Abstract
Based on digital karyotyping, we have identified a new, discrete amplified region at ch19p13.2 in a high-grade ovarian serous carcinoma. To further characterize this region, we determined the frequency and biological significance of ch19p13.2 amplification by analyzing 341 high-grade serous carcinomas from The Cancer Genome Atlas (TCGA) and found an increased DNA copy number at this locus in 18% of cases. We correlated the DNA and RNA copy number by analyzing the TCGA dataset for all amplified genes and detected 7 genes within ch19p13.2 that were significantly correlated (R ≥0.54) and were, in fact, listed as the top 100 potential “driver” genes at a genome-wide scale. Interestingly, one of the 7 genes, NACC1, encoding NAC1 was previously reported to be involved in the development of tumor recurrence in ovarian serous carcinoma and to play a causal role in the development of paclitaxel resistance. Therefore, we selected NACC1 for validation in an independent cohort. Based on fluorescence in situ hybridization, we found that 35 (20%) of 175 high-grade serous carcinomas had an increased DNA copy number at the NACC1 locus, and those amplified cases were associated with early disease recurrence within 6 months (p= 0.013). A significantly high level of NAC1 protein expression based on immunohistochemistry was detected in amplified tumors as compared to non-amplified tumors (p< 0.005). In summary, our data suggest that amplification at the ch19p13.2 NACC1 locus, leading to NAC1 overexpression, is one of the molecular genetic alterations associated with early tumor recurrence in ovarian cancer.
INTRODUCTION
Epithelial ovarian cancer is the most aggressive gynecologic malignancy. Ovarian cancer is composed of a diverse group of tumors that can be classified according to their distinctive morphologic and molecular features (1). Among them, high-grade serous carcinoma represents the major tumor type associated with frequent tumor recurrence and high mortality. In contrast to other subtypes, high-grade serous carcinomas are highly aggressive, evolve rapidly, and almost always present at advanced stage. Several studies have analyzed global DNA copy number alterations specifically in different types of ovarian epithelial carcinomas (2–8). The results from these reports indicate that high-grade serous carcinomas are characterized by higher levels of sub-chromosomal gains and losses than clear cell carcinoma, low-grade serous carcinomas, and their precursor lesions, serous borderline tumors. This finding suggests that chromosomal instability is more pronounced in high-grade serous carcinomas than other types of ovarian cancer.
To identify new cancer-associated genes that may participate in the pathogenesis of ovarian high-grade serous carcinoma, we have previously applied digital karyotyping (9) and, subsequently, SNP arrays to analyze somatic genome-wide DNA copy number alterations in purified ovarian high-grade serous carcinoma samples (2, 4, 10). As a result, in addition to several previously known amplified chromosomal regions containing CCNE1, AKT2, and PIK3CA loci, we identified several new amplified loci including chromosome (ch)11q13.5 harboring Rsf-1 (HBXAP) (11), chr19p13.12 harboring NOTCH3 (12), and regions at chr12p13, chr8q24 and chr19p13.2. Those regions containing known tumor-associated genes were validated using dual color fluorescence in situ hybridization (FISH) in an independent set of ovarian carcinomas (4). The purpose of this report is to study a previously uncharacterized amplified region at ch19p13.2. Based on digital karyotypic analysis in a limited set of clinical samples, we detected a discrete amplicon located at ch19p13.2 which has not been previously reported in ovarian cancer. This amplicon appears to be relatively large, encompassing 1.92 Mb and containing at least 60 coding sequences, which poses challenges for further investigation to identify the cancer “driver” gene(s) within this amplified region. Several genome database resources such as The Cancer Genome Atlas (TCGA) have recently become available for public access, providing a hitherto unavailable opportunity for molecular genetic discovery in cancer. In this study, we take the advantage of the emerging dataset of the TCGA to simultaneously analyze mRNA and genomic DNA copy numbers for all genes within the digital karyotyping-defined ch9p13.2 amplicon in a large set of high-grade serous carcinomas. This approach enabled us to distinguish cancer “driver” genes from co-amplified “passenger” genes. We then validated our results using fluorescence in situ hybridization and immunohistochemistry in an independent set of ovarian carcinomas.
MATERIALS AND METHODS
Analysis of The Cancer Genome Atlas database
Copy number variations of the ch9p13.2 amplicon in 343 ovarian tumor samples and paired normal samples were characterized using the Affymetrix SNP6.0 array by the Broad Institute (Boston). The intensity and log2 ratios for each probe set on the SNP array were downloaded from The Cancer Genome Atlas Data Portal (http://tcga-data.nci.nih.gov/tcga/). Somatic copy number alterations were analyzed using the Circular Binary Segmentation (CBS) algorithm (13) with R package “DNACopy” (version 1.14.0, default setting). Regions with focal somatic copy number alterations were identified by GISTIC analysis in GenePattern (http://www.broadinstitute.org/cancer/software/ genepattern/) (14). Expression analysis of 377 ovarian tumor samples was performed using the Affymetrix Human Exon 1.0 ST Array by the Lawrence Berkeley Laboratory. The CEL files were downloaded from the TCGA Data Portal. The probe set signals were summarized as RMA scores with Affymetrix Power Tools (http://www.affymetrix.com/partners_programs/programs/developer/tools/ powertools.affx). The locations of all probe sets were obtained from the Affymetrix annotation file (HuEx-1_0-stv2.na29.hg18.probeset.csv). A total of 8,101 core probe sets (representing 590 genes) located at the regions with focal somatic copy number alterations were selected for expression-copy number correlation analysis. Copy number data and exon-array data were available for 341 cases; therefore this sample set was used for analysis in this study. For these samples, Pearson correlation coefficients were calculated using the expression of each probe set and its corresponding copy number across the 341 cases. The distribution of correlation coefficients of all 8,101 probe sets is bimodal, separating the genes (probe sets) with no correlation (correlation coefficient below 0.2) from those that show correlations between DNA copy number and RNA expression.
