Service, Inc.; 1987. [Google Scholar]
- 63.The International HapMap Consortium The international hapmap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 64.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. Dbsnp: The NCBI database of genetic variation. Nucleic Acids Research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, Meyer L, Hsu F, Hinrichs AS, Harte RA, Giardine B, Fujita P, Diekhans M, Dreszer T, Clawson H, Barber GP, Haussler D, Kent WJ. The UCSC genome browser database: Update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kong X, Murphy K, Raj T, He C, White PS, Matise TC. A combined linkage-physical map of the human genome. Am J Hum Genet. 2004;75:1143–1148. doi: 10.1086/426405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bruse S, Moreau M, Azaro M, Zimmerman R, Brzustowicz L. Improvements to bead-based oligonucleotide ligation SNP genotyping assays. Biotechniques. 2008;45:559–571. doi: 10.2144/000112960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Connell JR, Weeks DE. Pedcheck: a program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet. 1997;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sobel E, Sengul H, Weeks DE. Multipoint estimation of identity-by-descent probabilities at arbitrary positions among marker loci on general pedigrees. Hum Hered. 2001;52:121–131. doi: 10.1159/000053366. [DOI] [PubMed] [Google Scholar]
- 70.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am J Hum Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 71.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2, a data-handling program for facilitating genetic linkage and association analyses. Am J Hum Genet. 1999;65:A436. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- 73.Wigginton JE, Abecasis GR. Pedstats: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 74.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 75.Govil M, Segre AM, Vieland VJ. Mlip: Using multiple processors to compute the posterior probability of linkage. BMC Bioinformatics. 2008;9(suppl 6):S2. doi: 10.1186/1471-2105-9-S6-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seok SC, Evans M, Vieland VJ. Fast and accurate calculation of a computationally intensive statistic for mapping disease genes. J Comput Biol. 2009;16:659–676. doi: 10.1089/cmb.2008.0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang K, Huang J, Vieland VJ. The consistency of the posterior probability of linkage. Ann Hum Genet. 2000;64:533–553. doi: 10.1046/j.1469-1809.2000.6460533.x. [DOI] [PubMed] [Google Scholar]
- 78.Huang J, Vieland VJ. Comparison of ‘model-free’ and ‘model-based’ linkage statistics in the presence of locus heterogeneity: single data set and multiple data set applications. Human Heredity. 2001;51:217–225. doi: 10.1159/000053345. [DOI] [PubMed] [Google Scholar]
- 79.Vieland VJ, Wang K, Huang J. Power to detect linkage based on multiple sets of data in the presence of locus heterogeneity: comparative evaluation of model-based linkage methods for affected sib pair data. Hum Hered. 2001;51:199–208. doi: 10.1159/000053343. [DOI] [PubMed] [Google Scholar]
- 80.Logue MW, Vieland VJ. A new method for computing the multipoint posterior probability of linkage. Hum Hered. 2004;57:90–99. doi: 10.1159/000077546. [DOI] [PubMed] [Google Scholar]
- 81.Yang X, Huang J, Logue MW, Vieland VJ. The posterior probability of linkage allowing for linkage disequilibrium and a new estimate of disequilibrium between a trait and a marker. Hum Hered. 2005;59:210–219. doi: 10.1159/000086699. [DOI] [PubMed] [Google Scholar]
- 82.Bartlett CW, Vieland VJ. Accumulating quantitative trait linkage evidence across multiple datasets using the posterior probability of linkage. Genet Epidemiol. 2007;31:91–102. doi: 10.1002/gepi.20193. [DOI] [PubMed] [Google Scholar]
- 83.Huang Y, Bartlett CW, Segre AM, O'Connell JR, Mangin L, Vieland VJ. Exploiting gene ×gene interaction in linkage analysis. BMC Proc. 2007;1(Suppl 1):S64. doi: 10.1186/1753-6561-1-s1-s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Logue M, Vieland VJ, Goedken R, Crowe R. Bayesian analysis of a previously published genome screen for panic disorder reveals new and compelling evidence for linkage to chromsome 7. Am J Med Genet B (Neuropsychiat Genet) 2003;121B:95–99. doi: 10.1002/ajmg.b.20072. [DOI] [PubMed] [Google Scholar]
