Abstract
To better understand the role of exosomes in the trabecular meshwork (TM), the site of intraocular pressure control, the exosome proteome from primary cultures of human TM cell monolayers was analyzed. Exosomes were purified from urine and conditioned media from primary cultures of human TM cell monolayers and subjected to two dimensional HPLC separation and MS/MS analyses using the MudPIT strategy. Spectra were searched against a human protein database using Sequest. Protein profiles were compared to each other and the Exocarta database and the presence of specific protein markers confirmed by Western blot analyses of exosomes from aqueous humor and human TM cell strains (n=5) that were untreated, or exposed to dexamethasone and/or ionomycin. TM cell exosomes contained 108 of the 143 most represented exosome proteins in ExoCarta, including previously characterized markers such as membrane organizing and tetraspanin proteins. Several cell-specific proteins in TM exosomes were identified including myocilin, emilin-1 and neuropilin-1. All TM exosome proteins had flotation densities on sucrose gradients and release responses to ionomycin typical for exosomes. Taken together, TM exosomes have a characteristic exosome protein profile plus contain unique proteins, including the glaucoma-causing protein, myocilin; suggesting a role for exosomes in the control of intraocular pressure.
Keywords: Aqueous humor, Endosome, Glaucoma, Intraocular Pressure, Ocular hypertension, Myocilin, Multivesicular body, Urine
Introduction
The prevention of vision loss due to death of retinal ganglion cells in glaucoma, a leading cause of irreversible blindness in the World, is dependent upon effective intraocular pressure (IOP) control over time [1]. Elevated IOP (ocular hypertension) in glaucoma is a function of pathology in the conventional/trabecular outflow pathway, the primary efflux route for aqueous humor from the eye [2–4]. While the mechanisms that underlie the regulation of IOP in the trabecular meshwork (TM) are presently unknown, the linkage of glaucoma/ocular hypertension to several genetic loci provides critical molecular candidates for evaluation [5].
Mutations in the glaucoma locus GLC1A results in amino acid alterations in its protein product, myocilin (MYOC); causing glaucoma in every population studied to date [5–7]. Ocular hypertension caused by a dysfunction in TM drainage of aqueous humor appears to be a significant component in MYOC-associated glaucoma [8, 9]. While examining the structural, biochemical and cellular properties of MYOC in TM cells, we observed that MYOC is not secreted in a traditional manner, but exits in association with nanovesicles, called exosomes [10, 11]. Further examination showed that release of MYOC-associated exosomes from TM cells is enhanced in a calcium-dependent fashion, similar to that for other exosome releasing cell types [11] and consistent with the immediate stimulated release of MYOC recently observed by others [12]. Significantly, we also observed that MYOC-associated exosomes are a prominent component of human aqueous humor, suggesting a physiological importance of exosomes in the regulation of trabecular drainage and thus IOP [13].
In addition to aqueous humor, exosomes have been found recently in a variety of other biological fluids including urine, amniotic fluid, blood and breast milk; stimulating interest in exosome biology due to their diagnostic and therapeutic potential [14–16]. The genesis of exosomes results from the inward budding and scission of vesicles from endosomal membranes (also known as intraluminal vesicles) containing cellular proteins. Intraluminal vesicles (ILV) accumulate within the multivesiclular body (MVB) until the fusion of the MVB with the plasma membrane elicits their extracellular release. During endosomal maturation, ILVs sequester proteins that are important for their biogenesis, structure and trafficking. Moreover, it appears that ILV protein content is cell-type specific, reflecting the biological role of the parent cell. Hence depending upon the cell type, exosomes mediate antigen presentation, cell-cell communication, RNA and protein transfer and non-classical protein secretion [for reviews, see [17–19]].
To better understand the role of exosomes in TM biology, we purified exosomes from conditioned media of primary cultures of human TM cell monolayers, examined and then validated their protein expression pattern. Results were compared to exosomes purified from two human bodily fluids, aqueous humor and urine. Using complementary biochemical techniques, we observed that exosomes purified from TM cells and aqueous humor contained many proteins common to exosomes from other cell types, including the vast majority of those most frequently reported. We also observed that TM exosomes house unique proteins, including the glaucoma-associated protein, myocilin.
