Abstract
Discovery of emerging REGγ-regulated proteins has accentuated the REGγ-proteasome as an important pathway in multiple biological processes, including cell growth, cell cycle regulation, and apoptosis. However, little is known about the regulation of the REGγ-proteasome pathway. Here we demonstrate that REGγ can be SUMOylated in vitro and in vivo by SUMO-1, SUMO-2, and SUMO-3. The SUMO-E3 protein inhibitor of activated STAT (PIAS)1 physically associates with REGγ and promotes SUMOylation of REGγ. SUMOylation of REGγ was found to occur at multiple sites, including K6, K14, and K12. Mutation analysis indicated that these SUMO sites simultaneously contributed to the SUMOylation status of REGγ in cells. Posttranslational modification of REGγ by SUMO conjugation was revealed to mediate cytosolic translocation of REGγ and to cause increased stability of this proteasome activator. SUMOylation-deficient REGγ displayed attenuated ability to degrade p21Waf//Cip1 due to reduced affinity of the REGγ SUMOylation-defective mutant for p21. Taken together, we report a previously unrecognized mechanism regulating the activity of the proteasome activator REGγ. This regulatory mechanism may enable REGγ to function as a more potent factor in protein degradation with a broader substrate spectrum.
Keywords: REGγ, SUMO, modification, PIAS1
Introduction
The proteasome activator REGγ (also known as PA28γ, PSME3, Ki antigen) has been shown to promote degradation of intact cellular proteins such as the nuclear receptor cofactor SRC-3 and cyclin-dependent kinase inhibitor p21 1, 2, 3. Aberrant expression of REGγ is involved in cancer progression 4, 5. A recent comparative analysis of REGγ expression in mouse and human tissues revealed unique patterns of REGγ in specific cell types, suggesting additional undisclosed functions and biological importance of this molecule 6. How REGγ function is regulated within mammalian cells is unclear.
The Small Ubiquitin-related Modifier or SUMO proteins, including the ubiquitously expressed SUMO-1, SUMO-2, and SUMO-3, are a family of ubiquitin-like polypeptides, which can be covalently attached to target proteins. Posttranslational SUMO modification of proteins results in modification of their functions, such as nuclear-cytosolic transportation, protein-protein interactions, protein stability, and modulation of cellular processes such as response to stress or cell cycle progression. Similar to the ubiquitination process, SUMOylation also requires activating (E1), conjugating (E2), and ligating (E3) enzymatic steps. In contrast to the ubiquitin pathway, SUMOylation process is mediated by a single E2 enzyme, UBC9, and a limited number of E3 enzymes 7. Among the recognized SUMO ligases, proteins with Siz/PIAS RING-finger-like domains (SP-RING domains) are known as protein inhibitor of activated STAT (PIAS) proteins in vertebrates 8. The five vertebrate PIAS proteins (PIAS1, PIAS3, PIASxα, PIASxβ, and PIASy) have been implicated in many processes, including gene expression, signal transduction, and genome maintenance 9. Yet, their biological targets are not fully disclosed.
Here we report that the proteasome activator REGγ can be SUMOylated posttranslationally by PIAS1. SUMO modification of REGγ at specific sites results in cytoplasmic localization and increased stability of REGγ. SUMOylation-defective mutant REGγ is attenuated for its function in degrading its target protein, p21Waf//Cip1. These results reveal a previously unknown mechanism for the regulation of REGγ activity.
Results
Identification of PIAS1 as a REGγ interacting protein
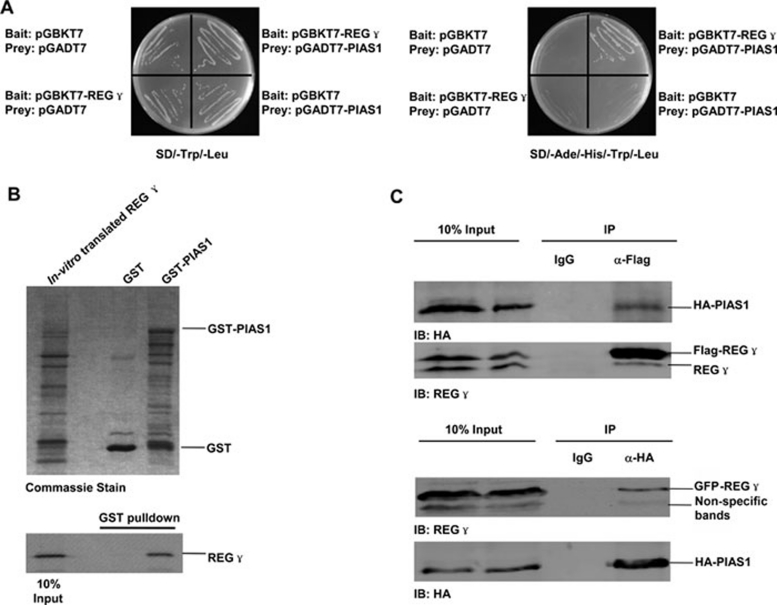
In an effort to search for REGγ interacting proteins, we performed yeast two-hybrid analysis using REGγ as the bait. One of the REGγ interacting proteins is PIAS1. We verified the physical interaction between REGγ and PIAS1 in yeast strain AH109 (Clontech) with a medium lacking Ade/His/Trp/Leu for quadruplicate selection of growth (Figure 1A). To further validate this interaction, we constructed GST-PIAS1 and purified GST and GST-PIAS1 proteins from bacteria (Figure 1B, upper panel). GST pull-down experiments were carried out in the presence of in vitro translated REGγ. Compared to GST alone, GST-PIAS1 can pull down a remarkable amount of REGγ (Figure 1B, lower panel). We then tested the intracellular capability of REGγ to interact with PIAS1. By overexpressing both REGγ and PIAS1 in 293T cells and immunoprecipitating REGγ, we detected overt co-immunoprecipitation of PIAS1 (Figure 1C, upper panel). We also performed reciprocal experiments by immunoprecipitating PIAS1 to detect co-immunoprecipitation of REGγ. Taken together, these experiments support that REGγ directly interacts with PIAS1.