Fluorescence in situ hybridization
A total of 175 ovarian high-grade serous carcinomas were analyzed for DNA copy number within the ch19p13.2 region using two-color fluorescence in situ hybridization (FISH). Stage III or IV tumor tissues from patients treated at the Johns Hopkins Hospital were originally retrieved from the Department of Pathology at the Johns Hopkins Hospital. The acquisition of the anonymous tissue specimens for this study was approved by the Johns Hopkins Institutional Review Board. The tissues were arranged in tissue microarrays to facilitate FISH and immunohistochemistry procedures. BAC clones (RP11-356L15 and CTD-2508D10) containing the genomic sequences of the ch19p13.2 amplicon were purchased from Bacpac Resources (Childrens' Hospital, Oakland, CA) and Invitrogen (Carlsbad, CA). Bac clones located at ch19p12 (CTD-2518O18) were used to generate reference probes. The method for FISH was previously described (15). The hybridization signals were counted by two individuals. Signal ratios of experimental probe/reference probe greater than 2.5 were considered as gain, and signal ratios of experimental probe/reference greater than 3.5 were considered as high-fold amplification. At least 50 nuclei were counted for each specimen.
Immunohistochemistry
The NAC1 mouse monoclonal antibody used for immunohistochemistry was purchased from Novus Biologicals (Littleton, CO). The specificity of the antibody was previous demonstrated (16). Paraffin sections from the same tissue microarrays as used for FISH were deparaffinized and pretreated with low pH citrate buffer in a microwave oven for antigen retrieval. Tissue sections were incubated with the anti-NAC1 antibody at a dilution of 1:100 at 4°C overnight. Visualization was performed using the EnVision™+ peroxidase system (DakoCytomation, Glostrup, Denmark). Positive controls consisted of an ovarian carcinoma shown to be positive in a pilot study. Negative controls were stained with an isotype-matched mouse myeloma protein. Immunoreactivity was scored by two investigators who were blinded to the patient clinical data. Nuclear localization was interpreted as positive staining. Staining intensity was scored on a scale of 0–3, corresponding to undetectable, weak, moderate, and intense immunoreactivity in tumor cells (16). At least 500 tumor cells were counted for each specimen.
RESULTS
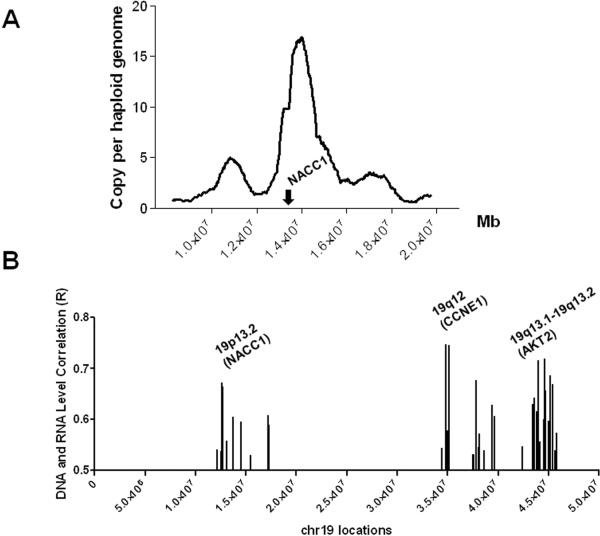
As a continuing effort to understand the molecular etiology of ovarian cancer, we have previously analyzed DNA copy number changes in different types of ovarian carcinoma. Based on digital karyotyping (9) in affinity-purified, high-grade ovarian serous carcinomas, we detected a discrete, novel amplified region located at ch19p13.2 in one of 7 specimens (Fig. 1A). This amplified locus was estimated to have ~17 copies of haploid genome and spanned 12,735,244 to 14,655,263, containing at least 60 genes. Based on this preliminary finding, we decided to determine the frequency of increased copy number at this region in a large set of high-grade serous carcinomas. First, we analyzed DNA copy number in 341 high-grade serous carcinomas from the TCGA ovarian cancer genome dataset. The results indicated that 18% of 341 high-grade serous carcinomas showed increased DNA copy numbers (>2.5) based on SNP arrays. If a higher cutoff value (>3.5) was applied, the fraction of amplified cases was 3%. To identify potential cancer “driver” genes within the ch19p13.2 amplicon, we correlated mRNA expression levels and DNA copy number at a genome wide scale in the same set of carcinomas. The rationale for this analysis is that, when amplified, “driver” genes almost always upregulate their expression, which enables their oncogenic functions. Based on this analysis, we listed the top 100 amplified genes with the highest correlation coefficient between DNA copy number and mRNA expression levels (as determined by Pearson correlation analysis (Table 1). Using this approach, we identified several amplicons harboring well-known amplified oncogenes, including CCNE1 (17), AKT2 (18), Pak1 (5), Rsf-1 (19) and Paf1 (20). Interestingly, chromosome 19 contained a very high number of candidate driver oncogenes. Among the top 100 genes, 54 genes were located at discrete subchromosomal regions of chromosome 19, indicating that frequent structural re-arrangement may occur in this chromosome in high-grade ovarian serous carcinoma (Fig. 1B). Importantly, the ch19p13.2 region harbored 7 genes which showed the most significant correlation between DNA copy number and RNA expression level.
