MicroRNAs (miRNAs) are small 21–25 nucleotide-long non-coding RNAs that have emerged as key negative post-transcriptional regulators of gene expression [1, 2]. Currently there are more than 700 mammalian miRNAs that can potentially target up to one-third of protein-coding human genes [1] involved in diverse physiological and pathological processes, including cancer [3, 4]. Indeed, aberrant levels of miRNAs have been reported in all major human malignancies [5, 6]. In tumors, altered expression of miRNAs has been demonstrated to inhibit tumor suppressor genes or inappropriately activate oncogenes and has been associated with every aspect of tumor biology, including tumor progression, invasiveness, metastasis, and acquisition of resistance by malignant cells to chemotherapeutic agents [3, 4, 7, 8]. These observations lead to the suggestion that aberrant expression of miRNAs may contribute to tumorigenesis [9]. However, most of the tumor-miRNA-related studies are based on expression analysis of miRNAs in tumors in comparison with corresponding adjacent normal tissues [4, 5, 6]. The altered expression of any given miRNA in neoplastic cells is not sufficient to address conclusively the role of these changes in tumorigenesis [10]. Additionally, despite the established biological significance of miRNA dysregulation in neoplastic cells, there is a lack of knowledge on the role of miRNAs during early stages of tumor development, especially if variations in the expression of specific miRNAs are associated with differences in the susceptibility to tumorigenesis.
In light of these considerations, the goals of this study were to: (1) define the role of miRNA dysregulation in early stages of liver carcinogenesis, and (2) determine how these alterations in miRNA expression may be mechanistically linked to the pathogenesis of liver cancer induced by dietary methyl deficiency.
Materials and Methods
Animals, Diets and Experimental Design
Male C57BL/6J and DBA/2J mice (Jackson Laboratory, Bar Harbor, Me., USA) were housed in sterilized cages in a temperature-controlled room (24°C) with a 12-hour light/dark cycle, and given ad libitum access to purified water and NIH-31 pelleted diet (Purina Mills, Richmond, Ind., USA). At 8 weeks of age, the mice from each strain were allocated randomly into 2 groups, 1 control and 1 experimental. The mice in the experimental group were maintained on a low methionine (0.18%) diet, lacking in choline and folic acid (Dyets Inc, Bethlehem, Pa., USA) for 12 weeks. The mice in the control group received a diet supplemented with 0.4% methionine, 0.3% choline bitartrate and 2 mg/kg folic acid. Diets were stored at 4°C and given ad libitum, with twice a week replacement. Five experimental and 5 control mice were sacrificed at 12 weeks after diet initiation. The livers were excised, frozen immediately in liquid nitrogen, and stored at −80°C for subsequent analyses. All animal experimental procedures were carried out in accordance with the animal study protocol approved by the National Center for Toxicological Research Animal Care and Use Committee.
RNA Extraction and miRNA Microarray Expression Analysis
Total RNA was extracted from the liver tissue using miRNAeasy Mini Kit (Qiagen, Valencia, Calif, USA) according to the manufacturer's instructions. The miRNA microarray analysis was performed by LC Sciences (Houston, Tex., USA), as reported previously in detail [11].
miRNA Expression Analysis by Quantitative Reverse Transcription Real-Time PCR
Total RNA (200 ng) was used for qRT-PCRs of the miR-29c, miR-34a, miR-122, miR-155, miR-192, miR-200b, miR-203 and miR-221, utilizing TaqMan miRNA assays (Applied Biosystems, Foster City, Calif, USA), according to the manufacturer's instructions. snoRNA202 was used as an endogenous control. The relative amount of each miRNA was measured using the 2−ΔΔCt method [12]. All qRT-PCR reactions were conducted in triplicate and repeated twice.
Gene Expression Analysis by qRT-PCR
Total RNA (10 μg) was reverse transcribed using random primers and a high-capacity cDNA archive kit (Applied Biosystems), according to the manufacturer's protocol. The expression of the α-smooth muscle actin (α-Sma) gene was measured by qRT-PCR, using Taqman® gene expression assay (Mm00725412_sl; Applied Biosystems).
Western Blot Analysis of Protein Expression
The levels of cyclin Gl (Ccngl), cyclogenase 2 (Cox2), E2F transcription factor 3 (E2f3), and CCAAT enhancer binding protein beta (C/ebp-ß) proteins were determined by Western immuno-blot analysis [13].