Figure 1.
REGγ interacts with PIAS1 in vitro and in vivo. (A) Interaction of REGγ and PIAS1 in yeast. AH109 was transformed with the indicated bait and prey plasmids as described in Materials and Methods. The transformants were then streaked on SD/-Trp/-Leu (Left) to indicate proper expression of each construct and a replicate on SD/-Ade/-His/-Trp/-Leu (right) selective plate demonstrated the physical interaction between REGγ and PIAS1. (B) REGγ interacts with PIAS1 in vitro. GST-PIAS1 or GST only was incubated with in vitro-translated REGγ as described in Materials and Methods. The relative amounts of GST and GST fusion proteins are shown in the Coomassie-stained gel (upper panel). Following 2-h incubation at 4 °C, the complex was pulled down by GST beads, and REGγ level was detected by autoradiograph (lower panel). (C) REGγ interacts with PIAS1 in cells. 293T cells were co-transfected with HA-PIAS1 and Flag-REGγ or GFP-REGγ for 48 h as described in Materials and methods. Immunoprecipitation was performed with whole cell lysates using anti-flag antibody, anti-HA or IgG (control). The immunocomplexes were separated by SDS-PAGE and detected by an antibody against REGγ or HA.
PIAS1 promotes SUMO-modification of REGγ in cells
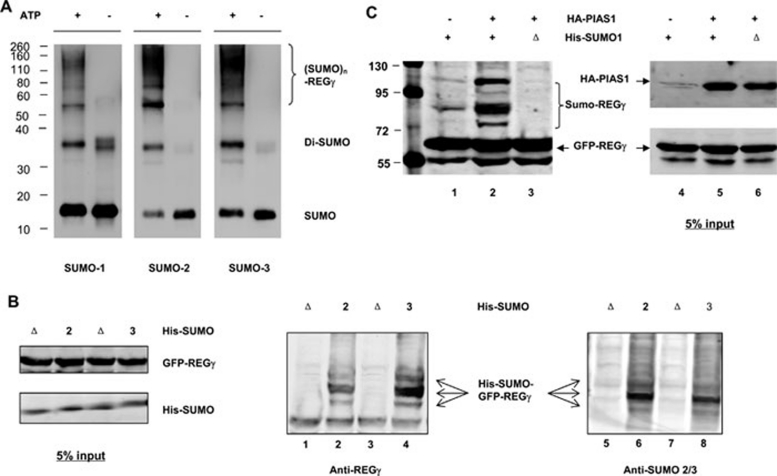
To test if REGγ can be SUMOylated, we first performed in vitro SUMOylation study in the presence of SUMO E1 (AOS1/UBA2), SUMO E2 (UBC9), and ATP. Figure 2A demonstrates that SUMO-1, -2, and -3 can be utilized in vitro to generate multiple forms of SUMOylated REGγ. To further verify REGγ SUMOylation in vivo, we performed cell-based analysis to enrich SUMO-REGγ conjugates by purification of 6His-SUMO under denaturing conditions, followed by western blot with anti-REGγ. To enhance the assay sensitivity, we used GFP-REGγ as the substrate, which does not form a homoheptamer. While GFP protein could not be SUMOylated (Supplementary information, Figure S1, lanes 3 and 4), GFP-REGγ was heavily SUMOylated in the presence of wild type (wt) His-SUMO-2 and His-SUMO-3 (Figure 2B, lanes 2 and 4; Supplementary information, Figure S1, lane 1), but not when conjugation-defective His-SUMO-2/3 (ΔGG) were coexpressed (Figure 2B, lanes 1 and 3). To validate if the SUMO-GFP-REGγ complexes detected in Figure 2B by western blot with a rabbit anti-REGγ were truly conjugates of SUMO proteins, the same membrane was probed with a mouse anti-SUMO 2/3. Similar patterns of SUMO-GFP-REGγ complexes were observed in Figure 2B, lanes 6 and 8, but not in the presence of conjugation-defective His-SUMO-2/3 (Figure 2B, lanes 5 and 7). We also observed that SUMO-1 can be conjugated to the REGγ molecule in cells (Supplementary information, Figure S2).
Figure 2.
REGγ is SUMOylated in vitro and in vivo. (A) SUMOylation of REGγ in vitro. In vitro SUMOylation assay was carried out as described in Materials and methods in a reaction buffer with (+) or without (−) ATP. The reaction mixtures were analyzed by western blot with antibodies to SUMO-1 and SUMO-2/3, respectively. (B) SUMOylation of REGγ in vivo. 293T cells were co-transfected with GFP-REGγ or GFP, along with Ubc9, His-SUMO-2, or conjugation-deficient His-SUMO-2 (ΔGG) mutant. SUMO substrates were purified using Ni-NTA-agarose beads and probed with REGγ antibodies as described in Materials and methods. The left panel displays the input of GFP-REGγ, His-SUMO-2/3, and defective His-SUMO-2/3 (ΔGG) as control. The middle panel shows the His-SUMO-GFP-REGγ conjugates detected by western blot using anti-REGγ. The right panel shows His-SUMO-GFP-REGγ conjugates (pointed with arrows) detected with anti-SUMO 2/3. No SUMO conjugation was observed in defective His-SUMO-2/3 (ΔGG) transfected controls. (C) PIAS1 enhances SUMOylation of REGγ. 293T cells were transfected with GFP-REGγ, His-SUMO1, or His-SUMO-1 (ΔGG) in the presence or absence of HA-PIAS1. SUMOylated protein was enriched with Ni-NTA-agarose beads and probed with REGγ antibody. The left panel displays marked increase of SUMOylated GFP-REGγ in the presence of HA-PIAS1, but no SUMOylation in His-SUMO-1 (ΔGG) transfected control. The input expressions of HA-PIAS1 and GFP-REGγ are shown in the right panel.