Fig. 1.
Amplification at the ch19p13.2 region harboring NACC1. A. A focused view of the digital karyotyping result demonstrates a discrete amplicon spanning from nucleotide position 12,735,244 to 14,655,263 in a high-grade serous carcinoma. NACC1 is located within the amplicon. B. Analysis of the TCGA ovarian cancer dataset reveals 100 top genes showing the highest correlation coefficient between DNA copy number and RNA expression levels. Among them, 54 genes are located on chromosome 19. Their physical map is shown. Of note, three amplified chromosomal regions, 19p13.2, 19q12, and 19q13.1–19q13.2 harbor NACC1, CCNE1 and AKT2, respectively.
Table 1.
The top 100 amplified potential “driver” genes in ovarian high-grade serous carcinomas*.
| Gene | Chr | Start | End | correlation | fCN> 2.5 | fCN> 3.5 |
|---|---|---|---|---|---|---|
| MRPS15 | chr1 | 36693996 | 36702545 | 0.54 | 0.157 | 0.017 |
| NDUFS5 | chr1 | 39264593 | 39272842 | 0.548 | 0.190 | 0.029 |
| NFYC | chr1 | 40950961 | 41009843 | 0.555 | 0.201 | 0.029 |
| CTPS | chr1 | 41221558 | 41250797 | 0.561 | 0.201 | 0.026 |
| ANKRD17 | chr4 | 74159477 | 74262083 | 0.544 | 0.055 | 0.020 |
| TAF2 | chr8 | 120812677 | 120914224 | 0.551 | 0.475 | 0.050 |
| DERL1 | chr8 | 124095009 | 124123533 | 0.674 | 0.484 | 0.055 |
| C8orf76 | chr8 | 124301438 | 124322767 | 0.545 | 0.487 | 0.061 |
| FAM91A1 | chr8 | 124849893 | 124894217 | 0.547 | 0.490 | 0.067 |
| TRMT12 | chr8 | 125532260 | 125534217 | 0.585 | 0.501 | 0.073 |
| RNF139 | chr8 | 125556194 | 125569280 | 0.609 | 0.501 | 0.073 |
| TATDN1 | chr8 | 125569937 | 125600266 | 0.573 | 0.504 | 0.073 |
| NDUFB9 | chr8 | 125620609 | 125631394 | 0.67 | 0.507 | 0.076 |
| SQLE | chr8 | 126080726 | 126103690 | 0.566 | 0.510 | 0.085 |
| KIAA0196 | chr8 | 126105711 | 126173112 | 0.563 | 0.510 | 0.082 |
| NSMCE2 | chr8 | 126173357 | 126448429 | 0.577 | 0.513 | 0.082 |
| FAM84B | chr8 | 127633871 | 127639562 | 0.534 | 0.539 | 0.085 |
| TRAPPC9 | chr8 | 140742588 | 141468678 | 0.53 | 0.487 | 0.061 |
| EIF2C2 | chr8 | 141610518 | 141714814 | 0.552 | 0.504 | 0.079 |
| PTK2 | chr8 | 141738135 | 142080483 | 0.594 | 0.519 | 0.093 |
| TSTA3 | chr8 | 144765938 | 144770015 | 0.592 | 0.464 | 0.055 |
| ZNF623 | chr8 | 144802992 | 144809553 | 0.54 | 0.466 | 0.055 |
| PUF60 | chr8 | 144970693 | 144983194 | 0.589 | 0.472 | 0.055 |
| EXOSC4 | chr8 | 145133522 | 145135551 | 0.526 | 0.469 | 0.055 |
| GPAA1 | chr8 | 145209555 | 145213055 | 0.546 | 0.469 | 0.055 |
| CYC1 | chr8 | 145222009 | 145224324 | 0.636 | 0.469 | 0.055 |
| MAF1 | chr8 | 145231324 | 145234469 | 0.554 | 0.472 | 0.055 |
| DGAT1 | chr8 | 145510769 | 145521317 | 0.54 | 0.478 | 0.050 |
| CPSF1 | chr8 | 145589295 | 145605344 | 0.544 | 0.481 | 0.055 |
| CYHR1 | chr8 | 145660034 | 145661276 | 0.61 | 0.481 | 0.055 |
| RPL8 | chr8 | 145986002 | 145988284 | 0.589 | 0.461 | 0.050 |
| ZNF7 | chr8 | 146025232 | 146039406 | 0.55 | 0.461 | 0.050 |
| C8orf33 | chr8 | 146277824 | 146281416 | 0.532 | 0.443 | 0.058 |
| UVRAG | chr11 | 75268572 | 77468762 | 0.538 | 0.187 | 0.012 |
| PRKRIR | chr11 | 75744310 | 75740093 | 0.56 | 0.184 | 0.023 |
