Abstract
Diffuse Optical Imaging (DOI) allows the recovery of the hemodynamic response associated with evoked brain activity. The signal is contaminated with systemic physiological interference which occurs in the superficial layers of the head as well as in the brain tissue. The back-reflection geometry of the measurement makes the DOI signal strongly contaminated by systemic interference occurring in the superficial layers. A recent development has been the use of signals from small source-detector separation (1 cm) optodes as regressors. Since those additional measurements are mainly sensitive to superficial layers in adult humans, they help in removing the systemic interference present in longer separation measurements (3 cm). Encouraged by those findings, we developed a dynamic estimation procedure to remove global interference using small optode separations and to estimate simultaneously the hemodynamic response. The algorithm was tested by recovering a simulated synthetic hemodynamic response added over baseline DOI data acquired from 6 human subjects at rest. The performance of the algorithm was quantified by the Pearson R2 coefficient and the mean square error (MSE) between the recovered and the simulated hemodynamic responses. Our dynamic estimator was also compared with a static estimator and the traditional adaptive filtering method. We observed a significant improvement (two-tailed paired t-test, p < 0.05) in both HbO and HbR recovery using our Kalman filter dynamic estimator compared to the traditional adaptive filter, the static estimator and the standard GLM technique.
Keywords: Diffuse Optical Imaging, hemodynamic response, multidistance measurements, adaptive filtering, Kalman filtering
1. Introduction
Diffuse optical imaging (DOI) is an experimental technique that uses near-infrared spectroscopy (NIRS) to image biological tissue (Villringer et al., 1993; Obrig and Villringer, 2003; Gibson et al., 2005; Hillman, 2007; Hoshi, 2007). The dominant chromophores in this spectrum are the two forms of hemoglobin: oxygenated hemoglobin (HbO) and reduced hemoglobin (HbR). In the past 15 years, this technique has been used for the noninvasive measurement of the hemodynamic changes associated with evoked brain activity (Villringer et al., 1993; Hoshi, 2007).
Compared with other existing functional imaging methods e.g., functional Magnetic Resonance Imaging (fMRI), Positron Emission Tomography (PET), Electroencephalography (EEG), and Magnetoencephalography (MEG), the advantages of DOI for studying brain function include good temporal resolution of the hemodynamic response, measurement of both HbO and HbR, nonionizing radiation, portability, and low cost. Disadvantages include modest spatial resolution and limited penetration depth.
The sensitivity of NIRS to evoked brain activity is also reduced by systemic physiological interference arising from cardiac activity, respiration, and other homeostatic processes (Obrig et al., 2000; Tonorov et al., 2000; Payne et al., 2009; Diamond et al., 2009). These sources of interference are called global interference or systemic interference. Part of the interference occurs both in the superficial layers of the head (scalp and skull) and in the brain tissue itself. However, the back-reflection geometry of the measurement makes NIRS significantly more sensitive to the superficial layers. As such, the NIRS signal is often dominated by systemic interference occurring in the skin and the skull.
Different methods have been used in the literature to remove the systemic interference from DOI measurements. Low pass filtering is widely used in the literature, as it is highly effective at removing cardiac oscillations (Franceschini et al., 2003; Jasdzewski et al., 2003). However, there is a significant overlap between the frequency spectrum of the hemodynamic response to brain activity and the spectrum of other physiological variations such as respiration, spontaneous low frequency oscillations and very low frequency oscillations. Frequency-based removal of these sources of interference can therefore result in large distortion and inaccurate timing for the recovered brain activity signal. As such, more powerful methods for global noise reduction have been developed. These include adaptive average waveform subtraction (Gratton and Corballis, 1995), subtraction of another NIRS source-detector (SD) channel performed over a non-activated region of the brain (Franceschini et al., 2003), principal component analysis (Zhang et al., 2005; Franceschini et al., 2006) and finally wavelet filtering (Lina et al., 2008; Matteau-Pelletier et al., 2009; Jang et al., 2009; Lina et al., 2010).
A recent development for removing global interference from NIRS measurements is to use additional optodes in the activated region with small SD separations that are sensitive to superficial layers only (Saager and Berger, 2008; Zhang et al., 2007a,b, 2009; Umeyama and Yamada, 2009; Yamada et al., 2009; Gregg et al., 2010). Making the assumption that the signal collected in the superficial layers is dominated by systemic physiology which is also dominant in the longer SD separation NIRS channel, those additional measurements can be used as regressors to filter systemic interference from the longer SD separations. Saager et al (2005) used additional optodes and a linear minimum mean square estimator (LMMSE) to partially remove the systemic interference in the signal. In a second step, the evoked hemodynamic response was estimated using a traditional block-average method over the different trials. The algorithm was further refined by Zhang et al (2007a; 2007b; 2009) to consider the non-stationary behavior of the systemic interference. They used an adaptive filtering technique together with additional small separation measurements to filter the systemic interference from the raw signal and then performed the block-average technique to estimate the hemodynamic response in a second step.
Although these methods greatly reduced global interference in NIRS data, the filtering of the systemic interference and the estimation of the hemodynamic response were performed in two steps, which might not be optimal. Previous studies have shown that the simultaneous estimation of the hemo-dynamic response and removal of the systemic interference using temporal basis functions (Kolehmainen et al., 2003; Prince et al., 2003) or auxiliary systemic measurements (Diamond et al., 2006) was possible using state-space modeling. Moreover, Diamond et al proposed a way to quantify the accuracy of such filtering methods. Real NIRS data collected over the head of human subjects at rest were used to generate realistic noise. A synthetic hemodynamic response was added over the real NIRS baseline time course and the response was then recovered from this noisy data set. The recovered response was then compared with the synthetic one used to generate the time course. This method for evaluating reconstruction algorithms has been reproduced by other groups (Lina et al., 2008; Matteau-Pelletier et al., 2009; Lina et al., 2010).
In the present study, we combined small separation measurements and state-space modeling for the estimation of the hemodynamic response and simultaneous global interference cancellation. We developed both a static and a dynamic estimator. We evaluated the performance of our algorithms using baseline data taken from 6 human subjects at rest and by adding a synthetic hemodynamic response over the baseline measurements. We finally compared our new methods with the adaptive filter (Zhang et al., 2007a) and the standard method using no small SD separation measurement.
2. Methods
2.1. Experimental data
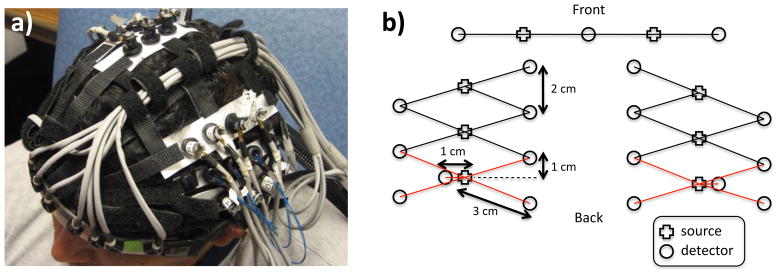
For this study, 6 healthy adult subjects were recruited. The Massachusetts General Hospital Institutional Review Board approved the study and all subjects gave written informed consent. Subjects were instructed to rest while simultaneous BOLD-fMRI and NIRS data were collected. Three 6-minute long runs were collected for each subject. Only the NIRS data was used in this study. The localization and the geometry of the NIRS probe used are shown in Fig. 1a) and b) respectively. Only the two 1 cm SD separation channels and the 8 closest neighbor (3 cm SD separation) channels were used in the analysis.