PIAS1 has been shown to have SUMO-E3 activity 9. Given the evidence for physical interaction between PIAS1 and REGγ, we investigated if PIAS1 can promote SUMO-modification of REGγ in vivo. GFP-REGγ was coexpressed with His-SUMO-1/His-SUMO-1 (ΔGG), HA-PIAS1 or a control vector, followed by enrichment of His-SUMO-REGγ conjugates with affinity chromatography. The amount of SUMOylated REGγ in the presence of exogenous PIAS1 was more than three-fold higher (Figure 2C, lanes 2) than that of the control sample (Figure 2C, lanes 1). To confirm that the slow migrating bands are SUMO-conjugated GFP-REGγ, the inactive His-SUMO-1 (ΔGG) was used as specificity control to show that no SUMOylation of REGγ occurred under such condition (Figure 2C, lane 3). We further tested other PIAS family members to determine the specificity of PIAS1 as an E3 ligase for REGγ. Cells co-expressing PIAS1, PIAS3, PIASxα, PIASxβ or PIASy were examined for SUMO conjugation of REGγ. Compared with control, only PIAS1 can promote robust SUMOylation of REGγ despite that PIASxβ also had a minor effect on REGγ modification (Supplementary information, Figure S3, lanes 2 and 4). These results indicate that PIAS1 functions as a specific SUMO-E3 for REGγ modification in vivo.
SUMO modification results in cytoplasmic translocation of REGγ
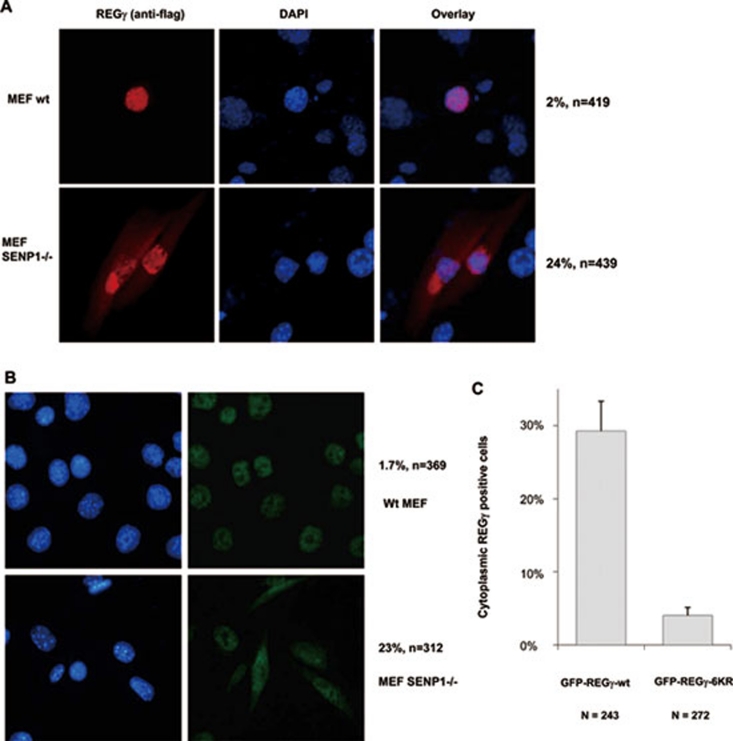
Protein SUMOylation has been known to regulate subcellular localization 9, 10. We explored the possibility that SUMOylated REGγ may also have altered subcellular localization patterns. We first overexpressed exogenous REGγ in wt MEF cells or in MEFs defective for SENP-1, which catalyzes deconjugation of SUMO-containing species. While the wt MEFs transfected with REGγ had ∼2% of cells with cytoplasmic localization (Figure 3A, upper panel), REGγ-transfected SENP-1−/− MEFs showed ∼24% of cells with cytoplasmic staining of REGγ by immunofluorescent image analysis (Figure 3A, lower panel). We further investigated the distribution of endogenous REGγ in wt and SENP-1−/− MEFs by immunostaining. Similar to exogenously expressed REGγ, endogenous REGγ also showed cytoplasmic expression in a significantly higher percentage of SENP-1−/− MEF cells (Figure 3B, lower panel) than in wt MEFs, which had less than 2% of cells with cytoplasmic REGγ staining (Figure 3B, upper panel). To further validate that cellular redistribution of REGγ is regulated by SUMO-modification, we generated K to R mutations in six predicted SUMO sites in REGγ (construct named REGγ-6KR based on the information in Supplementary information, Table S1). GFP-REGγ and GFP-REGγ-6KR were transfected into SENP-1−/− MEF cells to investigate the cytoplasmic expression profiles. In line with our prediction, wt GFP-REGγ displayed cytoplasmic staining in ∼30% of transfected cells (Figure 3C). In contrast, less than 5% of cells revealed cytoplasmic distribution in cells transfected with SUMOylation-deficient GFP-REGγ-6KR (Figure 3C). Furthermore, we fused SUMO to the C-terminus of REGγ. Compared with intact REGγ, both REGγ-SUMO-1 and REGγ-SUMO-2 fusion proteins exhibited significant nuclear export (Supplementary information, Figure S4A and S4B). Our data demonstrate that SUMOylation of REGγ clearly regulates cellular distribution of this proteasome activator.
Figure 3.