| C11orf30 | chr11 | 75835631 | 75939155 | 0.584 | 0.190 | 0.026 |
| PAK1 | chr11 | 76721458 | 76781171 | 0.581 | 0.213 | 0.029 |
| CLNS1A | chr11 | 77004989 | 77026476 | 0.638 | 0.222 | 0.029 |
| RSF1 | chr11 | 77055016 | 77056050 | 0.558 | 0.224 | 0.032 |
| C11orf67 | chr11 | 77209888 | 77261043 | 0.56 | 0.222 | 0.032 |
| INTS4 | chr11 | 77306760 | 77306854 | 0.542 | 0.227 | 0.032 |
| NDUFC2 | chr11 | 77457655 | 77468762 | 0.537 | 0.233 | 0.035 |
| ALG8 | chr11 | 77489671 | 77528282 | 0.612 | 0.233 | 0.035 |
| TM7SF3 | chr12 | 27018069 | 27058508 | 0.55 | 0.248 | 0.023 |
| MED21 | chr12 | 27066787 | 27073856 | 0.585 | 0.248 | 0.023 |
| CCDC91 | chr12 | 28410133 | 28703099 | 0.528 | 0.222 | 0.020 |
| ZNF136 | chr19 | 12134940 | 12159758 | 0.541 | 0.169 | 0.020 |
| ZNF564 | chr19 | 12497231 | 12500487 | 0.537 | 0.172 | 0.026 |
| ZNF791 | chr19 | 12582753 | 12601047 | 0.605 | 0.169 | 0.023 |
| C19orf56 | chr19 | 12639907 | 12640414 | 0.672 | 0.175 | 0.023 |
| MORG1 | chr19 | 12645066 | 12647612 | 0.549 | 0.175 | 0.023 |
| DHPS | chr19 | 12647536 | 12653522 | 0.617 | 0.178 | 0.023 |
| FBXW9 | chr19 | 12660756 | 12668379 | 0.541 | 0.178 | 0.023 |
| TNPO2 | chr19 | 12671568 | 12695172 | 0.603 | 0.178 | 0.023 |
| C19orf43 | chr19 | 12702580 | 12706471 | 0.563 | 0.181 | 0.020 |
| ASNA1 | chr19 | 12709344 | 12719395 | 0.664 | 0.181 | 0.020 |
| TRMT1 | chr19 | 13076853 | 13088417 | 0.55 | 0.216 | 0.032 |
| NACC1 | chr19 | 13108094 | 13112537 | 0.558 | 0.216 | 0.032 |
| STX10 | chr19 | 13115892 | 13122076 | 0.557 | 0.216 | 0.032 |
| CCDC130 | chr19 | 13723435 | 13735101 | 0.605 | 0.230 | 0.041 |
| C19orf53 | chr19 | 13746303 | 13750255 | 0.561 | 0.233 | 0.041 |
| DNAJB1 | chr19 | 14486600 | 14490026 | 0.585 | 0.224 | 0.029 |
| NDUFB7 | chr19 | 14537895 | 14543869 | 0.595 | 0.227 | 0.029 |
| AKAP8 | chr19 | 15464337 | 15490603 | 0.529 | 0.251 | 0.029 |
| USE1 | chr19 | 17187243 | 17191604 | 0.608 | 0.207 | 0.023 |
| NR2F6 | chr19 | 17203747 | 17216995 | 0.542 | 0.210 | 0.023 |
| C19orf62 | chr19 | 17239287 | 17251013 | 0.589 | 0.210 | 0.023 |
| UQCRFS1 | chr19 | 34390062 | 34395865 | 0.544 | 0.222 | 0.070 |
| POP4 | chr19 | 34797996 | 34798411 | 0.747 | 0.257 | 0.111 |
| C19orf12 | chr19 | 34883970 | 34891160 | 0.555 | 0.262 | 0.117 |
| CCNE1 | chr19 | 34994774 | 35007039 | 0.578 | 0.268 | 0.117 |
| C19orf2 | chr19 | 35125351 | 35198176 | 0.745 | 0.262 | 0.111 |
| ZNF507 | chr19 | 37529996 | 37542468 | 0.532 | 0.181 | 0.035 |
| ZNF420 | chr19 | 37569382 | 37620662 | 0.531 | 0.172 | 0.020 |
| ANKRD27 | chr19 | 37779935 | 37781120 | 0.676 | 0.187 | 0.038 |
| CCDC123 | chr19 | 38061790 | 38154709 | 0.545 | 0.175 | 0.029 |
| RHPN2 | chr19 | 38161367 | 38173389 | 0.572 | 0.175 | 0.026 |
| PEPD | chr19 | 38569954 | 38594491 | 0.539 | 0.163 | 0.023 |
| LSM14A | chr19 | 39355265 | 39355508 | 0.628 | 0.178 | 0.023 |
| UBA2 | chr19 | 39611147 | 39652626 | 0.607 | 0.175 | 0.023 |
| ZNF585B | chr19 | 42368828 | 42369771 | 0.547 | 0.178 | 0.023 |
| ZNF383 | chr19 | 42409209 | 42418811 | 0.546 | 0.178 | 0.020 |
| SPINT2 | chr19 | 43447250 | 43474812 | 0.63 | 0.190 | 0.029 |
| PSMD8 | chr19 | 43557200 | 43566216 | 0.642 | 0.184 | 0.029 |