Materials and Methods
Human trabecular meshwork cell cultures
Human TM cell strains were isolated and characterized by techniques developed in our laboratory as previously described [20]. Cells were seeded onto T75 culture flasks and maintained at confluence to differentiate for at least 14 days prior to treatment in low glucose DMEM (Invitrogen, CA) containing penicillin (100 units/mL), streptomycin (100 mg/mL) and glutamine (0.29 mg/mL). Media was supplemented with 10% fetal bovine serum (Gemini Bio-Products, CA) for the first week and then lowered to 1% for the second week to facilitate differentiation. For exosome collection after differentiation, cells were fed with 10 ml of media supplemented with 0.1% exosome-free FBS as described previously [11] for all experiments. Blood-borne exosomes were removed from FBS by centrifugation (100,000g for 1 hour) prior to supplementing media
Human samples
Urine was collected in the Vanderbilt General Clinical Research Center from healthy subjects after each provided written informed consent. All samples were collected during vehicle (control) treatment during low dietary sodium intake (20 mmol Na+/day). Approximately 200 mL of urine from each subject was collected into a protease cocktail, then stored at −80ºC until preparation for exosome analysis as described below (<7 months). Aqueous humor (~200 μl/eye) was collected from human cadaveric eyes (donor age= 60–100 years) free of known ocular disease (except cataract) within 35 hours of death and frozen at −80oC until analysis (<3 months). Samples were pooled from both eyes of 10 individual donors.
Exosome fractionation
Aqueous humor from cadaveric human eyes, urine collected from human patients or media collected from human TM cells with and without treatment were centrifuged for 1 hour at 10,000g at 4ºC to remove cell debris. Exosomes were then isolated by centrifuging at 100,000g (using SW41 rotor) for 1 hour at 4ºC. Pellets were resuspended in cold PBS (10 ml) and centrifuged again at 100,000g to minimize sticking and trapping of non-exosomal materials. Pellets were solubilized in Laemmli buffer to yield a 100X concentration of original sample volume. For cultured cells, samples (100 μl) were taken from conditioned media before and after each centrifugation step and combined with equal volume of 2X Laemmli buffer to monitor purification process. All samples were boiled and stored at −20ºC.
Sample Preparation
Lyophilized exosomes were suspended in 20 μl PBS, 3.0 μl DTT and 7.5 μl 4× lithium dodecylsulfate (LDS) buffer (10% glycerol, 141 mM Tris base, 106 mM Tris HCl, 2% LDS, 0.51 mM EDTA, 0.22 mM SERVA® Blue G250, 0.175 mM Phenol Red, pH 8.5). The samples were heated for 10 min at 85°C and each sample was loaded on two lanes on a NuPAGE Novex 10% Bis Tris gel. The gels were run at a constant 180 volts for 10 minutes and the dye front ran approximately 2.5 cm into the gel. The gel was fixed for 10 minutes (50% methanol 10% acetic acid and 40% water), stained with Colloidal Coomassie Blue (Invitrogen) for three hours and the background stain removed with deionized water overnight. The entire stained protein regions were excised, chopped into 1 mm cubes, and placed in 1.5 ml Eppendorf tubes containing 150 μl of 100 mM ammonium bicarbonate. Samples were reduced with 10 μl 45 mM DTT for 20 min at 55°C and alkylated with 10 μl 100 mM iodoacetamide for 20 min at room temperature in the dark. Samples were destained with 100 μL 50% acetonitrile (ACN) and 50 mM ammonium bicarbonate. The gel pieces were then dehydrated with 100% acetonitrile, air dried for 5 min, and digested with trypsin (10 ng/μl and a total of 220 ng) in a ratio of 1:50 overnight at 37°C. The peptides were extracted with two rounds of 60% acetonitrile and 0.1% trifluoroacetic acid and were then lyophilized to dryness. They were resolubilized in 40 μL of 0.1% formic acid and analyzed by LC-MS/MS.