SUMO modification regulates cytosolic translocation of REGγ. (A) Cytosolic translocation of exogenous REGγ in SUMOylation-active cells. Wild-type and SENP-1−/− MEF cells were transiently transfected with pcDNA5/FRT/TO Flag-REGγ (1 μg). Twenty-four hours after transfection, immunostaining was performed with anti-DDK antibody (Flag) as described in Materials and methods to examine the expression and distribution of REGγ (red). DAPI staining indicates the location of nuclei. The percentage of transfected cells with cytoplasmic expression of REGγ is shown on the right. (B) SUMOylation enhances cytoplasmic translocation of endogenous REGγ. Wild-type and SENP-1−/− MEF cells were fixed and immunostaining was performed using anti-REGγ as in (A) to visualize cellular localization of endogenous REGγ. The percentage of REGγ-positive cells with cytoplasmic translocation is shown on the right. (C) SUMOylation-deficient REGγ has diminished cytoplasmic localization. SENP-1−/− MEF cells were transfected with 1 μg of GFP-REGγ or GFP-REGγ-6KR (mutation in all six predicated SUMO sites). The cytoplasmic vs nuclear fluorescent patterns in transfected cells expressing GFP fusion proteins were scored for statistical analysis. Data are shown as mean ± SD of three independent experiments (P < 0.01).
SUMOylation of REGγ occurs on multiple sites
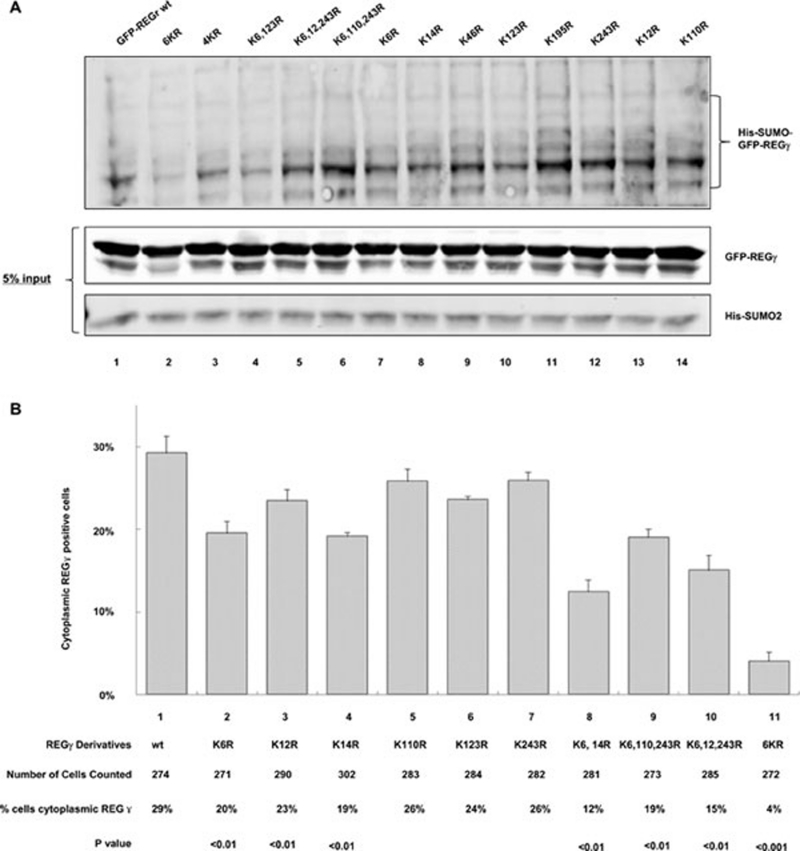
Since the covalent SUMO modification of target proteins occurs via lysine residues 11, 12, we searched potential SUMOylation sites in REGγ sequence with bioinformatic analysis. Our analysis with SUMOplot indicated six candidate SUMO modification sites bearing the highest scores (Supplementary information, Table S1), while additional sites with high score were found using program SUMOsp2.0 (Supplementary information, Table S2). All these high-score sites are evolutionarily conserved among vertebrates and some are conserved in worm and insect (Supplementary information, Figure S5). To estimate the contribution of each lysine in SUMO modification, we generated the individual mutation of each lysine (K) to arginine (R) and generated constructs containing simultaneous mutation of all K to R at positions 123, 6, 14, 243, 46, and 195 (named 6KR, note that the individual K-R mutation at position 6 is K6R). While most REGγ constructs with individual K mutation did not reveal significantly quantifiable difference in SUMO conjugation patterns compared with wt by western analysis, double or triple mutations mostly revealed obvious SUMOylation deficiency (Figure 4A and Supplementary information, Figure S6). Simultaneous mutation of all six lysines in REGγ significantly attenuated SUMO modification of REGγ in vivo, yet SUMOylation of REGγ was not completely eradicated (Figure 4A, lanes 1 and 2), indicating additional sites being SUMOylated. These results suggest that REGγ SUMOylation occurs in multiple sites.
Figure 4.
SUMOylation of REGγ occurs on multiple sites. (A) REGγ SUMOylation occurs on multiple sites. GFP-REGγ, GFP-REGγ-6KR, or GFP-REGγ derivatives containing one or combinational mutations were co-expressed with His-SUMO-2 in 293T cells. SUMO conjugates were purified and examined as described in Materials and methods. GFP-REGγ-6KR (lane 2) had markedly reduced SUMO conjugates than GFP-REGγ (lane 1). The 4KR refers to combinational mutations in K6, K12, K110, and K243. The input levels of His-SUMO-2 and GFP-REGγ are shown in the lower panels. (B) Effect of SUMOylation site mutation(s) in cytosolic translocation of REGγ. Flag-REGγ and Flag-REGγ with single or combinatorial K mutation at putative SUMO site(s) were transfected into SENP-1−/− MEF cells. Twenty-four hours after transfection, cytoplasmic localization of transfected REGγ derivatives was examined by immunostaining with anti-DDK (Flag) antibody and scored for statistical analysis. Data are shown as mean ± SD of three independent experiments along with the number of cells counted and percentage of positive cells in the cytoplasm, as well as P values.