| EIF3K | chr19 | 43806612 | 43819425 | 0.615 | 0.201 | 0.029 |
| ACTN4 | chr19 | 43830180 | 43912972 | 0.581 | 0.201 | 0.032 |
| ECH1 | chr19 | 43997942 | 44014245 | 0.715 | 0.195 | 0.035 |
| SIRT2 | chr19 | 44061065 | 44061778 | 0.535 | 0.204 | 0.050 |
| SARS2 | chr19 | 44097752 | 44113153 | 0.556 | 0.201 | 0.050 |
| SAMD4B | chr19 | 44539177 | 44565771 | 0.6 | 0.198 | 0.052 |
| PAF1 | chr19 | 44568126 | 44573344 | 0.718 | 0.198 | 0.052 |
| MED29 | chr19 | 44573856 | 44582453 | 0.657 | 0.195 | 0.052 |
| RPS16 | chr19 | 44615726 | 44618471 | 0.556 | 0.195 | 0.052 |
| SUPT5H | chr19 | 44628301 | 44659054 | 0.641 | 0.190 | 0.052 |
| TIMM50 | chr19 | 44662966 | 44672833 | 0.657 | 0.181 | 0.052 |
| FBL | chr19 | 45016951 | 45023256 | 0.597 | 0.166 | 0.041 |
| PSMC4 | chr19 | 45168950 | 45179137 | 0.686 | 0.152 | 0.035 |
| AKT2 | chr19 | 45430106 | 45483092 | 0.668 | 0.128 | 0.023 |
| SERTAD3 | chr19 | 45638677 | 45640331 | 0.539 | 0.120 | 0.023 |
| SHKBP1 | chr19 | 45774650 | 45789026 | 0.573 | 0.111 | 0.020 |
These genes showed the best correlation between DNA and copy number and RNA expression levels with R> 0.526 from the TCGA ovarian cancer dataset. Different chromosomes are indicated by different color highlights. Genes within the chr19p13.2 amplicon are in red fonts.
Chr: chromosome
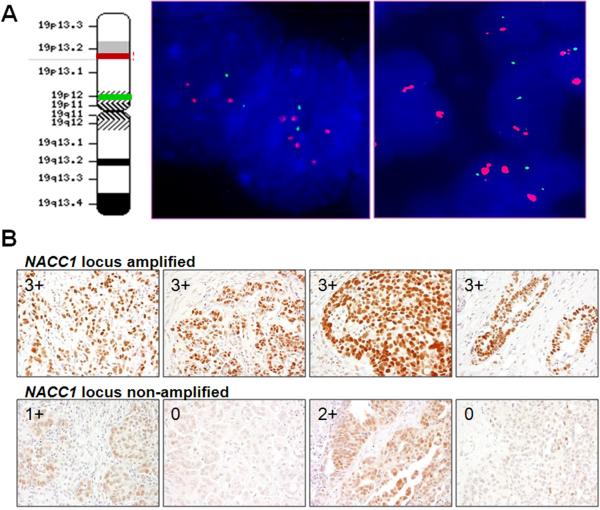
To validate the above results obtained from in silica analysis, we applied two-color FISH and immunohistochemistry to an independent set of tumors composed of 175 high-grade serous carcinomas collected at our institution. We focused on one of the 7 candidate “driver” genes at ch19p13.2, NACC1, because as compared to the other 6 genes, the biological role of NACC1 has been previously reported. NAC1, encoded by NACC1, is a nuclear protein involved in stemness of embryonic stem cells (21) and in the pathogenesis of human cancer (16). NAC1 has been demonstrated to participate in the development of chemoresistant recurrent tumors (22, 23). In this study, we further determined if amplification of the NACC1 locus was related to early disease recurrence in high-grade ovarian serous carcinomas. We designed FISH probes that hybridized to the NACC1 coding region and found that 35 (20%) of 175 carcinomas demonstrated gene copy number gain (> 2.5) at ch19p13.2. Furthermore, 8 (5%) of 175 tumors exhibited high level amplification (>3.5) in DNA copy number at this locus. An example of FISH in a high-grade serous carcinoma is shown in Fig. 2. Immunohistochemistry was used to estimate semi-quantitatively the protein expression levels of NAC1 based on immunostaining intensity in the same set of 175 tumor tissues. We observed that amplified tumors exhibited a significantly higher level of NAC1 expression than those without amplification (p< 0.005, Fisher's exact test). As shown in Table 2, the percentage of tumors with an immunostaining score of 3+ was 63%, 26%, and 13% in specimens showing high amplification, low level gain, and no amplification at the NACC1 locus, respectively. NAC1 immunoreactivity in representative amplified and non-amplified tumors is shown in Fig. 2B.
Fig. 2.