Figure 1.
a) Position of the probe over the head of the subjects b) Geometry of the optical probe. Two different SD separations were used: 1 cm and 3 cm. The NIRS channels used for the analysis are shown in red.
Changes in optical density for each SD pair were converted to changes in hemoglobin concentrations using the Beer-Lambert relationship (Cope and Deply, 1998; Deply et al., 1988; Boas et al., 2004) and the SD distances illustrated in Fig. 1b). A pathlength correction factor of 6 and a partial volume correction factor of 50 were used for all SD pairs (Huppert et al., 2006a,b).
2.2. Synthetic hemodynamic response
To compare the performance of our two algorithms with existing algorithms, a synthetic hemodynamic response was generated using a modified version of a three compartment biomechanical model (Huppert et al., 2007, 2009; Huppert, 2007). Each parameter of the model was set to the middle of its physiological range (Huppert et al., 2007) which results in an HbO increase of 15 μM and an HbR decrease of 7 μM. The amplitude of this synthetic response was of the same order as real motor responses on humans using NIRS and those specific pathlength and partial volume correction factors (Huppert et al., 2006b). These synthetic HbO and HbR responses were then added to the unfiltered concentration data with an inter-stimulus interval taken randomly from a uniform distribution (10–35 s) for each individual trial. Over the six-minute data series, we added either 10, 30 or 60 individual evoked responses. The resulting HbO and HbR time courses were then highpass filtered at 0.01 Hz to remove any drifts and lowpass filtered at 1.25 Hz to remove the instrument noise. The filter used was a 3rd order Butterworth-type filter.
Four different methods were then used to recover the simulated hemodynamic response added to our baseline data. The first two were taken directly from literature and consisted of the standard General Linear Model (GLM) without using a small SD separation measurement and the adaptive filtering (AF) method developed by Zhang et al (Zhang et al., 2007a). The third one was a simultaneous static deconvolution and regression and will be called the static estimator (SE) here for simplicity. The last one was a dynamic Kalman filter estimator (KF).
2.3. Signal modeling
For all the methods used in this study, the discrete-time hemodynamic response h at sample time n was reconstructed with a set of temporal basis functions
| (1) |
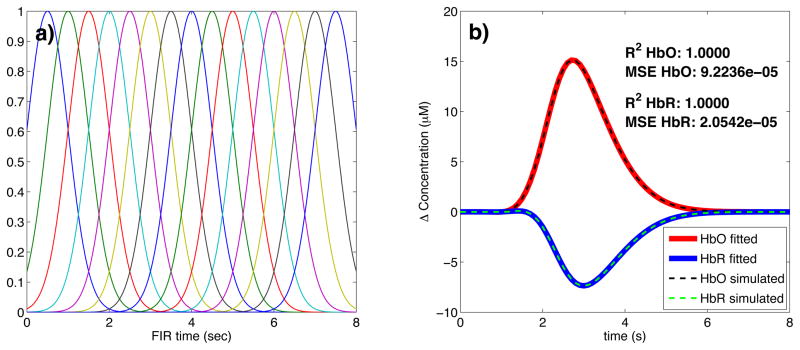
where bi [n] are normalized Gaussian functions with a standard deviation of 0.5 s and their means separated by 0.5 s over the regression time as shown in Fig. 2a). Nw is the number of Gaussian functions used to model the hemodynamic response and was set to 15 in our work. Using this set, the noise-free simulated HbO response was fit with a Pearson R2 of 1.00 and a mean square error (MSE) of 9.2 × 10−5 and the noise-free simulated HbR response was fit with an R2 of 1.00 and an MSE of 2.1 × 10−5. The MSE was lower for HbR only because the amplitude of the simulated HbR response was lower. These fits are shown in Fig. 2b). The weights for the temporal bases wi were estimated using the four different methods described in the following sections.
Figure 2.
a) Temporal basis set used in the analysis. The finite impulse response (FIR) of the temporal basis functions ranged from 0 to 8 s after the onset of the simulated response. b) Noise-free simulated responses (dotted lines) overlapped with the responses recovered with a least-square fit (continuous lines) using the temporal basis set. The R2 and the MSE of the fit are indicated for both HbO and HbR.
For the standard block average estimator, we modeled the concentration signal in the 3 cm separation channel y3 [n] by
| (2) |
u [n] is called the onset vector and is a binary vector taking the value 1 when n corresponds to a time where the stimulation starts and 0 otherwise.
For our static simultaneous estimator and our dynamic Kalman filter simultaneous estimator, we modeled the signal in the 3 cm separation channel y3 [n] by a linear combination of the 1 cm separation signal y1 [n] and the hemodynamic response h [n] by
| (3) |
Na is the number of time points taken from the 1 cm separation channel to model the superficial signal in the 3 cm separation channel. This value was set to 1 in our work for all three estimators using short SD separation measurements but could be any integer in principle. The ai’s are the weights used to model the superficial signal in the 3 cm separation channel from the linear combination of the 1 cm separation signal. The states to be estimated by the static and the Kalman filter estimators were the weights for the superficial contribution ai and the weights for the temporal bases wi. All those weights were assumed stationary in the case of the static estimator, and time-varying in the case of the Kalman filter estimator.
The motivation for Eq. 3 is that the residual between the 3 cm channel and the 1 cm channel corresponds to the hemodynamic response of the brain. This is well justified when the brain activation is detected only in the 3 cm separation channel and when the systemic physiology pollutes both the 1 cm and the 3 cm separation channels. It is a reasonable assumption for cognitive NIRS measurements performed on an adult head. In this case, the hemodynamic response is expected to occur only in the brain tissue and the 1 cm separation channel does not reach the cerebral cortex, making the 1 cm measurement sensitive to scalp and skull fluctuations only. This would also be justified for cognitive measurements on babies by reducing the separation of the 1 cm signal to ensure that this channel remains insensitive to brain hemodynamics. However, our assumption would be violated for specific stimuli (e.g. the Valsalva maneuver) for which the hemodynamic response occurs more globally across the head. Other scenarios that could be troublesome would be if the systemic physiology occurs only in the brain tissue (e.g. an activation-like oscillation a few seconds after the true stimulus response) or if the interference is phase-locked with the stimulus. In this case, the systemic physiology could potentially be modeled by our temporal basis set (overfitting).