To elucidate which of these prospective SUMO sites are important for SUMO modification of REGγ, we used a more quantifiable approach based on the fact that SUMOylated REGγ can be exported to the cytoplasm. Transient transfection of each REGγ construct with a single lysine mutation was scored for the percentage of cells with cytoplasmic localization of REGγ in transfected SENP-1−/− MEF cells. Averaged results from three independent experiments suggested that compared to wt, REGγ derivatives with K6R, K14R, and K12R mutations had significantly reduced capacity in cytoplasmic localization (Figure 4B), suggesting that each of these sites can be SUMOylated. To further examine the contribution of K12, K110, and K243 in REGγ SUMOylation, we generated combination of triple mutations. Triple mutations in K6, K110, and K243R revealed no significant difference with that of K6R mutation alone, suggesting a less important role for K110 and K243 in SUMO modification. Compared with the K6 mutation, further reduction of cytoplasmic localization associated with REGγ triple mutations at K6, K12, and K243R reflected a combinatorial contribution for K6 and K12 to SUMO modification of REGγ. Combined mutation of K6 and K14 resulted in an even lower percentage in nuclear export than the triple mutation at K6, K12, and K243R. Consistent with SUMO-conjugation analysis, REGγ with multiple mutations (REGγ-6KR) expressed in SENP-1−/− MEF cells displayed a dramatic reduction in cytoplasmic localization (Figure 4A, lane 2 and Figure 4B, lane 11), supporting that SUMOylation of REGγ occurs at multiple sites.
SUMOylation enhances the stability of REGγ protein in cells
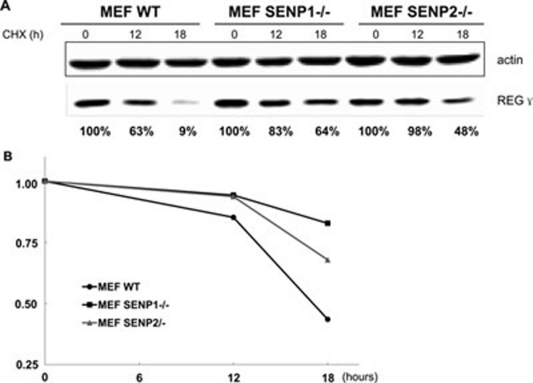
A known consequence of SUMO modification is alteration of the stability of target proteins 9. To test the effect of SUMO modification on REGγ stability, we examined the decay rate of endogenous REGγ in wt, and SENP-1 and SENP-2 knockout MEFs. Following treatment with cycloheximide (CHX), an inhibitor of protein biosynthesis in eukaryotic organisms, for 12 and 18 h, the protein level of REGγ was evaluated by western blot (Figure 5A). Results from three independent experiments were quantitated and averaged results were plotted in Figure 5B. Increased stability of REGγ in SENP-1−/− MEFs was also observed with longer period of treatment by CHX (Supplementary information, Figure S7). Our results indicate that posttranslational modification of REGγ by SUMOylation enhances the half-life of this proteasome activator in cells.
Figure 5.
SUMOylation augments REGγ stability. (A) REGγ stability is increased in SENP-1−/− and SENP-2−/− MEFs. WT, SENP-1−/− and SENP-2−/− MEF cells were treated with cycloheximide (100 μg / ml) for the indicated time. An equal amount of total protein was loaded for western blotting analysis. (B) Quantitative analysis of REGγ stability in SENP null MEFs. Experiments were performed as described in (A). Relative REGγ level was plotted against the time course following cycloheximide treatment. Data are shown as mean of three independent experiments (standard deviation of the last time point is shown).
SUMOylation-defective mutant of REGγ has attenuated capacity to degrade p21
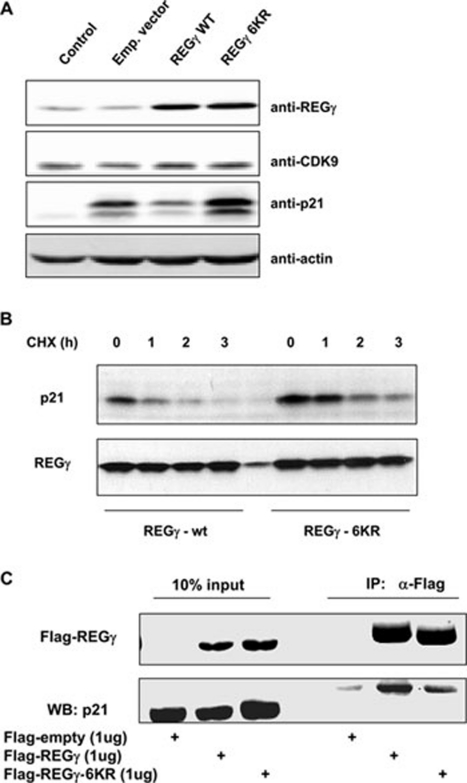
p21 has been reported as a direct substrate of REGγ-proteasome 1, 2. To understand the biological consequence of REGγ SUMOylation, we analyzed the ability of SUMOylation-deficient REGγ-6KR to promote the degradation of p21. When wt REGγ, REGγ-6KR, or an empty vector was co-expressed with p21 for 24 h to examine the activity of REGγ in p21 degradation, we found a significant reduction in the capacity of REGγ-6KR to degrade p21 (Figure 6A). To further validate the effect of REGγ-6KR on p21 stability, we performed similar transient transfection experiments and treated cells with CHX for different periods of time to prevent de novo synthesis of proteins. We found faster decay of p21 in cells co-expressing wt REGγ and much slower degradation of p21 in cells transfected with REGγ-6KR, suggesting an effect of REGγ-6KR on p21 protein stability (Figure 6B). Consistent with this, we found that hyper-SUMOylated REGγ in SENP-1−/− cells can promote endogenous p21 turnover. Despite the overall p21 levels in SENP-1−/− cells being higher, likely due to SUMOylation-mediated regulation of proteins like p53, we observed a faster decay rate of p21 in SENP-1−/− cells (Supplementary information, Figure S8). Finally, we examined the potential mechanism involved in SUMOylation-mediated regulation of REGγ activity. To test the impact of REGγ SUMOylation on substrate binding, we expressed Flag-tagged REGγ or SUMOylation-deficient REGγ-6KR with p21 in 293T cells. Following immunoprecipitation and western blotting analysis, we observed significantly reduced binding of SUMOylation-deficient REGγ-6KR to p21. This may reflect the reduced ability of REGγ-6KR in recruiting p21 to the REGγ-proteasome for subsequent degradation.