DNA copy number at the NACC1 locus and NAC1 immunoreactivity from representative high-grade serous carcinomas. A. Two-color fluorescence in situ hybridization in two high-grade serous carcinomas shows increased signal of the ch19p13.2 probe (red fluorescence) whereas the control probe (green fluorescence) that hybridizes to a region near the ch19p12 does not shown any gain of intensity. The case on the left panel shows discrete red probe signals whereas the case on the right shows the homogeneously staining regions. The ideogram on the left illustrates the locations which hybridize to the ch19p13.2 probe (red bar) and to the ch19p12 control probe (green bar). B: Immunohistochemistry of NAC1. High-grade ovarian serous carcinomas with high levels of ch19p13.2 amplification show intense immunostaining (3+) of NAC1 in the nuclei (top panels). High-grade ovarian serous carcinomas without detectable alteration of DNA copy number at the ch19p13.2 show variable immunostaining intensity with staining scores ranging from 0 to 2+ (bottom panel). Immunostaining scores are indicated in each photomicrograph.
Table 2.
Correlation of NACC1 DNA copy number and staining intensity in 175 high-grade serous carcinomas.
| 3+ | 2+ | 1+ | 0 | Total | |
|---|---|---|---|---|---|
| NACC1 amplification (copy number>3.5) | 5 | 1 | 2 | 0 | 8 |
| NACC1 gain (3.5> copy number > 2.5 | 7 | 6 | 9 | 5 | 27 |
| Non-amplified | 18 | 45 | 33 | 45 | 140 |
| Total | 30 | 52 | 44 | 50 | 175 |
Fisher's exact test p= 0.0047
Next, we asked whether NACC1 amplification was associated with amplification of other chromosomal loci, including CCNE1, RSF1, NOTCH3, AKT2, and PIK3CA, which were frequently amplified in high-grade serous carcinoma (4). The FISH results from the above genes were available in 146 cases and they were used for the correlation study. Based on FISH data and analysis using a 2×2 contingency table, we found that among these genes amplification of only the NOTCH3 locus (ch19p13.12) was significantly correlated with amplification at the ch19p13.2 locus (p = 0.0044) (Table 3). Among 175 high-grade serous carcinomas, we identified a subset of 52 primary tumors from patients whose follow-up information was available. These patients were selected also based on their similar clinical treatment outcome including optimal cytoreductive surgery (residual tumor< 0.5 cm) followed by a similar regimen of carboplatin/paclitaxel therapy at the Johns Hopkins Hospital, Baltimore, Maryland. We observed that an increase in DNA copy number in the NACC1 locus significantly correlated with earlier recurrence (within 6 months after diagnosis) compared with those without amplification (p= 0.013) (Table 4).
Table 3.
Co-amplification of NACC1 and NOTCH3 loci.
| NOTCH3 amplified | NOTCH3 non-amplified | Total | |
|---|---|---|---|
| NACC1 amplified | 15 | 10 | 25 |
| NACC1 non-amplified | 34 | 87 | 121 |
| Total | 49 | 97 | 146 |
Fisher's exact test p= 0.0044
Table 4.
Correlation of NACC1 locus amplification and the time to first recurrence.
| Recur within 6 months | Recur after 6 months | Total | |
|---|---|---|---|
| NACC1 amplified | 6 | 10 | 16 |
| NACC1 non-amplified | 5 | 31 | 36 |
| Total | 11 | 41 | 52 |
Fisher's exact test p= 0.013
DISCUSSION
In this study, we provide new evidence that DNA copy number at the ch19p13.2 subchromosomal region is increased in approximately one fifth of high-grade serous carcinomas. This finding is based on two large independent cohorts, using two independent techniques including SNP arrays and FISH. The frequency of ch19p13.2 amplification is comparable to that at the CCNE1, NOTCH3, RSF1, AKT2, and PIK3CA loci which were found to be amplified in 36%, 32%, 16%,14%, and 11% of high-grade serous carcinomas, respectively (4). However, it is uncertain whether ch19p13.2 represents a discrete amplicon or a continuum of a larger region of DNA copy number gain in ch19p, perhaps involving the whole arm. The significant co-amplification event of ch19p13.2 and the NOTCH3 locus (ch19p13.12) suggests such a possibility given the proximal location of both loci. Nevertheless, based on our FISH and immunohistochemistry analysis of the same tumor samples, NAC1 expression likely depends on amplification within the ch19p13.2 amplicon. The above results have several important implications with respect to molecular genetic changes and pathogenesis of tumor recurrence in ovarian cancer.
To determine the significance of amplified genes in human cancer, we applied an approach based on the rationale that a tumor-driving gene, when amplified, is almost always over-expressed, which activates the oncogenic pathway, while co-amplified “passenger” genes that are unrelated to tumor pathogenesis may or may not be over-expressed (24). Therefore, we analyzed all genes within all amplified regions detected by the TCGA ovarian cancer dataset to correlate DNA and transcript copy numbers within the same tumor samples. As a result, we have identified 100 genes with the most significant correlation between DNA copy numbers and RNA expression levels and have found that several ovarian cancer-associated oncogenes were on the list, including CCNE1 (17), AKT2 (18), Pak1 (5), Rsf-1 (19), and Paf1 (20). This finding, together with the data showing CCNE1 as the most frequently amplified region (copy number > 3.5) in ovarian serous carcinoma, supports the robustness of this approach in identifying potential cancer driver genes within amplicons. From this perspective, we found that NACC1 was listed with an R value as high as 0.558. This finding suggests that NACC1 is one of the genes that contribute to tumor progression in ovarian cancer in which ch19p13.2 amplification is found. Of note, analysis of the 100 top gene list reveals that the majority of the amplified ovarian cancer driver genes are located on three chromosomes, 8, 11, and 19, a finding consistent with our previous FISH study that profiled all amplicons in ovarian serous carcinoma (4). This information may provide a potential roadmap to study the pathogenesis of ovarian serous carcinoma in the future.