LC-MS/MS Analysis
The resulting peptide mixtures were analyzed via MudPIT [21]. Briefly, peptides were loaded with a pressure cell (New Objective) onto a biphasic pre-column using an Upchurch M-520 filter union. This 150-μm fused silica microcapillary column was packed with 4 cm of 5-μm C18 reverse-phase resin (Jupiter, Phenomenex) followed by 4 cm of strong cation-exchange resin (Luna SCX, Phenomenex). Once loaded, it was placed in-line with a 100 μm × 20 cm, C18 packed emitter tip column (Jupiter C18, 3 μm, 300 Å, Phenomonex) coupled to an LTQ ion trap mass spectrometer equipped with an Eksigent NanoLC-AS1 Autosampler 2.08, an Eksigent NanoLC-1D plus HPLC pump, and Nanospray source. Multidimensional separations were accomplished using 5 μl pulses of ammonium acetate in 0.1% formic acid (25, 50, 75, 100, 150, 200, 250, 300, 500, 750, 1000 mM) delivered by an autosampler. Each salt pulse was followed by a 115 min reversed phase gradient beginning with 0.1% formic acid in water and increasing to 40% ACN with 0.1% formic acid.
LTQ Parameters
Centroided MS/MS scans were acquired using an isolation width of 2 m/z, an activation time of 30 ms, an activation Q of 0.250 and 30% normalized collision energy using 1 microscan and max injection 150 ms for each MS/MS scan. The tune parameters were as follows: spray voltage of 2.43 kV, a capillary temperature of 200ºC. The MS/MS spectra of the peptides were collected using a data-dependent scanning protocol in which one full MS spectrum was followed by five sequential MS/MS spectra. Dynamic exclusion was enabled with a repeat count of 1, a list length of 150 masses or 1 min, and an exclusion width of 3.5 m/z units.
Data Analysis
The custom ScanSifter algorithm read tandem mass spectra stored as centroided peak lists from. RAW files and transcoded them to DTA files. Spectra that contained fewer than 25 peaks or that had less than 2e1 measured total ion current were not converted to DTA files. DTA files for singly charged precursor ions were created if 90% of the total ion current occurred below the precursor ion m/z ratio, and all other spectra were processed to form both doubly and triply charged DTA files. DTA files were searched against the UniprotKB (August, 2010 release) database using the TurboSEQUEST v.27 (rev. 12) algorithm (Thermo Electron, San Jose, CA). The searches were performed allowing for the following differential modifications: +57 on cysteine (for carboxyamidomethylation from iodoacetamide) and +16 on methionine (oxidation). The database was concatenated with the reverse sequences of all proteins in the database to allow determination of false-positive rates. Sequest search result files were imported into ProteoIQ software (NuSep, Inc) for comparative proteomics analysis. Results were filtered using the following criteria: Sequest XCorr values: 1.5 for 1+ ions, 2.5 for 2+ ions, and 3.0 for ≥ 3+ ions, a minimum of two peptides per protein, an overall false discovery rate maximum of 5.0, and an average of two spectral counts per protein in each urine or TM sample.
Western blotting
Proteins in exosome preparations or whole cell lysates were separated by 10% SDS/PAGE and transferred electrophoretically to nitrocellulose. Membranes were blocked in Tris-buffered saline containing 2% tween (TBS-T) and 5% non-fat, dry milk for one half hour. Immunoglobulins that specifically recognize candidate proteins were added to blocking buffer. Polyclonal rabbit anti- myocilin IgGs (1:3000) were produced in our laboratory [22], while the rest were purchased commercially. Antibodies against annexin-A1 (1:1000), flotillin-1 (1:1000) and GAPDH (1:1500) were obtained from Cell Signaling. Antibodies that recognize Annexin-5 (1:3000), CD9 (1:100), neuropilin-1 (1:1000), Syntenin-1(1:1000) and CD81 (1:500) were from Abcam. Antibodies specific for Cyclophilin B (1:2500) were from Alexis, Emilin-1 (1:1000) was from Santa Cruz and Annexin 2 (1:2500) from BD transduction laboratories. After overnight incubations, membranes were washed in TBS-T (4 X 15 minutes). Horseradish peroxidase conjugated secondary antibodies (1:5000, Santa Cruz, CA) were incubated in TBS-T with 5% nonfat dry milk for 1 hour. Membranes were then washed in TBS-T (4 X 15 minutes), incubated with Hyglo chemiluminescence reagent (Denville, NJ) and exposed to X-Ray film (Genesee, CA) to visualize protein/antibody complexes. Densitometry of protein bands was performed following image capture using a bioimaging system (Syngene, Frederick, MD) and processed with GeneTools (Syngene). Only captured protein bands in the linear range of the X-ray film were analyzed.