Statistical Analysis
Results are presented as mean ± SD. Statistical analyses were conducted by 1-way ANO VA, using treatment and weeks as fixed factors. Pair-wise comparisons were conducted by the Student-Newman-Keuls test, p values <0.05 were considered significant.
Results and Discussion
Dysregulation of miRNAs in the Livers of C57BL/6J Mice Fed a Methyl-Deficient Diet
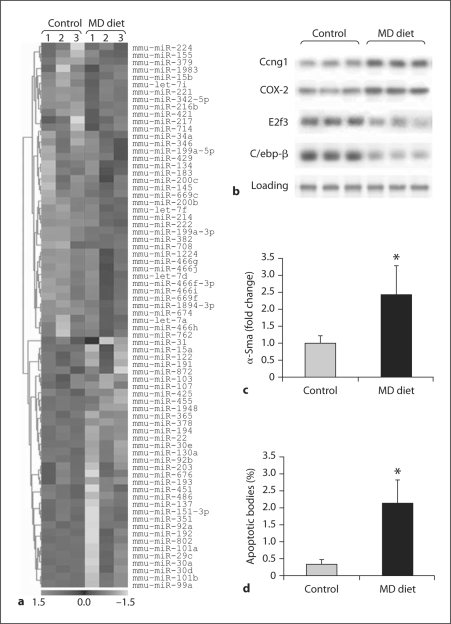
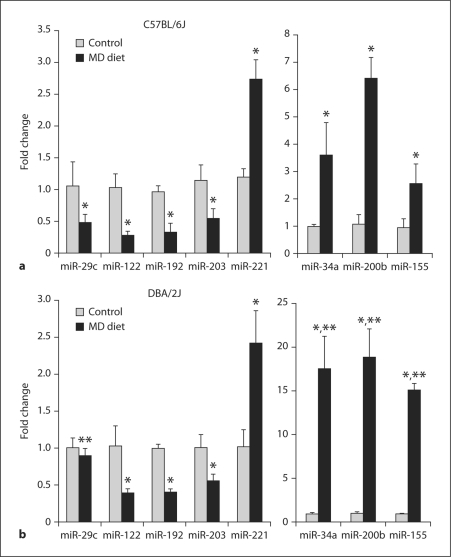
miRNA microarrays were used to analyze the miRNA expression profiles in the livers of control C57BL/6J mice and C57BL/6J mice fed a methyl-deficient diet that causes a liver pathological state similar to human nonalcoholic fatty liver disease [14]. We identified 74 miRNAs (40 up-regulated and 34 down-regulated) that were differentially expressed (p < 0.05), including miR-15a, miR-29c, miR-30a, miR-34a, miR-lOla, miR-107, miR-122, miR-155, miR-200b, miR-200c, miR-221, miR-222 and miR-224 in the livers of the C57BL/6J methyl-deficient mice (fig. 1a). The results obtained by miRNA microarray analysis were confirmed by qRT-PCR (fig. 2a).
Fig. 1.
Dysregulation of miRNA expression in the livers of C57BL/6J mice fed a methyl-deficient diet for 12 weeks, a Hierarchical clustering of the differentially expressed miRNA genes (as determined by ANOVA) in the livers of control and methyl-deficient (MD) mice. Rows show miRNA, while columns show independent biological replicates. For each miRNA red indicates high expression levels and green indicates low expression levels. Each miRNA listed is significantly differentially expressed (p < 0.05; n = 3). b Western blot analysis of Ccngl (miR-122), COX-2 (miR-101a), E2f3 (miR-34a and miR-200b) and Cebp/β (miR-155) proteins in the livers of control and methyl-deficient mice, c qRT-PCR analysis of α-Sma gene in the livers of control and methyl-deficient mice (mean ± SD; n = 5). d Apoptotic cell death in the livers of control and methyl-deficient mice as detected by TUNEL assay (mean ± SD; n = 5).
Fig. 2.
qRT-PCR analysis of differentially expressed miRNAs in the livers of control C57BL/6J (a) and DBA/2J mice (b) and mice fed a methyl-deficient diet (MD) for 12 weeks. * Significantly different from control mice. ** Significantly different from C57BL/6J methyl-deficient mice (mean ± SD; n = 5).