The association of the ch19p13.2 amplification and shorter disease relapse time may be related to NAC1 overexpression. NAC1, encoded by NACC1, belongs to the BTB/POZ domain gene family and contains the BTB/POZ domain, which is responsible for homodimerization and heterodimerization with other BTB/POZ proteins as well as the BEN domain that mediate protein-DNA and protein-protein interactions during chromatin organization and transcription (25). The role of NAC1 in the development of human cancer has recently emerged. NAC1 is significantly overexpressed in several types of human cancers including ovarian high-grade serous carcinoma (16, 22, 26, 27), endometrial carcinoma (28), and cervical carcinoma (29). Like several BTB/POZ family members, NAC1 proteins homo-dimerize through the BTB domain. Induced expression of a NAC1 deletion mutant (N130) containing exclusively the BTB domain attenuates the tumor-promoting functions of NAC1 (16). On the other hand, over-expression of full-length NAC1 is sufficient to enhance tumorigenicity of ovarian surface epithelial cells and NIH3T3 cells in athymic nu/nu mice (16). More recently, we observed that enforced expression of NAC1 conferred drug resistance, and NAC1 knockdown by shRNA sensitized paclitaxel cytotoxicity in ovarian cancer cells in vitro (22). NAC1 contributed to the development of drug resistance through multiple mechanisms including upregulating fatty acid synthase (30) and negatively regulating the components of the Gadd45 tumor suppressor pathway including Gadd45α and its binding protein, Gadd45gip1 (22, 26). The above findings may explain the clinical observation that upregulation of NAC1 immunoreactivity in primary ovarian tumors is associated with aggressive clinical behavior and tumor recurrence in ovarian cancer patients (16, 27). We did not attempt to analyze the clinical correlation of NACC1 amplification using the TCGA dataset because of concerns about the difference in treatment regimens and patient populations from the various institutions that contributed to the dataset.
The frequent co-amplification of the NACC1 and NOTCH3 is of interest. Our previous studies showed amplification of the NOTCH3 locus in 32% of ovarian high-grade serous carcinomas (12), and Notch3 overexpression is related to the recurrence of ovarian cancer and confers drug resistance (31). Ovarian cancer cells which had amplified and overexpressed Notch3 were dependent on Notch3 signaling for cellular survival and growth. Thus, it is likely that increased DNA copy number in both genes may contribute to the resistance of carboplatin and paclitaxel that are routinely used in treating advanced stage ovarian cancer patients. Interestingly, we have recently demonstrated that inactivation of the Notch3 pathway led to inhibition of NAC1 expression, indicating that Notch3 signaling may regulate the expression of NAC1 (31). Further studies are required to confirm the molecular cross talk between these pathways in the development of chemoresistance.
In summary, based on analysis of the TCGA ovarian cancer dataset and our FISH result, we were able to demonstrate that amplification at the NACC1 locus was one of the frequent molecular genetic alterations in ovarian high-grade serous carcinomas. NAC1 overexpression may be, in part, attributed to the increase in DNA copy number, explaining why amplification at the NACC1 locus is related to early tumor recurrence in ovarian cancer. Future studies should aim at fine mapping the ch19p13.2 amplified region and assessing the potential of other genes at the ch19p13.2 locus to contribute to the aggressive behavior of ovarian serous carcinomas that harbor this amplification.
Grant acknowledgement
This work was supported by an NIH/NCI grant (CA103937).
REFERENCES
- 1.Cho KR, Shih IM. Ovarian cancer. Annu Rev Pathol Mech Dis. 2009;4:287–313. doi: 10.1146/annurev.pathol.4.110807.092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuo K, Mao T, Feng Y, et al. DNA copy number profiles in affinity-purified ovarian clear cell carcinoma. Clin Cancer Res. 2010;16:1997–2008. doi: 10.1158/1078-0432.CCR-09-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo KT, Guan B, Feng Y, et al. Analysis of DNA Copy Number Alterations in Ovarian Serous Tumors Identifies New Molecular Genetic Changes in Low-Grade and High-Grade Carcinomas. Cancer Res. 2009;69:4036–42. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama K, Nakayama N, Jinawath N, et al. Amplicon profiles in ovarian serous carcinomas. Int J Cancer. 2007;120:2613–7. doi: 10.1002/ijc.22609. [DOI] [PubMed] [Google Scholar]
- 5.Schraml P, Schwerdtfeger G, Burkhalter F, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163:985–92. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meinhold-Heerlein I, Bauerschlag D, Hilpert F, et al. Molecular and prognostic distinction between serous ovarian carcinomas of varying grade and malignant potential. Oncogene. 2005;24:1053–65. doi: 10.1038/sj.onc.1208298. [DOI] [PubMed] [Google Scholar]
- 7.Mayr D, Kanitz V, Anderegg B, et al. Analysis of gene amplification and prognostic markers in ovarian cancer using comparative genomic hybridization for microarrays and immunohistochemical analysis for tissue microarrays. Am J Clin Pathol. 2006;126:101–9. doi: 10.1309/n6x5mb24bp42kp20. [DOI] [PubMed] [Google Scholar]