Figure 6.
SUMOylation-deficient REGγ has reduced activity in p21 degradation. (A) REGγ-6KR influences expression of p21. H1299 cells were transfected with REGγ, REGγ-6KR, an empty vector, or a mock control along with p21 for 24 h. Expression of the protein of interest was examined by western blotting with indicated antibodies. CDK9 serves as a control for specificity. (B) SUMOylation-defective REGγ has attenuated proteolytic activity. Experiments were performed as in (A) and cells were treated with cycloheximide for the indicated time. The expression of p21 and dynamic decay rate were determined by western blot analysis. The expression of REGγ and REGγ-6KR was equivalent in the two groups of samples examined. (C) SUMOylation-deficient REGγ-6KR has attenuated affinity for p21. In vivo interaction between p21 and wt or SUMOylation-defective REGγ was examined by co-expressing these constructs and by subsequent immunoprecipitation analysis using anti-Flag antibody. Expression of a Flag-empty vector was used as negative control. To prevent degradation of the substrate p21, cells were treated with MG132 for 4 h before collection.
Discussion
Along with the discovery of the first mammalian target of REGγ-proteasome 3, identification of increasing numbers of cellular proteins proteolytically regulated by REGγ 1, 2, 13 has established the newly recognized alternative proteasome pathway. Yet, the regulatory input that may alter the biological function of REGγ was not previously described. Here we show that SUMOylation of REGγ can occur in vitro and in vivo. This posttranslational modification can be enhanced in the presence of PIAS1. SUMO modification of REGγ mainly occurs at three lysine residues, which results in cytoplasmic distribution and stabilization of REGγ. We also demonstrated that the SUMOylation-defective mutant of REGγ had attenuated capacity to degrade its target protein, p21.
To our knowledge, this study is the first report describing the molecular details of REGγ SUMOylation, which can be facilitated by PIAS1. Our discovery that REGγ physically associates with PIAS1 is consistent with a previously unpublished result mentioned in a review article 14. Despite the SUMOylation of proteins being known to occur in the presence of SUMO-E1, E2, and SUMO in the absence of a SUMO-E3 15, recent studies have highlighted the importance of SUMO-E3, such as PIAS1, in numerous cases of SUMO modification of biologically important substrate proteins 16, 17, 18. In our studies, we found that SUMO-1, -2, and -3 can all be conjugated to REGγ both in vitro and in vivo, suggesting that PIAS1-dependent and -independent processes are involved in REGγ SUMOylation. Consistent with this notion, we observed that equivalent REGγ SUMOylation occurred in both SENP-1 and SENP-2 knockout MEFs.
Although SUMO modification in some proteins occurs at a single site, SUMO conjugation via multiple sites was known to happen 19, 20. Our mutational analysis indicates that at least three lysines within REGγ function as SUMO-acceptor residues. Notably, three of these residues, K6, K12, and K14, are located within the N-terminal region, which partially contributes to distinct functions between REGγ and REGα/β 4. It is possible that SUMO modification in these three amino-acid residues is specific only to REGγ, but not to REGα/β.
SUMO modification has been well known for regulation of cellular distribution of conjugated proteins. SUMO modification has been mostly described for its impact on nuclear import of target proteins. Nevertheless, SUMOylation also regulates nuclear export of modified proteins 21. In our study, we found marked increase of cytoplasmic localization of SUMOylated REGγ, which normally stays in the nucleus. Therefore, SUMO modification enables REGγ to reach substrates in the cytoplasm under certain conditions and increases the spectrum of REGγ-mediated protein degradation. In fact, we have analyzed the role of cytoplasmic REGγ in coxsackieviral infection since coxsackievirus replication takes place exclusively in the cytoplasm. Relevant work is being published elsewhere.
Furthermore, we discovered that REGγ SUMOylation enhanced its stability and that SUMOylation-deficient REGγ had attenuated function in p21 degradation. Increased stability of endogenous REGγ in SUMOylation super-active SENP-1−/− and SENP-2−/− cells is likely due to reduction of ubiquitination at the SUMO conjugated sites. Consistent with this prediction, we found that SUMOylation-deficient REGγ is also more stable, probably due to ubiquitination deficiency as well (data not shown). Although increased stability of REGγ may contribute to the rapid turnover of p21, enhanced REGγ activity may play a more important role. The biological significance of REGγ SUMOylation is highlighted by the observation that SUMOylation-defective REGγ mutant shows significantly reduced ability to degrade p21. The observation of weakened physical interaction between the SUMOylation-defective REGγ mutant and p21 further validated our hypothesis that perhaps posttranslational modification of REGγ is required for this proteasome activator to fully interact with its substrate proteins or a specific target. Further experiments are required to determine whether other types of posttranslational modifications occur in K6, K14, and K12, which may also contribute to the regulation of REGγ function.