2.4. Standard General Linear Model
For this first method, and only for this one, the 1 cm SD separation channels were not used. The pre-filtered concentrations from the 3 cm SD separation were further lowpass filtered at 0.5 Hz using a 3rd order Butter-worth filter. Re-expressing Eq. 2 in matrix form, we get
| (4) |
where y3 is simply the length Nt time course vector y3 [n]
| (5) |
The columns of U are the linear convolution of the onset vector u [n] with each temporal basis function bi [n]
| (6) |
and w is the vector containing the weights for the temporal basis wi
| (7) |
The estimates of the weights ŵ are found by inverting Eq. 4 using the Moore-Penrose pseudoinverse
| (8) |
and the hemodynamic response is finally reconstructed with the estimates of the temporal basis weights ŵi obtained from ŵ.
When the GLM was used without any other estimator (i.e. not as the last step of the adaptive filter or the Kalman filter), we included a 3rd order polynomial drift as a regressor. This procedure is used regularly in fMRI analysis. In this case, the matrix U is expanded
| (9) |
where D is an Nt by 4 drift matrix given in the Appendix A. The estimates of the weights ŵ are found by inverting
| (10) |
2.5. Adaptive filtering
The adaptive filtering technique was taken directly from (Zhang et al., 2007a). Only the salient points are outlined here. The HbO and the HbR responses were recovered independently and the adaptive filter was used for both. The two pre-filtered concentration signals at 1 cm (y1) and 3 cm (y3) were first normalized with respect to their respective standard deviation. This was to ensure that the standard deviation of the two signals used in the computation were close 1 to accelerate the convergence of the algorithm (Zhang et al., 2007a). The output of the filter, e [n], is then given by
| (11) |
where the coefficient of the filter, wk;n, is updated via the Widrow-Hoff least mean square algorithm (Haykin, 2001a):
| (12) |
In our study, w was initialized at and μ was set to 1×10−4 as in (Zhang et al., 2007a). After trying different values for Na, we identified Na = 1 as the value minimizing the MSE between our simulated and recovered hemodynamic responses. The output e [n] was then multiplied by the original standard deviation of y3 to rescale it back to its original scale. The output of the filter was then further lowpass filtered at 0.5 Hz and the hemodynamic response was finally estimated using the standard GLM method (with no drift) by substituting y3 by e in Eq. 8
| (13) |
where e is simply the length Nt time course vector e [n]
| (14) |
and again the hemodynamic response is finally reconstructed with the estimates of the temporal basis weights ŵi obtained from ŵ.
2.6. Static estimator
Our static estimator is an improved version of the linear minimum mean square estimator (LMMSE) developed by Saager et al (Saager and Berger, 2005, 2008). In their work, they used the small separation signal and an LMMSE to estimate the contribution of the superficial signal in the large separation signal. This superficial contamination was then removed from the large separation signal and the hemodynamic response was then estimated from the residual (large separation signal without the superficial contamination). In our study, we simultaneously removed the contribution of the superficial signal in the 3 cm separation signal and estimated the hemodynamic response.
Eqs. 3 and 1 can be re-expressed in matrix form
| (15) |
where y3 is the vector representing the signal in the 3 cm channel and is given by Eq. 5, x is the concatenation of the wi’s and ai’s
| (16) |
and A is the concatenation of the Nt by Nw matrix U given by Eq. 6 and the Nt by Na matrix Y
| (17) |
Where
| (18) |
The first Nw columns of A are the linear convolution of the onset vector u [n] with each temporal basis function bi [n] and the last Na columns of A are simply the signal from the 1 cm separation channel y1 [n] delayed by one more sample in each column. In order to compare the different estimators on the same footing, Na was set to 1 for all three estimators using short SD separations. A more explicit expression for A is given in Appendix A. The estimates of the weights x̂ are found by inverting Eq. 15 using the Moore-Penrose pseudoinverse
| (19) |
and the hemodynamic response is finally reconstructed with the estimates of the temporal basis weights ŵi obtained from x̂. This reconstructed response was further lowpass filtered at 0.5 Hz.
2.7. Kalman filter estimator
For our dynamic Kalman filter estimator, Eqs. 3 and 1 need to be re-express in state-space form:
| (20) |
| (21) |
where w [n] and v [n] are the process and the measurement noise respectively. x [n] is the sample n of x given by Eq. 16, I is an Nw + Na by Nw + Na identity matrix and C [n] is an Nw + Na by 1 vector whose entries correspond to the nth row of A in Eq. 17. The estimate x̂ [n] at each sample n is then computed using the Kalman filter (Kalman, 1960) followed by the Rauch–Tung–Striebel smoother (Rauch et al., 1965). The Kalman filter recursions require initialization of the state vector estimate x̂ [0] and estimated state covariance P [0]. In our study, the initial state vector estimate x̂ [0] was set to the values obtained using our static estimator and the initial state covariance estimate P [0] was set to an identity matrix with diagonal entries of 1×10−1 for the temporal basis states and 5×10−4 for the superficial contribution state. The Kalman filter algorithm was run a first time to estimate the initial state covariance and then run a second time. The initial covariance estimate for the second run was set to the final covariance estimate of the first run. Running the filter twice makes the method less sensitive to the initial guess P [0]. Statistical covariance priors must also be specified for the state process noise cov (w) = Q and the measurement noise cov (v) = R. The process noise determines how big the states are allowed to vary at each time step. If this value is small, the estimator will approach the static estimator. If it is large, the state will be allowed to vary significantly over time. In this work, the process noise covariance only contained nonzero terms on the diagonal elements. Those diagonal terms were set to 2.5×10−6 for the temporal basis state and 5×10−6 for the superficial contribution states. This imbalance in state update noise was also used by Diamond et al (2006) and caused the functional response model to evolve more slowly than the superficial contribution model. Practically, the measurement noise determine how well we trust the measurements during the recovery procedure. In our study, the measurement noise covariance was set to an identity matrix scaled by 5×10−2. Different values have been tried for the process noise and the measurement noise covariances. Changing the value of Q and R over two orders of magnitude did not result in notable performance changes and we could have drawn all the same conclusions presented in this paper using these alternative Q and R values. The values for Q and R presented above were empirically determined to minimize the MSE between the recovered and the simulated hemodynamic response. The algorithm was then processed with the following prediction-correction recursion (Gelb, 1974).
Since the state update matrix is the identity matrix in Eq. 20, the state vector x and state covariance P are predicted with
| (22) |
| (23) |
The Kalman gain K is then computed
| (24) |
and the state vector x and state covariance P predictions are corrected with the most recent measurements y3 [n]
| (25) |
| (26) |
After the Kalman algorithm was applied twice, the Rauch–Tung–Striebel smoother was applied in the backward direction. With the identity matrix as the state-update matrix in Eq. 20, the algorithm is given by (Haykin, 2001b):
| (27) |
The complete time course of the estimated hemodynamic response ĥ [n] was then reconstructed for each sample time n using the final state estimates x̂ [n|Nt] and the temporal basis set contained in C [n]
| (28) |
This reconstructed hemodynamic response time course ĥ [n] was further low-pass filtered at 0.5 Hz and the standard GLM estimator (with no polynomial drift) was then applied
| (29) |
where U is the matrix defined in Eq. 6 and
| (30) |
to obtain the final weights ŵi used to reconstructed the final estimate of the hemodynamic response. We observed that these last filtering and averaging steps further improved the estimate of the hemodynamic response compared to reconstructing the hemodynamic response from the final state estimates of the smoother.