- 8.Nowee ME, Snijders AM, Rockx DA, et al. DNA profiling of primary serous ovarian and fallopian tube carcinomas with array comparative genomic hybridization and multiplex ligation-dependent probe amplification. J Pathol. 2007;213:46–55. doi: 10.1002/path.2217. [DOI] [PubMed] [Google Scholar]
- 9.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci U S A. 2002;99:16156–61. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakayama K, Nakayama N, Davidson B, et al. Homozygous Deletion of MKK4 in Ovarian Serous Carcinoma. Cancer Biol Ther. 2006;5:630–4. doi: 10.4161/cbt.5.6.2675. [DOI] [PubMed] [Google Scholar]
- 11.Shih Ie M, Sheu JJ, Santillan A, et al. Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Proc Natl Acad Sci U S A. 2005;102:14004–9. doi: 10.1073/pnas.0504195102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JT, Li M, Nakayama N, et al. Notch-3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–8. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- 13.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 14.Reich M, Liefeld T, Gould J, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 15.Wang TL, Diaz LA, Jr., Romans K, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci U S A. 2004;101:3089–94. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayama K, Nakayama N, Davidson B, et al. A BTB/POZ protein, NAC-1, is related to tumor recurrence and is essential for tumor growth and survival. Proc Natl Acad Sci U S A. 2006;103:18739–44. doi: 10.1073/pnas.0604083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farley J, Smith LM, Darcy KM, et al. Cyclin E expression is a significant predictor of survival in advanced, suboptimally debulked ovarian epithelial cancers: a Gynecologic Oncology Group study. Cancer Res. 2003;63:1235–41. [PubMed] [Google Scholar]
- 18.Cheng JQ, Godwin AK, Bellacosa A, et al. AKT2, a putative oncogene encoding a member of a subfamily of protein- serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci U S A. 1992;89:9267–71. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi JH, Sheu JJ, Guan B, et al. Functional analysis of 11q13.5 amplicon identifies Rsf-1 (HBXAP) as a gene involved in paclitaxel resistance in ovarian cancer. Cancer Res. 2009;69:1407–15. doi: 10.1158/0008-5472.CAN-08-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhary K, Deb S, Moniaux N, Ponnusamy MP, Batra SK. Human RNA polymerase II-associated factor complex: dysregulation in cancer. Oncogene. 2007;26:7499–507. doi: 10.1038/sj.onc.1210582. [DOI] [PubMed] [Google Scholar]
- 21.Wang JRS, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–8. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 22.Jinawath N, Vasoontara C, Yap KL, et al. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene. 2009;28:1941–8. doi: 10.1038/onc.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishibashi M, Nakayama K, Yeasmin S, et al. A BTB/POZ gene, NAC-1, a tumor recurrence-associated gene, as a potential target for taxol resistance in ovarian cancer. Clin Cancer Res. 2008;14:3149–55. doi: 10.1158/1078-0432.CCR-07-4358. [DOI] [PubMed] [Google Scholar]
- 24.Hogarty MD, Brodeur GM. Gene amplification in human cancers: biological and clinical significance. In: Vogelstein B, Kinzler Kw, editors. The genetic basis of human cancer. 2 edn. McGraw-Hill; New York: 2002. pp. 115–28. [Google Scholar]
- 25.Abhiman S, Iyer LM, Aravind L. BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics. 2008;24:458–61. doi: 10.1093/bioinformatics/btn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakayama K, Nakayama N, Wang T-L, Shih I-M. NAC-1 Controls Cell Growth and Survival by Repressing Transcription of Gadd45GIP1, a Candidate Tumor Suppressor. Cancer Res. 2007;67:8058–64. doi: 10.1158/0008-5472.CAN-07-1357. [DOI] [PubMed] [Google Scholar]
- 27.Davidson B, Berner A, Trope CG, Wang TL, Shih Ie M. Expression and clinical role of the bric-a-brac tramtrack broad complex/poxvirus and zinc protein NAC-1 in ovarian carcinoma effusions. Hum Pathol. 2007;38:1030–6. doi: 10.1016/j.humpath.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Ishikawa M, Nakayama K, Yeasmin S, et al. NAC1, a potential stem cell pluripotency factor expression in normal endometrium, endometrial hyperplasia and endometrial carcinoma. Int J Oncol. 2010;36:1097–103. doi: 10.3892/ijo_00000591. [DOI] [PubMed] [Google Scholar]
- 29.Yeasmin S, Nakayama K, Ishibashi M, et al. Expression of the bric-a-brac tramtrack broad complex protein NAC-1 in cervical carcinomas seems to correlate with poorer prognosis. Clin Cancer Res. 2008;14:1686–91. doi: 10.1158/1078-0432.CCR-07-4085. [DOI] [PubMed] [Google Scholar]
- 30.Ueda SM, Yap KL, Davidson B, et al. Expression of Fatty Acid Synthase Depends on NAC1 and Is Associated with Recurrent Ovarian Serous Carcinomas. J Oncol. 2010;2010:285191. doi: 10.1155/2010/285191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JT, Chen X, Trope CG, et al. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to Carboplatin. Am J Pathol. 2010;177:1087–94. doi: 10.2353/ajpath.2010.100316. [DOI] [PMC free article] [PubMed] [Google Scholar]