Functions of Dysregulated miRNAs
Dysregulated miRNAs are known to affect cell proliferation, apoptosis, lipid metabolism, oxidative stress, DNA methylation and inflammation. These processes are substantially compromised in pathological states associated with hepatocarcinogenesis. Specifically, it is well-established that altered lipid metabolism, oxidative stress, apoptosis and epigenetic alterations may directly trigger hepatic steatosis, a condition that has been shown to progress to hepatocellular carcinoma [15, 16, 17].
Among the down-regulated miRNAs, miR-15a, miR-30a, miR-101a and miR-122 are of particular interest. Previously, we and other investigators have demonstrated a substantial down-regulation of liver-specific miR-122 during liver carcinogenesis and in primary hepatocellular carcinomas [18, 19, 20, 21]. Recently, a significant decrease in miR-122 expression has been observed in individuals with non-alcoholic steato-hepatitis [22]. The down-regulation of miR-122 in the livers of C57BL/6J mice fed a methyl-deficient diet was accompanied by increased level of Ccngl protein (fig. 1b). The altered expression of CCNG1 [19] and other confirmed targets of miR-122, such as fatty acid synthase [22, 23], sterol regulatory element-binding protein-lc [22, 23], cationic amino acid transporter (CAT1; SLC7A1) [24], and BCL-W, an anti-apoptotic member of BCL2 family member [25], has frequently been observed during hepatocarcinogenesis and has been attributed to the pathogenesis of liver cancer.
Feeding C57BL/6J mice a methyl-deficient diet for 12 weeks resulted in decreased expression of miR-lOla and miR-lOlb (fig. 1a). One of the confirmed targets for miR-lOla is Cox-2 [26], which is substantially up-regulated in the livers of mice exposed to the methyl-deficient diet (fig. 1b). The increased expression of COX-2 has been detected during human and rodent liver tumor development [27, 28] and is currently considered as an attractive target for chemoprevention during early stages of hepatocarcinogenesis. Additionally, recent evidence has demonstrated that miR-101 targets FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene [29], a key component of the liver oncogenic network [30].
Another down-regulated miRNA in the livers of mice fed the methyl-deficient diet is miR-15a, one of the first miRNA's discovered to be dysregulated in cancer [31]. miR-15a targets multiple oncogenic pathways, including BCL2, cyclin Dl (CCNDl) and WNT3A signaling [31], a pathway that triggers the activation of hepatic stellate cells and progression of hepatic fibrosis [32]. miR-107 [20] and let-7a and let-7d [33], which are down-regulated (miR-107) and up-regulated (let-7a and let-7d) in the livers of methyl-deficient mice (fig. 1a), have also been associated with the pathogenesis of hepatic steatosis, fibrosis and hepatocarcinogenesis. Indeed, figure 1c shows an increase in expression of the α-Sma gene, a marker of hepatic stellate cell activation and fibrosis development [34] in the livers of mice fed the methyl-deficient diet.
miR-34a, miR-155, miR-200b and miR-221 were the most up-regulated miRNAs among the differentially expressed miRNAs in the livers of methyl-deficient C57BL/6J mice (figs. 1a and 2). The transcription factor E2f3, a critical regulator of the p53 network, is one of the targets for these miRNAs as reported in Targetscan 5.1 (www.targetscan.org) and in other reports [35, 36]. Furthermore, there is a solid connection between miR-34 and the p53 apoptotic pathway [37, 38, 39], which plays a pivotal role in the pathogenesis of liver injury regardless of its etiology, and especially in non-alcoholic hepatosteatitis [40, 41]. Figure 1d shows the increased apoptosis in the livers of C57BL/6J mice fed a methyl-deficient diet. Additionally, recent evidence has demonstrated the importance of miR-34a, not only in apoptosis, but also in non-apoptotic cell death in vivo [42].
The over-expression of miR-155 and miR-221 has been frequently detected during tumor development [43, 44]. The up-regulation of these miRNAs has been associated with activation of the extracellular signal-regulated (ERK) and phosphati-dylinositol 3-kinase (PI3)-AKT pathways, 2 pathways frequently disturbed during liver tumorigenesis. Furthermore, the results of a recent study have demonstrated that miR-221 targets and down-regulates pro-apoptotic BCL2-modifying factor during human hepatocarcinogenesis [45]. It is well-established that one of the hallmarks of the carcinogenic process is a dysregulation of cell proliferation and apoptosis [46]. In this context, the altered expression of miR-34a, miR-155, miR-200b and miR-221 in the livers of methyl-deficient mice illustrates the critical role of miRNA in the disruption of the delicate balance between cell division and apoptosis during carcinogenesis.