Materials and Methods
Cell lines and cell culture
HEK293T and H1299 cells were purchased from ATCC and maintained at Cell Culture Core in our Department. The SENP-1−/−, SENP-2−/− MEFs 22, SW1116 NS, and SW1116 SiSENP1 cells were kindly provided by Dr Jingke Chen from Shanghai Jiao Tong University School of Medicine. All cells were cultured under standard conditions described by the ATCC.
Plasmids and reagents
The cDNAs for SUMO-1, SUMO-2, and SUMO-3 were subcloned into pCNDA3.0 with N-terminally translational fusion of His-tags. SUMO-1ΔGG (deletion of C-terminal diglycine motif), SUMO2ΔGG, and SUMO3ΔGG were generated by PCR mutagenesis. REGγ and Flag-REGγ from pCMV-Tag2B were subcloned into pCNDA5/FRT/TO. GFP-REGγ was kindly provided by Domenico Delia at the Fondazione IRCCS Istituto Nazionale Tumori, Milano, Italy. Point mutants of REGγ were created by PCR-based site-directed mutagenesis (Promega). Constructs expressing REGγ-SUMO-1 and REGγ-SUMO-2 fusion proteins were generated by two-step PCR strategy. GST-PIAS1 was generated by cloning a PCR product amplified from pXF3H-HA-PIAS1 23 into pGEX-4T-1.
Anti-HA and anti-GFP were purchased from Santa Cruz (sc-7392 and sc-9996, respectively). Anti-β-actin was acquired from Sigma (A5441) and anti-DDK (also known as anti-Flag) from Origene (TA5011). REGγ antibody was purchased from Invitrogen (38-3800) and monoclonal antibody against p21 was from BD Pharmingen (556431). Anti-Flag M2 Affinity Gel and Anti-HA Affinity Gel were from Sigma (F2426 and E6779, respectively). CHX was prepared as a 100-mg/ml stock solution dissolved in DMSO.
Yeast two-hybrid analysis
The full-length human REGγ cDNA fragment was inserted in frame into the Gal4 DNA-binding domain (DBD) vector pGBKT7 and a human liver cDNA library (CLONTECH Laboratories, Inc.) was fused with the transcriptional activation domain of GAL4. We performed a medium-stringency scale procedure to screen REGγ against human liver cDNA following the instructions of the manufacturer and characterized positive clones by sequence analysis.
GST pull-down assays
GST or GST-PIAS1 was purified as described 3 and pre-incubated with glutathione-sepharose beads. Then 35S-Met-labeled, in vitro-translated REGγ protein produced with a TNT kit (Promega) was incubated with the beads-bound GST derivatives at 4 °C for 2 h. After extensive washing of the protein-bound beads with a buffer containing 20 mM Tris-HCl, pH 7.4, 200 mM NaCl, and 0.5% Triton X-100, PIAS1-bound REGγ was separated by SDS-PAGE and visualized by autoradiography.
Immunoprecipitation
To test in vivo interactions between REGγ and PIAS1, 293T cells were transfected with Flag-REGγ (2 μg) and HA-PIAS1 (4 μg) or HA-PIAS1 (2 μg) along with GFP-REGγ (4 μg) in 6-cm dishes. Forty-eight hours after transfection, cells were harvested in ice-cold PBS and dispersed in lysis buffer (150 mM NaCl, 25 mM Tris, pH 6.8, 1% NP40). The lysates were centrifuged at 15 000× g for 20 min at 4 °C. The cleared supernatant was mixed with 5 μl of Anti-Flag M2 Affinity Gel or Anti-HA Affinity Gel and rotated for 4 h at 4 °C. The immunoprecipitates were washed three times with NTN buffer (25 mM Tris, pH 6.8, 150 mM NaCl, 0.1% NP40). The washed beads were boiled with SDS sample buffer and subjected to SDS-PAGE analysis.
Immunofluorescence
Wild-type and SENP-1−/− MEFs were transfected with REGγ derivative constructs, fixed, permeabilized, and immunoblotted as described previously 3. Alexa Fluor 568 goat anti-mouse and Alexa Fluor 488 goat anti-rabbit secondary antibodies (Invitrogen) were used in epifluorescence microscopy. All image files were digitally processed for presentation with Image-Pro Plus and Adobe Photoshop.
In vitro SUMOylation analysis
The in vitro SUMOylation assay was carried out with a kit from Biomol (UW8955) using purified recombinant REGγ (Biomol, PW9875) as a substrate. Briefly, purified REGγ protein was incubated in a SUMOylation buffer with SUMOylation E1 (Aos1/Uba2), E2 (Ubc9), and SUMO-1, -2, or -3 in the presence or absence of Mg2+-ATP at 30 °C for 1 h. The reaction products were separated by SDS-PAGE and analyzed by western blot using anti-SUMO antibodies.