Sucrose gradients
For analysis of equilibration density of marker proteins, purified exosome fractions were resuspended in 1ml 2.6M sucrose in 20mM Tris-HCl pH 7.2. Linear gradients ranging from 2 to 0.25 M Sucrose in 20mM Tris-HCl pH 7.2 were layered on top of the exosome preparation. Gradients were centrifuged at 100,000g for 15 hours. Equal fractions (~1ml) were taken sequentially from the top of the gradient. Sucrose concentrations of fractions were analyzed by refractometery and the remaining fraction samples were solubilized in 2X Laemmli buffer and stored at 7minus;20oC.
Treatments
Cells were exposed to dexamethasone (Sigma, MO) in exosome-free media at a final concentration of 100 nM or remained untreated as control for 5 days. Media was changed after 48 hours and collected from the final 72 hrs of treatment. For experiments testing effect of a calcium ionophore (Sigma), ionomycin was added to conditioned media at a final concentration of 100 μM for final 3 hours before collection and at the end of a 5 day dexamethasone supplementation. In a previous study, we found that preconditioning cell monolayers with dexamethasone resulted in more consistent ionomycin responses [11]. Conditioned media was collected (10 ml) and exosomes were isolated.
Results
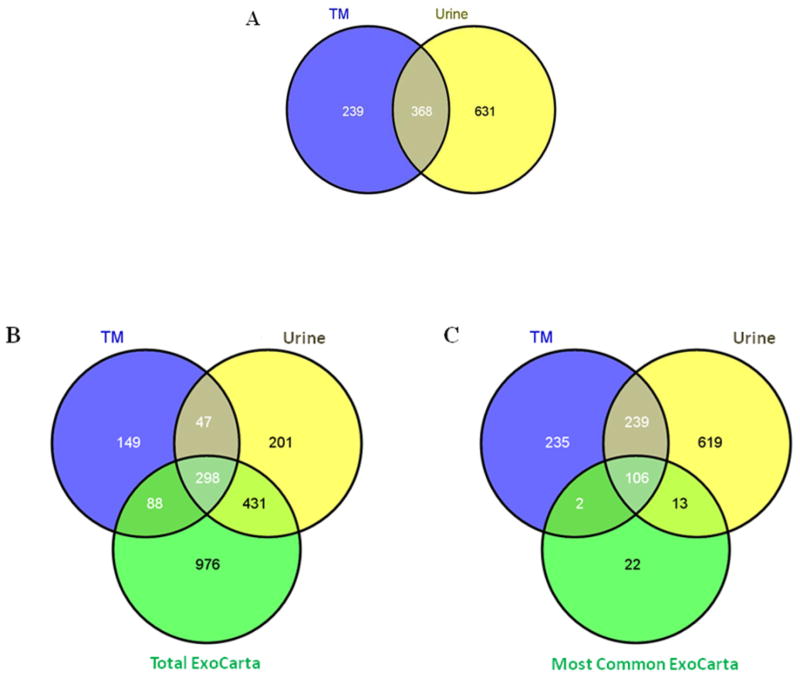
In order to determine whether exosomes produced by human TM cells contained proteins typical of exosomes [23], we prepared an exosome fraction from conditioned media of mature monolayers of primary cultures of human trabecular meshwork cells using differential centrifugation steps and subjected purified samples to LC-MS/MS analyses. We tested cell strains obtained from two different human donors, with one of the cell strains tested at two different passages, giving three samples that were analyzed and compared to exosomes that were purified from human urine as a control using identical methods. We identified 607 proteins that were present in exosomes prepared from TM cells, compared to 999 proteins identified in urine exosomes. Direct comparison of UniProt accession numbers revealed that 368 of these proteins were common between the two different types of samples. When compared to an exosome protein database [ExoCarta-[23]], remarkably we observed that 298 proteins were observed in all three datasets and that TM cell preparations contained 108 of the most frequently observed 143 proteins found in exosomes purified from other cell types and bodily fluids (figure 1). Note that the total protein count is slightly lower than reported above due to redundancy in Gene Symbols, e.g. HLA-A isoforms) that is removed in the software used for Venn diagram generation.