In a previous study [17], we demonstrated that feeding DBA/2 J mice a lipogenic methyl-deficient diet resulted in more prominent pathomorphological and molecular changes in the livers, including DNA hypomethylation, a greater severity of steatosis and necrosis, and oval cell proliferation, as compared to C57BL/6J mice. Interestingly, we detected strain-specific significant differences in the expression of miR-29c, miR-34a, miR-155 and miR-200b in the livers of C57BL/6J (fig. 2a) and DBA/2J methyl-deficient mice (fig. 2b). Specifically, the expression of miR-34a, miR-155 and miR-200b in the livers of DBA/2J mice fed the methyl-deficient diet was, respectively, 4.9, 5.9 and 3.0 times greater than in methyl-deficient C57BL/6J mice. Likewise, the livers of C57BL/6J mice were characterized by a more pronounced down-regulation of miR-29c. The aberrant expression of these miRNAs is associated with an altered DNA methylation status (miR-29c), increased cell death (miR-34a and miR-200b), and liver steatosis and fibrosis (miR-155). miR-155, which was the most differentially expressed miRNA in the livers of DBA/2J and C57BL/6J mice fed the methyl-deficient diet, activates the AKT signaling pathway [47], triggering oval cell proliferation [48], a fundamental event in hepatocarcinogenesis.
In conclusion, these findings demonstrate that alterations in expression of miRNAs are a prominent event during early stages of liver carcinogenesis induced by methyl deficiency and strongly suggest that differences in the susceptibility to liver carcinogenesis may be determined by the variations in miRNA expression response. More importantly, our data provide a mechanistic link between alterations in microRNA expression and the pathogenesis of liver cancer.
Disclaimer
The views presented in this chapter do not necessarily represent those of the US Food and Dru£ Administration.
Footnotes
Originally published: Simopoulos AP Milner JA (eds): Personalized Nutrition.
World Rev Nutr Diet. Basel, Karger, 2010, vol 101, pp 123–130
A.S.-D. and V.T contributed equally to this work.
Starlard-Davenport et al.: Dietary Methyl Deficiency, miRNA Expression and Liver Carcinogenesis
References
- 1.Bartell DP. MicroRNAs: genomics, biogenesis, and 15 function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Guarnieri DJ, DiLeone RJ. MicroRNAs: a new class of gene regulators. Ann Med. 2008;40:197–208. doi: 10.1080/07853890701771823. [DOI] [PubMed] [Google Scholar]
- 3.Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go along way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garzón R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Ma L, Teruya-Feldstein J, Weinberg RA. Tumor 19 invasion and metastasis initiated by microRNA- 10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 8.Zheng T, Wang J, Chen X, Liu LX. Role of microRNA in anticancer drug resistance. Int J Cancer. 2010;126:2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- 9.Osada H, Takahashi T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis. 2007;28:2–12. doi: 10.1093/carcin/bgl185. [DOI] [PubMed] [Google Scholar]
- 10.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and onco-genes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 11.Pogribny IP, Tryndyak VP, Boyko A, et al. Induction of microRNAome deregulation in rat liver by long- 22 term tamoxifen exposure. Mutat Res. 2007;619:30–37. doi: 10.1016/j.mrfmmm.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR 23 and the 2–ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Pogribny IP, Muskhelishvili L, Tryndyak VP, Beland FA. The tumor-promoting activity of 2-acetylamin-ofiuorene is associated with disruption of the p53 24 signaling pathway and the balance between apoptosis and cell proliferation. Toxicol Appl Pharmacol. 2009;235:305–311. doi: 10.1016/j.taap.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 14.Anstee QM, Goldin RD. Mouse models in nonalcoholic fatty liver and steatohepatitis research. Int JExpPathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell GC, Larter CZ. Nonalcoholic fatty liver disease: from steatosis to cirrhosis. Hepatology. 2006;43:S99–S112. doi: 10.1002/hep.20973. [DOI] [PubMed] [Google Scholar]