2.8. Statistical analysis
Only specific channels based on the following criteria were kept in the analysis. The raw hemoglobin concentrations were bandpass filtered with a 3rd order Butterworth-type filter between 0.01 Hz and 1.25 Hz (Zhang et al., 2007b). The Pearson correlation coefficient R2 between each 1 cm HbO channel and its 4 closest neighbor 3 cm HbO channels (before adding the synthetic hemodynamic response) were then computed and the SD pairs for which R2 < 0.1 were discarded for the analysis. The mean R2 across the selected channels was 0.47 for HbO and 0.22 for HbR. We also computed the Pearson correlation coefficient after adding the synthetic hemodynamic response and similar results were obtained. The mean differences between the R2’s computed before and after adding the synthetic response was 0.01 for HbO and 0.003 for HbR, with the highest value obtained before adding the synthetic response to the real data. Those small differences emphasize the fact that the signals were dominated by systemic physiology in our simulations. This result also suggests that no resting state measurement is required to select the channels which would benefit from the small separation measurement since the correlation can be estimated from the time course containing brain activation. Zhang et al (2009) showed that the adaptive filter method was working well when the correlation between the short and the long separation channel for HbO was greater than 0.6. We used 0.1 in this work to include more channels in the analysis and to show that our state-space method was working well when the initial correlation was lower than 0.5. Using this criterion, 94 out of the 144 possible channels (6 subjects × 3 runs × 8 channels) were kept for further analysis. This represented 65 % of the original data set. The numbers of channels kept for each of the subjects were 16, 14, 13, 17, 19 and 15 respectively. The signal to noise ratio (SNR) for each channel was computed as the amplitude of the simulated hemodynamic response divided by the standard deviation of the time course of the signal. The mean SNR across the selected channels was 0.45 for HbO and 0.38 for HbR.
We used two different metrics to compare the performance of the different algorithms. The first one was the Pearson correlation coefficient R2 between the true synthetic hemodynamic response and the recovered response given by each algorithm. This metric was used to access the level of oscillation in the recovered hemodynamic response created by the global interference not removed by the algorithms and still contaminating the signal. Since the R2 coefficient is scale invariant, it could not give any information about the accuracy of the amplitude of the recovered hemodynamic response. To overcome this problem, we also used the mean square error (MSE) as a metric to compare the performance of the different algorithms.
Since the random position of the trials across the same time course can greatly affect the accuracy of the recovered hemodynamic response, we repeated the procedure 30 times with 30 different random onset time instances for each of the 94 selected channels. The mean and the standard deviation of the 2820 R2 coefficients (94 channels × 30 instances) for each algorithm were then computed after applying the Fisher transformation
| (31) |
and the results were then inverse transformed. The mean and the standard deviation of the 2820 MSEs were also computed. This procedure was repeated independently for 10, 30 and 60 trials in each six-minute data series. The different algorithms were compared together by computing two-tailed paired t-tests on their MSEs and Fisher transformed R2 coefficients.
3. Results
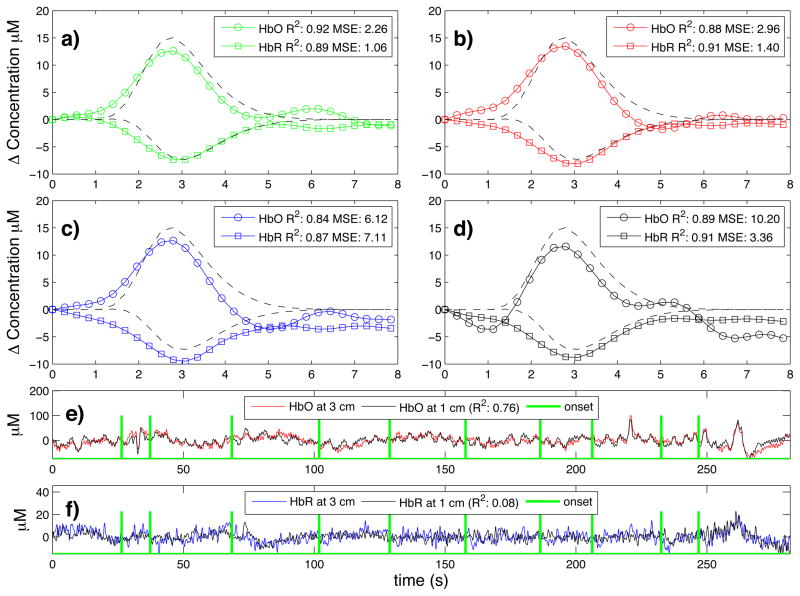
Typical time courses of the recovered hemodynamic response overlapped with the true simulated response are shown in Fig. 3a) to d) for the four algorithms tested. The SNR for this particular simulation was 0.33 for HbO and 0.81 for HbR. The R2’s and the MSEs for HbO and HbR are shown in the legend of each individual panel. Those individual results were obtained from a single simulation with 10 trials. The time courses for this specific simulation are shown in panel e) for HbO and f) for HbR. Both the initial 1 cm channel and the 3 cm channel containing the added synthetic hemodynamic responses are shown as well as the position of the 10 individual onset times. The R2 between the initial 1 cm channel and the initial 3 cm channel (no response added) is also shown in the legend of panel e) and f) for HbO and HbR respectively. All concentrations are expressed in micromolar (μM) units.
Figure 3.
a) to d) Typical time courses of the recovered hemodynamic responses overlapped with the simulated hemodynamic response. For these specific traces, the SNR was 0.33 for HbO and 0.81 for HbR. R2 coefficients and MSEs between the recovered (circles) and the simulated (dashed) response are shown in the legends. a) Kalman filter estimator b) Static estimator c) Adaptive filter d) Standard GLM with 3rd order drift. e) HbO and f) HbR time courses of the 3 cm channel (with synthetic responses added) overlapped with the 1 cm channel. The positions of the onset time are also shown and the correlation coefficients between the 1 cm and the 3 cm channels (before adding synthetic responses) are indicated in parenthesis.