In vivo SUMOylation assay
To generate and purify His-tagged SUMO conjugates in vivo, 293T cells were transfected with plasmids encoding GFP-REGγ, His-SUMO, or in combination with HA-PIAS1. Cells were harvested 36 h later and 5% of the cells were dispersed in lysis buffer (150 mM NaCl, 25 mM Tris-HCl, pH 6.8, 25 mM Tris-HCl, pH 8.0, 1% NP40) as input. The rest was lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, 0.01 M Tris-HCl, pH 8.0, 5 mM imidazole, 10 mM β-mercaptoethanol). Lysates in buffer A were mixed with 20 μl of Ni-NTA-agarose beads (Qiagen) and incubated at 4 °C overnight. The beads were then washed sequentially with buffer A, buffer B (8 M urea, 0.1 M Na2HPO4/NaH2PO4, pH 8.0, 0.01 M Tris-HCl, pH 8.0, 10 mM β-mercaptoethanol), buffer C (8 M urea, 0.1 M Na2HPO4/NaH2PO4, pH 6.3, 0.01 M Tris-HCl, pH 6.3, 10 mM β-mercaptoethanol) +0.2% Triton X-100 and buffer C alone for 5 min each round. Washed beads were then incubated with 40 μl elution buffer (200 mM imidazole, 0.15 M Tris-HCl, pH 6.7, 30% glycerol, 5% SDS, 0.72 M β-mercaptoethanol) at room temperature. The input and the eluates were analyzed by western blotting.
Data collection and statistical analysis
The intensity of the western blot results was analyzed by densitometry using Bio-Rad Quantity One 4.4.0 software and normalized to the band with the least intensity, which was arbitrarily set as 1. The results were expressed as the mean ± standard deviation (SD). Statistical analysis was performed using the two-tailed, paired Student's t test. A P value of less than 0.05 was considered statistically significant.
Acknowledgments
This work was supported by the National Institutes of Health (1R01CA131914). This manuscript was also funded in part by the National Natural Science Foundation of China (30811120435, 30870503, 81071657), the Science and Technology Commission of Shanghai Municipality (06DZ22923, 08PJ14047, 10JC1404200, 09ZZ41), and the National Basic Research Program (2009CB918402, 2011CB504200).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
GFP can not be SUMOylated.
REGγ can be SUMOylated by SUMO-1, -2, and -3.
PIAS1 is the major SUMO-E3 ligase for REGγ.
REGγ-SUMO fusion proteins are capable of cytoplasmic translocation.
Evolutionary view of the putative SUMOylation sites in REGγ.
REGγ SUMOylation occurs on multiple sites.
SUMO modification enhances REGγ stability.
Endogenous p21 turnover is increased in hyper-SUMOylated cell.
Predicted REGγ SUMOylation sites with SUMOplot
Predicted REGγ SUMOylation sites with SUMOsp2.0
References
- Chen X, Barton LF, Chi Y, Clurman BE, Roberts JM. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26:843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Amazit L, Long W, Lonard DM, Monaco JJ, O'Malley BW. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol Cell. 2007;26:831–842. doi: 10.1016/j.molcel.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Li X, Lonard D, Jung S, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin-and ATP-independent manner by the REG [gamma] proteasome. Cell. 2006;124:381–392. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Mao I, Liu J, Li X, Luo H. REGgamma, a proteasome activator and beyond. Cell Mol Life Sci. 2008;65:3971–3980. doi: 10.1007/s00018-008-8291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler M, Rollinger W, Mantovani-Endl L, et al. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol Cell Proteomics. 2006;5:2092. doi: 10.1074/mcp.M600118-MCP200. [DOI] [PubMed] [Google Scholar]
- Yu G, Zhao Y, He J, et al. Comparative analysis of REG{gamma} expression in mouse and human tissues. J Mol Cell Biol. 2010;2:192–198. doi: 10.1093/jmcb/mjq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET. SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem. 2009;284:8223–8227. doi: 10.1074/jbc.R800050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dasso M. SUMOylation and deSUMOylation at a glance. J Cell Sci. 2009;122:4249. doi: 10.1242/jcs.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Yurchenko V, Xue Z, Sadofsky M. SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol Cell Biol. 2006;26:1786. doi: 10.1128/MCB.26.5.1786-1794.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 2008;27:852–864. doi: 10.1038/emboj.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15:27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Miyauchi Y, Yogosawa S, Honda R, Nishida T, Yasuda H. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J Biol Chem. 2002;277:50131–50136. doi: 10.1074/jbc.M208319200. [DOI] [PubMed] [Google Scholar]
- Tan JA, Song J, Chen Y, Durrin LK. Phosphorylation-dependent interaction of SATB1 and PIAS1 directs SUMO-regulated caspase cleavage of SATB1. Mol Cell Biol. 2010;30:2823–2836. doi: 10.1128/MCB.01603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Lin D, Huang Y, Jeng J, et al. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of SUMOylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Figueroa-Romero C, Iniguez-Lluhi JA, Stadler J, et al. SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J. 2009;23:3917–3927. doi: 10.1096/fj.09-136630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD, Irvin BJ, Nucifora G, Luce KS, Hiebert SW. Small ubiquitin-like modifier conjugation regulates nuclear export of TEL, a putative tumor suppressor. Proc Natl Acad Sci USA. 2003;100:3257–3262. doi: 10.1073/pnas.0637114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584–595. doi: 10.1016/j.cell.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Melchior F, Feng X, Lin X. Regulation of Smad4 sumoylation and transforming growth factor-beta signaling by protein inhibitor of activated STAT1. J Biol Chem. 2004;279:22857–22865. doi: 10.1074/jbc.M401554200. [DOI] [PubMed] [Google Scholar]
- Verger A, Perdomo J, Crossley M. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP can not be SUMOylated.
REGγ can be SUMOylated by SUMO-1, -2, and -3.
PIAS1 is the major SUMO-E3 ligase for REGγ.
REGγ-SUMO fusion proteins are capable of cytoplasmic translocation.
Evolutionary view of the putative SUMOylation sites in REGγ.
REGγ SUMOylation occurs on multiple sites.
SUMO modification enhances REGγ stability.
Endogenous p21 turnover is increased in hyper-SUMOylated cell.
Predicted REGγ SUMOylation sites with SUMOplot
Predicted REGγ SUMOylation sites with SUMOsp2.0