- 16.Erickson SK. Nonalcoholic fatty liver disease. J Lipid Res. 2009;50:S412–S416. doi: 10.1194/jlr.R800089-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pogribny IP, Tryndyak VP, Bagnyukova TV, et al. Hepatic epigenetic phenotype predermines individual susceptibility to hepatic steatosis in mice fed a lipogenic methyl-deficient diet. J Hepatol. 2009;51:176–186. doi: 10.1016/j.jhep.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutay H, Bai S, Datta J, et al. Down-regulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Gramantieri L, Ferrracin M, Fornari F, et al. Cyclin Gl is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 20.Ladeiro Y, Couchy G, Balabaud C, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical fetures and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 21.Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung O, Puri P, Eicken C, et al. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology. 2008;48:1810–1820. doi: 10.1002/hep.22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuyoshi H, Yasui K, Harano Y, et al. Analysis of hepatic genes involved in the metabolism of fatty acids and iron in non-alcoholic fatty liver disease. Hepatol Res. 2009;39:366–373. doi: 10.1111/j.1872-034X.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- 24.Chang J, Nicolas E, Marks D, et al. miR-122, a mammalian liver-specific microRNA, is processed from her mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 25.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka T, Haneda S, Imakawa K, Sakai S, Nagaoka K. A microRNA, miR-lOla, controls mammary gland development by regulating cyclooxygenase-2 expression. Differentiation. 2009;77:181–187. doi: 10.1016/j.diff.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Denda A, Kitayama W, Murata A, et al. Increased expression of cycloogenase-2 protein during rat hepatocarcinogenesis caused by a choline-deficient, L-amino acid defined diet and chemopreventive efficacy of a specific inhibitor, nimesulide. Carcinogenesis. 2002;23:245–256. doi: 10.1093/carcin/23.2.245. [DOI] [PubMed] [Google Scholar]
- 28.Sung YK, Hwang SY, Kim JO, et al. The correlation between cyclooxygenase-2 expression and hepatocellular carcinogenesis. Mol Cells. 2004;17:35–38. [PubMed] [Google Scholar]
- 29.Li S, Fu H, Wang Y, et al. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–1202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 30.Caselmann WH. Transactivation of cellular gene expression by hepatitis B viral proteins: a possible molecular mechanism of hepatocarcinogenesis. J Hepatol. 1995;22:34–37. [PubMed] [Google Scholar]
- 31.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2009;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 32.Myung Sj, Yoon JH, Gwak GY, et al. Wnt signaling enhances the activation and survival human hepatic stellate cells. FEBS Lett. 2007;581:2954–2958. doi: 10.1016/j.febslet.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 33.Mott JL. MicroRNAs involved in tumor suppressor and oncogene pathways: implications for hepatobil-iary neoplasia. Hepatology. 2009;50:630–637. doi: 10.1002/hep.23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefkowitch JH. Hepatobiliary pathology. CurrOpin Gastroenterol. 2006;22:198–208. doi: 10.1097/01.mog.0000218955.55688.af. [DOI] [PubMed] [Google Scholar]
- 35.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 36.Tazawa H, Tsuchiya N, Izumiya M, Nakagawa H. Tumor-suppressive miR34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Nati Acad Sei USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 39.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRTl regulates apoptosis. Prie Nati Acad Sei USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wieckowska A, Zein NN, Yerian LM, et al. In vivo assessment of liver cell apoptosis as a novel bio-marker of disease severity in nonalcoholic fatty liver disease. Hepatology. 2006;44:27–33. doi: 10.1002/hep.21223. [DOI] [PubMed] [Google Scholar]
- 41.Farrell GC, Larter CZ, Hou JY, et al. Apoptosis in experimental NASH is associated with p53 activation and TRAIL receptor expression. J Gastroenterol Hepatol. 2009;;24:443–452. doi: 10.1111/j.1440-1746.2009.05785.x. [DOI] [PubMed] [Google Scholar]
- 42.Kato M, Paranjape T, Ullrich R, et al. The miR-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gramantieri L, Fornari F, Callegari E, et al. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 45.Gramantieri L, Fornari F, Ferracin M, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073–5081. doi: 10.1158/1078-0432.CCR-09-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 47.Yamanaka Y, Tagawa H, Takahashi N, et al. Aberrant overexpression of microRNAs activate AKT signaling via downregulation of tumor suppressors in NK-celllymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 48.Okano J, Shiota G, Matsumoto K, et al. Hepatocyte growth factor exerts a proliferative effect on oval cells through the PI3/AKT signaling pathway. Biochem Biophys Res Commun. 2003;309:298–304. doi: 10.1016/j.bbrc.2003.04.002. [DOI] [PubMed] [Google Scholar]