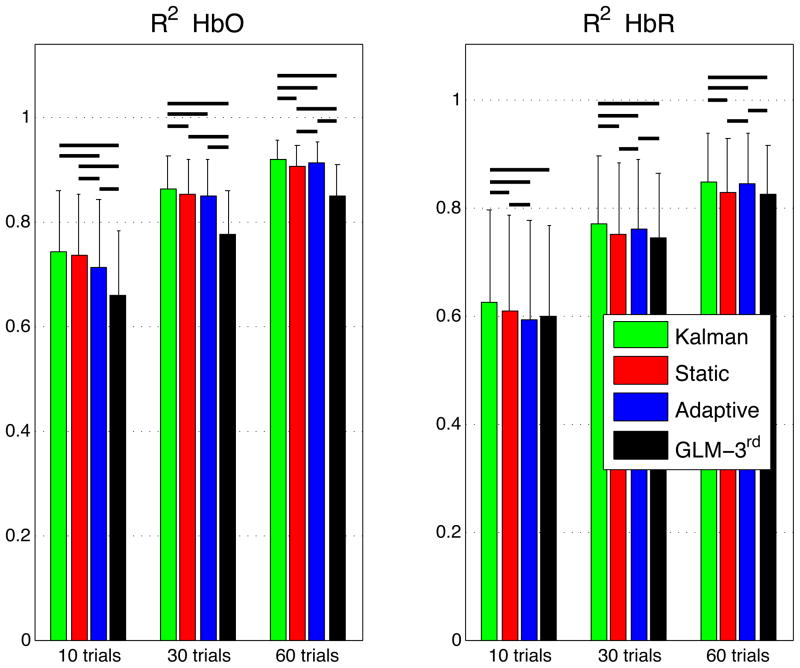
The summary R2 statistics over all subjects, all channels and all instances are shown in a bar graph in Fig. 4 for both HbO and HbR. These values represent the Pearson R2 coefficients computed between the recovered and the simulated hemodynamic responses. The bars represent the mean and the error bars represent the standard deviation. Both the mean and the standard deviation were computed on the Fisher transformed values and then inverse transformed. Two-tailed paired t-tests on the Fisher transformed values were performed between all the different estimators and statistical significance at the level p < 0.05 is illustrated by a black line over the bars for which a significant difference was observed. In our three simulations using 10, 30 and 60 trials respectively, the R2’s for HbO and HbR obtained using our Kalman filter dynamic estimator were significantly higher (p < 0.05) than the ones obtained using the adaptive filter. Moreover, the R2’s obtained were higher with the Kalman filter than with the static estimator. These differences were significant (p < 0.05) except in our 10 trial simulation for HbO.
Figure 4.
Pearson R2 coefficients between simulated and recovered hemodynamic responses. The bars represent the means and the error bars represent standard deviations computed accross all subjects, all channels and all intances. The means and the standard deviation were computed in the Fisher space and then inverse transformed. Two-tailed paired t-tests were performed on the Fisher transformed R2’s. Statistical differences (p < 0.05) between the four algorithms are indicated by black horizontal lines over the corresponding bars.
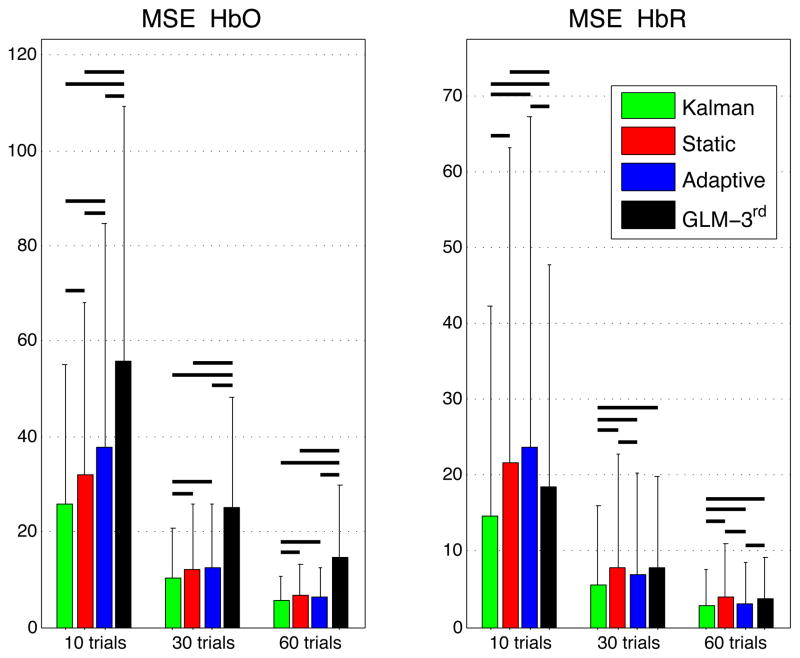
Similarly, the summary MSE statistics over all subjects, all channels and all instances are shown in Fig. 5. These values represent the mean square error computed between the recovered and the simulated hemodynamic responses. The bars represent the mean while the error bars represent the standard deviation. Two-tailed paired t-tests were performed between all the different estimators and statistical significance at the level p < 0.05 is illustrated by a black line over the bars for which a significant difference was observed. The MSEs obtained for HbO and HbR in our three simulations (10, 30 and 60 trials) were significantly lower (p < 0.05) with our Kalman filter estimator than with the adaptive filter. Futhermore, the MSEs obtained with the Kalman filter were also lower (p < 0.05) than the ones obtained with the static estimator for both HbO and HbR in our three simulations.
Figure 5.
Mean squared errors (MSE) between simulated and recovered hemodynamic responses. The bars represent the means and the error bars represent the standard deviations computed accross all subjects, all channels and all instances. Two-tailed paired t-tests were performed between the four estimators and statistical differences at the level p < 0.05 are indicated by black horizontal lines over the corresponding bars.
Table 1 summarizes the statistical analysis over all the subjects, all the channels and all the instances for both HbO and HbR and for the simulations with 10, 30 and 60 trials. Each algorithm was compared to every other. The values shown are the p-values obtained from a two-tailed paired t-test. Statistical differences at the level p < 0.05 are indicated with bold script. These p-values were computed from the data summarized in the bar graphs shown in Figs. 4 and 5.
Table 1.
Cross-comparison of the different algorithms. P-values for the two-tailed paired t-tests accross all subjects, all channels and all intances are shown. For the R2 coefficients, the tests were performed on the Fisher transformed values
| 10 trials |
30 trials |
60 trials |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| KF | SE | AF | KF | SE | AF | KF | SE | AF | |
| R2 HbO | |||||||||
| SE | 6e-02 | - | - | 3e-03 | - | - | 2e-07 | - | - |
| AF | 2e-07 | 6e-06 | - | 1e-03 | 5e-01 | - | 4e-04 | 2e-03 | - |
| GLM | 3e-15 | 2e-13 | 4e-07 | 8e-16 | 2e-12 | 4e-13 | 6e-15 | 1e-10 | 5e-14 |
| R2 HbR | |||||||||
| SE | 5e-05 | - | - | 2e-09 | - | - | 1e-08 | - | - |
| AF | 2e-07 | 2e-03 | - | 3e-05 | 5e-03 | - | 4e-02 | 6e-10 | - |
| GLM | 5e-04 | 2e-01 | 4e-01 | 7e-04 | 3e-01 | 4e-02 | 1e-04 | 6e-01 | 7e-04 |
| MSE HbO | |||||||||
| SE | 4e-06 | - | - | 3e-05 | - | - | 4e-06 | - | - |
| AF | 2e-05 | 7e-03 | - | 1e-06 | 7e-01 | - | 2e-05 | 6e-02 | - |
| GLM | 2e-12 | 2e-09 | 5e-05 | 1e-13 | 7e-11 | 1e-11 | 4e-10 | 3e-08 | 2e-09 |
| MSE HbR | |||||||||
| SE | 2e-05 | - | - | 4e-05 | - | - | 1e-04 | - | - |
| AF | 3e-05 | 2e-01 | - | 2e-04 | 4e-03 | - | 7e-03 | 7e-05 | - |
| GLM | 6e-08 | 3e-02 | 1e-02 | 3e-07 | 9e-01 | 9e-02 | 4e-06 | 1e+00 | 5e-04 |
Bold face indicates significant difference at the p < 0.05 level. KF: Kalman filter estimator, SE: Static estimator, AF: Adaptive filter, GLM: Standard GLM with 3rd order drift.