- 85.Bartlett CW, Vieland VJ, Bartlett J, Bell JT, Bhattacharjee S, Clerget-Darpoux F, Bush WS, Edwards TL, Gao G, Halder I, Huang Y, Kotti S, Larkin EK, Li H, Motsinger AA, Mukhopadhyay N, Namkung J, Park T, Ritchie MD, Stein CM, Zhou JY. Discussing gene-gene interaction: warning – translating equations to English may result in jabberwocky. Genet Epidemiol. 2007;31(Suppl 1):S61–67. doi: 10.1002/gepi.20281. [DOI] [PubMed] [Google Scholar]
- 86.Vieland VJ. Bayesian linkage analysis, or: how I learned to stop worrying and love the posterior probability of linkage. Am J Hum Genet. 1998;63:947–954. doi: 10.1086/302076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Logue MW, Vieland VJ, Goedken RJ, Crowe RR. Bayesian analysis of a previously published genome screen for panic disorder reveals new and compelling evidence for linkage to chromosome 7. Am J Med Genet B Neuropsychiatr Genet. 2003;121B:95–99. doi: 10.1002/ajmg.b.20072. [DOI] [PubMed] [Google Scholar]
- 88.Baddeley A. Working memory and language: an overview. J Commun Disord. 2003;36:189–208. doi: 10.1016/s0021-9924(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 89.Harlaar N, Hayiou-Thomas ME, Dale PS, Plomin R. Why do preschool language abilities correlate with later reading? A twin study. J Speech Lang Hear Res. 2008;51:688–705. doi: 10.1044/1092-4388(2008/049). [DOI] [PubMed] [Google Scholar]
- 90.Bailey PJ, Snowling MJ. Auditory processing and the development of language and literacy. Br Med Bull. 2002;63:135–146. doi: 10.1093/bmb/63.1.135. [DOI] [PubMed] [Google Scholar]
- 91.Bishop DV, Snowling MJ. Developmental dyslexia and specific language impairment: same or different? Psychol Bull. 2004;130:858–886. doi: 10.1037/0033-2909.130.6.858. [DOI] [PubMed] [Google Scholar]
- 92.Larkin RF, Snowling MJ. Comparing phonological skills and spelling abilities in children with reading and language impairments. Int J Lang Commun Disord. 2008;43:111–124. doi: 10.1080/13682820601178584. [DOI] [PubMed] [Google Scholar]
- 93.Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Genetic variant BDNF (val66met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Newbury DF, Bishop DV, Monaco AP. Genetic influences on language impairment and phonological short-term memory. Trends Cogn Sci. 2005;9:528–534. doi: 10.1016/j.tics.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 95.Tallal P. Improving language and literacy is a matter of time. Nat Rev Neurosci. 2004;5:721–728. doi: 10.1038/nrn1499. [DOI] [PubMed] [Google Scholar]
- 96.Catts H, Adlorf S, Weismer S. Language deficits in poor comprehenders: a case for the simple view of reading. J Speech Hear Res. 2006;49:278–293. doi: 10.1044/1092-4388(2006/023). [DOI] [PubMed] [Google Scholar]
- 97.Catts HW, Adlof SM, Hogan TP, Weismer SE. Are specific language impairment and dyslexia distinct disorders? J Speech Lang Hear Res. 2005;48:1378–1396. doi: 10.1044/1092-4388(2005/096). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Archibald LM, Gathercole SE. Short-term and working memory in specific language impairment. Int J Lang Commun Disord. 2006;41:675–693. doi: 10.1080/13682820500442602. [DOI] [PubMed] [Google Scholar]
- 99.Baddeley A, Wilson BA. A developmental deficit in short-term phonological memory: implications for language and reading. Memory. 1993;1:65–78. doi: 10.1080/09658219308258225. [DOI] [PubMed] [Google Scholar]
- 100.Leonard LB, Ellis Weismer S, Miller CA, Francis DJ, Tomblin JB, Kail RV. Speed of processing, working memory, and language impairment in children. J Speech Lang Hear Res. 2007;50:408–428. doi: 10.1044/1092-4388(2007/029). [DOI] [PubMed] [Google Scholar]
- 101.Montgomery JW, Evans JL. Complex sentence comprehension and working memory in children with specific language impairment. J Speech Lang Hear Res. 2009;52:269–288. doi: 10.1044/1092-4388(2008/07-0116). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS. Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science. 2000;288:678–682. doi: 10.1126/science.288.5466.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