4. Discussion
4.1. Simultaneous filtering and estimation
One of the salient features of our Kalman filter estimator is that it filters the global interference and simultaneously estimates the hemodynamic response. This feature resulted in a more accurate recovery of the hemo-dynamic response with our Kalman filter estimator compared to the adaptive filter, for which the filtering and the estimation were performed in two distinct steps. Independent regression of the small separation channel potentially removes contributions of the hemodynamic response in the signal which lead to an underestimation of the hemodynamic response thereafter. Our Kalman filter estimator avoids this pitfall. Compared to the adaptive filter, our Kalman filter estimator showed significant improvements at the p < 0.05 level in both HbO and HbR recoveries for our 10, 30 and 60 trial simulations. Those improvements were observed in both Pearson R2 and MSE metrics.
4.2. Dynamic versus static estimation
The systemic interference present in NIRS data is non-stationary. This has been nicely shown by Lina et al (2008) who performed a detailed wavelet analysis of resting NIRS data with blood pressure, respiratory and heart rate data acquired simultaneously on awake human subjects. The amplitude of the systemic physiology measured by the 1 cm and the 3 cm channel depends on the respective pathlength of the light for each channel. Systemic physiology could alter the optical properties of the tissue over time. As a result, a sustained change in absorption could modify the pathlength of the light independently in the 1 cm and the 3 cm channel, modifying at the same time the relative amplitude of the systemic physiology detected in each channel. This feature of the systemic interference explains why our Kalman filter, which is a dynamic estimator, performed better than the static estimator. Using our Kalman filter estimator, improvements in the HbO and HbR recovery were observed in both the Pearson R2 and the MSE metrics compared to the static estimator. All these improvements were significant at the p < 0.05 level except for the HbO Pearson R2 improvement which was not significant in our 10 trial simulation.
4.3. HbO versus HbR
In their wavelet analysis, Lina et al (2008) also showed that the HbO time courses were more contaminated by global interference than the HbR time courses. As such, the correlation between the 1 cm and 3 cm channel should be higher for HbO than HbR, and filtering methods using 1 cm SD separations should work better for HbO than for HbR. In our data, the mean initial Pearson R2 correlation between the 1 cm and 3 cm signals were higher for HbO than HbR (0.47 vs 0.22). Comparing our Kalman filter estimator with the standard block average estimator, the p-values obtained in the t-tests performed on the Fisher transformed Pearson R2’s and the MSEs were at least five orders of magnitude lower for HbO than HbR. This indicates that the improvements observed with our Kalman filter were more prominent for HbO than HbR. This better performance in the recovery of HbO over HbR using a small separation method was also reported by Zhang et al (2009) using their adaptive filter.
4.4. Impact of initial correlation
In the case where the systemic physiology present in the 3 cm separation did not correlate with the systemic physiology present in the 1 cm channel, the performance of the Kalman filter was similar to the standard GLM. In this case, the model cannot reproduce the data and the ai coefficients in Eq. 3 converge to zero. As such, the wi’s estimated by the Kalman filter are very close to the ones obtained using the GLM. An important point is that in the case of low initial R2 coefficients (0.1 < R2 < 0.2), taking into account the 1 cm channel with the Kalman filter did not decrease the performance of the recovery compared to the GLM. On the other hand, the performance of the adaptive filter for (0.1 < initial R2 < 0.2) was worst than the GLM. This counter-performance of the adaptive filter for poor initial correlation between the short and the long channel was also reported by Zhang et al (2009). These findings suggest that the Kalman filter can be used even if the correlation between the 1 cm and the 3 cm channel is low as opposed to the adaptive filter. In the worst case, the Kalman filter will be as good as the standard GLM. However, the higher the initial correlation between the 1 cm and the 3 cm channel is, the more significant is the improvement using a small separation measurement. This is illustrated by the larger improvement obtained for HbO than HbR when using a small separation measurement together with our Kalman filter.
4.5. Technical notes
The MSEs obtained in our simulations and presented in Fig. 5 were lower for HbR than HbO. This occurred because the amplitude of the simulated HbR response was lower than the simulated HbO response which resulted in lower MSEs for HbR. This is illustrated for noise-free data in Fig. 2b.
For all the results presented in this paper, a single time point was taken from the 1 cm channel to regress the 3 cm channel. In practice, this value could be any integer. A simple phase shift (delay) between the 3 cm and 1 cm channel would be taken into account by using multiple time points from the 1 cm. In this case, all the a’s in Eq. 3 would converge to zero except for one a at the value of i corresponding to the shift between the two signals in terms of number of sample points. Different values for Na were tested during our simulations. With the adaptive filter, we obtained better results using a single point than using 100 points as in Zhang et al (2007a). Using 100 points results in overfitting the signal which removes more of the hemodynamic response contribution than using a single point. This is another pitfall of the non-simultaneous recovery and filtering feature of the adaptive filter which is avoided with our Kalman filter. Finally, we did not observe any improvement when using multiple points with our Kalman filter, suggesting that no delays were present in our data between the 1 cm and the 3 cm channel.
The Gaussian temporal basis functions used in this work allow us to model different hemodynamic responses with different shapes and components. This includes a potential initial dip and post-stimulus undershoot, responses with a double bump and negative responses. It is also easy to use additional Gaussian functions to extend this method for longer stimuli, making the temporal basis set used in the present work very general and less restrictive. However, as stated in section 2.3, the drawback for using a more general set is the potential overfitting of phase-locked systemic physiology. This could be avoided using a more restrictive temporal basis set such as a gamma-variant function and its derivatives (Huppert et al., 2008; Abdelnour and Huppert, 2009; Hu et al., 2010; Glover, 1999), and at the same time could potentially reduce the number of parameters to estimate.
We tested different values for the separation between the basis and also different values for the width of the Gaussians. The values of 0.5 second for both the separation and the width presented in this paper resulted in the lowest MSEs between the recovered and the simulated responses and highest R2’s. The separation between our temporal basis Gaussians and their widths was three times lower than the values used by Diamond et al (2006).
In order to compare the four methods used in this work on the same footing, we used temporal basis functions for each estimator. For the standard GLM estimator, the adaptive filter and the Kalman filter, we have also tried to replace the final step of using the GLM with a temporal basis set by a simple block average without using any temporal prior. For all these three estimators, using temporal basis functions in the final step further improved the recovery of both HbO and HbR. The MSEs between the recovered and the simulated hemodynamic response were lower when temporal basis were used than when a simple block average without temporal basis was applied. Similarly, the R2’s computed between the recovered and the simulated responses were higher when temporal basis were used in the final block average step. This result raises the importance of using temporal priors to reduce the dimensionality of the estimation problem.
As stated in section 2.7, changing the state process noise and the measurement noise priors over two orders of magnitude did not affect the performance of our Kalman estimator. For HbO, no differences could be observed (two-tailed paired t-test, p < 0.05) between the MSEs recovered using values for the process noise or the measurement noise ten times lower or higher than the ones presented in section 2.7. For HbR, small differences in the MSEs were observed but these results did not change any conclusions drawn in this paper. The MSEs recovered with our Kalman filter in this case were still the lowest of the four estimators.
4.6. Future directions
As mentioned in Zhang et al (2009), an important question is whether an additional short separation optode is required for each longer separation op-tode or whether a single one is sufficient. Although the systemic interference is thought to be global in the brain, it might be reflected differently in the NIRS data collected over different regions of the head. Sources of variation include blood vessel size which might affect the amplitude of the recovered response but also blood vessel length and geometry which might give rise to phase mismatches between different NIRS channels. Studies using multiple small SD separation optodes at different locations over the head should be performed in the future to address this question.
5. Conclusion
In summary, we filtered the global interference present in NIRS data by using additional small separation optodes and we simultaneously estimated the hemodynamic response using a dynamic algorithm. Our dynamic Kalman filter performed better than the traditional adaptive filter, the static estimator and the standard block average estimator for both HbO and HbR recovery. These results were consistent with the fact that dynamic estimation better captures the non-stationary behavior of the systemic interferences in NIRS and that the simultaneous filtering and estimation prevents underestimation of the hemodynamic response. The algorithm is easily implementable and suitable for a wide range of NIRS studies.
Acknowledgments
This work was supported by NIH grants P41-RR14075 and R01-EB006385. L. Gagnon was supported by the Fonds Quebecois sur la Nature et les Technologies and by the IDEA-squared program at MIT. We acknowledge fruitful discussions with Sol Diamond, Emery Brown, Patrick Purdon, Lino Becerra and Dana Brooks. We would also like to thank Michele Desjardins for critical reading of the manuscript.
Appendix A
The explicit expression for D in Eq. 9 is given by
The dimension of the matrix D is Nr by 4. Each column is normalized by its highest value to keep the matrix G well conditioned and to avoid numerical errors during the inversion in Eq.10.
The explicit expression for A in Eq. 17 is given by
Nb is the length of each temporal basis function and was 80 in our work due to the 10 Hz temporal resolution and 8 s FIR for our temporal basis functions. The vertical dimension of matrix A corresponds to Nt, the total number of time points in the entire time course. The number of copies of the temporal basis functions corresponds to the number of trials (or stimuli) in the specific time course (i.e. if the run contained 10 trials, then 10 copies of the temporal basis set will appear in the corresponding A matrix).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelnour AF, Huppert TJ. Real-time imaging of humain brain function by near-infrared spectroscopy using an adaptive general linear model. NeuroImage. 2009;46:133–143. doi: 10.1016/j.neuroimage.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boas D, Dale AM, Franceschini MA. Diffuse optical imaging of brain activation: Approaches to optimizing image sensitivity, resolution and accuracy. Neuroimage. 2004;23:S275–S288. doi: 10.1016/j.neuroimage.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Cope M, Deply DT. System for long-term measurement of cerebral blood flow and tissue oxygenation on newborn infants by infrared transillumination. Med Biol Eng Comput. 1998;26:289–294. doi: 10.1007/BF02447083. [DOI] [PubMed] [Google Scholar]
- Deply DT, Cope M, van der Zee P, Arridge S, Wray S, Wyatt J. Estimation of optical pathlength through tissue from direct time of flight measurement. Phys Med Biol. 1988;33:1433–1442. doi: 10.1088/0031-9155/33/12/008. [DOI] [PubMed] [Google Scholar]
- Diamond S, Huppert TJ, Kolehmainen V, Franceschini MA, Kaipo JP, Arridge SR, Boas DA. Dynamic physiological modeling for functional diffuse optical tomography. Neuroimage. 2006;30:88–101. doi: 10.1016/j.neuroimage.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond SG, Perdue KL, Boas DA. A cerebrovascular response model for functional neuroimaging including dynamic cerebral autoregulation. Math Biosci. 2009;220 (2):102–117. doi: 10.1016/j.mbs.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini M, Fantini S, Thompson J, Culver J, Boas D. Hemodynamic evoked response of the sensorimotor cortex measured noninvasively with near-infrared optical imaging. Psychophysiology. 2003;40 (4):584–560. doi: 10.1111/1469-8986.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini M, Joseph DK, Huppert TJ, Diamond SG, Boas D. Diffuse optical imaging of the whole head. J Biomed Opt. 2006;11 (5):054007. doi: 10.1117/1.2363365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb A. Applied Optimal Estimation. MIT Press; 1974. [Google Scholar]
- Gibson A, Hebden J, Arridge S. Recent advances in diffuse optical imaging. Phys Med Biol. 2005;50:R1–R43. doi: 10.1088/0031-9155/50/4/r01. [DOI] [PubMed] [Google Scholar]
- Glover GH. Deconvolution of impulse response in event-related bold fmri. NeuroImage. 1999;9 (4):416–429. doi: 10.1006/nimg.1998.0419. [DOI] [PubMed] [Google Scholar]
- Gratton G, Corballis PM. Removing the heart from the brain: compensation for the pulse artifact in the photon migration signal. Psychophysiology. 1995;32 (3):292–299. doi: 10.1111/j.1469-8986.1995.tb02958.x. [DOI] [PubMed] [Google Scholar]
- Gregg NM, White BR, Zeff BW, Berger AJ, Culver JP. Brain specificity of diffuse optical imaging: improvements from superficial signal regression and tomography. Font in Neuroenergetics. 2010;2 (14):1–8. doi: 10.3389/fnene.2010.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haykin S. Adaptive Filter Theory. Prentice-Hall; Upper Saddle River, NJ: 2001a. [Google Scholar]
- Haykin S. Kalman filtering and neural Networks. John Wiley & Sons; New York: 2001b. [Google Scholar]
- Hillman EMC. Optical brain imaging in vivo: techniques and applications from animal to man. J Biomed Opt. 2007;12 (5):051402. doi: 10.1117/1.2789693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi Y. Functional near-infrared spectroscopy: current status and future prospects. J Biomed Opt. 2007;12 (6):062106. doi: 10.1117/1.2804911. [DOI] [PubMed] [Google Scholar]
- Hu X-S, Hong K-S, Ge SS, Jeong M-Y. Kalman estimator-and general linear model-based on-line brain activation mapping by near-infrared spectroscopy. BioMedical Engineering OnLine. 2010;9(82) doi: 10.1186/1475-925X-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert T, Allen MS, Benay H, Jones PB, Boas D. A multi-compartment vascular model for inferring baseline and functional changes in cerebral oxygen metabolism and arterial dilation. J Cereb Blood Flow Metab. 2007;27:1262–1279. doi: 10.1038/sj.jcbfm.9600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert T, Hoge R, Dale AM, Franceschini M, Boas D. Quantitative spatial comparison of diffuse optical imaging with blood oxygen level-dependent and arterial spin labeling-based functional magnetic resonance imaging. J Biomed Opt. 2006a;11 (6):064018. doi: 10.1117/1.2400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert T, Hoge R, Diamond S, Franceschini M, Boas D. A temporal comparison of bold, asl, and nirs hemodynamic responses to motor stimuli in adult humans. NeuroImage. 2006b;29:368–382. doi: 10.1016/j.neuroimage.2005.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert TJ. PhD thesis. Harvard University; 2007. Hemodynamic-based inference of cerebral oxygen metabolism. [Google Scholar]
- Huppert TJ, Allen MS, Diamond SG, Boas DA. Estimating cerebral oxygen metabolism from fmri with a dynamic multicompartment windkessel model. Hum Brain Mapp. 2009;30 (5):1548–1567. doi: 10.1002/hbm.20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert TJ, Diamond SG, Boas DA. Direct estimation of evoked hemoglobin changes by multimodality fusion imaging. J Biomed Opt. 2008;13 (5):054031. doi: 10.1117/1.2976432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang KE, Tak S, Jung J, Jang J, Jeong Y, Ye JC. Wavelet minimum description length detrending for near-infrared spectroscopy. Vol. 14. 2009. p. 034004. [DOI] [PubMed] [Google Scholar]
- Jasdzewski G, Strangman G, Wagner J, Kwong KK, Poldorack RA, Boas DA. Differences in the hemodynamic response to event-related motor and visual paradigms as measured by near- infrared spectroscopy. Neuroimage. 2003;20 (1):479–488. doi: 10.1016/s1053-8119(03)00311-2. [DOI] [PubMed] [Google Scholar]
- Kalman RE. A new approach to linear filtering and prediction problems. Transactions of the ASME–Journal of Basic Engineering. 1960;82 :35–45. Series D. [Google Scholar]
- Kolehmainen V, Prince S, Arridge SR, Kaipo JP. State- estimation approach to the nonstationary optical tomography problem. J Opt Soc Am A. 2003;20 (5):876–889. doi: 10.1364/josaa.20.000876. [DOI] [PubMed] [Google Scholar]
- Lina JM, Dehaes M, Matteau-Pelletier C, Lesage F. Complex wavelets applied to diffuse optical spectroscopy for brain activity detection. Optics Express. 2008;16 (2):1029–1050. doi: 10.1364/oe.16.001029. [DOI] [PubMed] [Google Scholar]
- Lina JM, Matteau-Pelletier C, Dehaes M, Desjardins M, Lesage F. Wavelet-based estimation of the hemodynamic responses in diffuse optical imaging. Med Imag Anal. 2010;14:606–616. doi: 10.1016/j.media.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Matteau-Pelletier C, Dehaes M, Lesage F, Lina JM. 1/f noise in diffuse optical imaging and wavelet-based response estimation. IEEE Trans Med Imag. 2009;28:415–422. doi: 10.1109/TMI.2008.2006524. [DOI] [PubMed] [Google Scholar]
- Obrig H, Neufang M, Wenzel R, Kohl M, Steinbrink J, Einhaupl K, Villringer A. Spontaneous low frequency oscillations of cerebral hemodynamics and metabolism in human adults. Neuroimage. 2000;12 (6):623–639. doi: 10.1006/nimg.2000.0657. [DOI] [PubMed] [Google Scholar]
- Obrig Y, Villringer A. Beyond the visibleimaging the human brain with light. J Cereb Blood Flow Metab. 2003;23:1–18. doi: 10.1097/01.WCB.0000043472.45775.29. [DOI] [PubMed] [Google Scholar]
- Payne S, Selb J, Boas DA. Effects of autoregulation and co2 reactivity on cerebral oxygen transport. Ann Biomed Eng. 2009;37 (11):2288–2298. doi: 10.1007/s10439-009-9763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince S, Kolehmainen V, Kaipo JP, Franceschini MA, Boas DA, Arridge SR. Time-series estimation of biological factors in optical diffusion tomography. Phys Med Biol. 2003;48:1491–1504. doi: 10.1088/0031-9155/48/11/301. [DOI] [PubMed] [Google Scholar]
- Rauch HE, Tung F, Striebel CT. Maximum likelihood estimates of linear dynamic systems. AIAA Journal. 1965;3 (8):1445–1450. [Google Scholar]
- Saager RB, Berger AJ. Direct characterization and removal of interfering absorption trends in two-layer turbid media. J Opt Soc Am A. 2005;22 (9):1874–1882. doi: 10.1364/josaa.22.001874. [DOI] [PubMed] [Google Scholar]
- Saager RB, Berger AJ. Measurement of layer-like hemodynamic trends in scalp and cortex: implications for physiological baseline suppression in functional near-infrared spectroscopy. J Biomed Opt. 2008;13 (3):034017. doi: 10.1117/1.2940587. [DOI] [PubMed] [Google Scholar]
- Tonorov V, Franceschini M, Filiaci M, Fantini S, Wolf M, Michalos A, Gratton E. Near-infrared study of fluctuations in cerebral hemodynamics during rest and motor stimulation: temporal analysis and spatial mapping. Med Phys. 2000;27 (4):801–815. doi: 10.1118/1.598943. [DOI] [PubMed] [Google Scholar]
- Umeyama S, Yamada T. Monte carlo study of global interference cancellation by multidistance measurement of near-infrared spectroscopy. J Biomed Opt. 2009;14 (6):064025. doi: 10.1117/1.3275466. [DOI] [PubMed] [Google Scholar]
- Villringer A, Hock C, Schleinkofer L, Dirnagl U. Near infrared spectroscopy (nirs): A new tool to study hemodynamic changes during activation of brain function in human adults. Neurosci Lett. 1993;154:101–104. doi: 10.1016/0304-3940(93)90181-j. [DOI] [PubMed] [Google Scholar]
- Yamada T, Umeyama S, Matsuda K. Multidistance probe arrangement to eliminate artifacts in functional near-infrared spectroscopy. J Biomed Opt. 2009;16 (4):06434. doi: 10.1117/1.3275469. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Brooks DH, Franceschini MA, Boas DA. Eigenvector-based spatial filtering for reduction of physiological interference in diffuse optical imaging. J Biomed Opt. 2005;10 (1):011014. doi: 10.1117/1.1852552. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Brown EN, Strangman GE. Adaptative filtering for global interference cancellation and real-time recovery of evoked brain activity: a monte carlo simulation study. J of Biomed Opt. 2007a;12 (4):044014. doi: 10.1117/1.2754714. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Brown EN, Strangman GE. Adaptative filtering to reduce global interference in evoked brain activity detection: a human subject case study. J of Biomed Opt. 2007b;12 (6):064009. doi: 10.1117/1.2804706. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Strangman G, Ganis G. Adaptive filtering to reduce global interference in non-invasive nirs measures of brain activation: How well and when does it work? Neuroimage. 2009;45:788–794. doi: 10.1016/j.neuroimage.2008.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]