Figure 1. Venn Diagrams showing protein profile overlap between TM and urine exosomes and ExoCarta database.
Panel A compares TM (blue) and urine (yellow) proteins identified by Sequest/ProteoIQ analysis using UniProt accession numbers. Panel B compares the TM (blue) and urine (yellow) proteins to those in the entire ExoCarta database (green) using Gene Symbols. Panel C compares the 143 most commonly observed exosomal proteins in ExoCarta database with expression profile observed for TM and urine exosome proteome. Total TM and urine proteins that met the search criteria were compared to the ExoCarta database using the Venn diagram generator found at: http://bioinfogp.cnb.csic.es/tools/venny/index.html
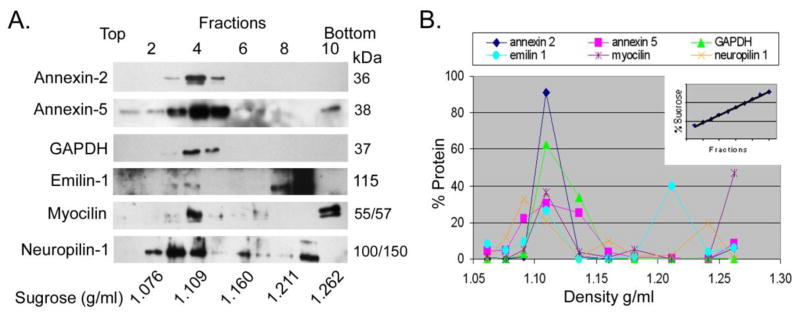
The fifty most abundant proteins identified in TM cell preparations are shown in Table 1, are compared to proteins found in urine exosomes and include commonly identified exosome proteins from other cells such as those in the lipid organizer class (annexins-1,2,5,6), tetraspanin (syntenin-1) plus other commonly observed exosomal markers such as GAPDH, β-actin, moesin, heat shock 70kDa protein-8, enolase and pyruvate kinase. All proteins found in TM exosomes are shown in Supplemental Table 1. Included in this list are additional common exosome markers including; CD-9, CD-63, CD81, cofilin-1, 14-3-3 proteins epsilon and zeta, phosphoglycerate kinase, lactadherin, peptidyl-prolyl cis-trans isomerase A, programmed cell death 6-interacting protein, elongation factor 1 alpha 1, and fructose-bisphosphate aldolase A. We also identified proteins that appeared abundantly and were TM cell specific, such as emilin-1 and myocilin. To validate protein candidates obtained from proteomic studies, we again prepared an exosome fraction from conditioned media of mature TM cell monolayers and this time subjected the samples to sedimentation gradient densitometry. We then collected gradient fractions and separated proteins using SDS-PAGE followed by western blot analysis using antibodies specific for several of the proteins identified by MS. Figure 2 shows that annexin-2, annexin-5, GAPDH, emilin-1, myocilin and neuropilin-1 all equilibrate at a density about ~1.10 g/ml, characteristic of exosomal proteins [19].
Table 1.
50 most abundant proteins in trabecular meshwork exosomes
| Gene Symbol | uniProt Id | Sequence Name | TM Normalized SC | Urine Normalized SC |
|---|---|---|---|---|
| FN1 | P02751 | Fibronectin | 2447.53 | 31.75 |
| HSPG2 | P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein | 420.76 | 56.74 |
| COL12A1 | Q99715 | Collagen alpha-1(XII) chain | 308.47 | 1.69 |
| MFGE8 | Q08431 | Lactadherin | 244.3 | 4.72 |
| A2M | P01023 | Alpha-2-macroglobulin | 267.99 | 19.23 |
| ANXA2 | P07355 | Annexin A2 | 103.57 | 80.76 |
| COL1A2 | P08123 | Collagen alpha-2(I) chain | 96.4 | 1.7 |
| EMILIN1 | Q9Y6C2 | EMILIN-1 | 98.39 | 0 |
| ANXA6 | P08133 | Annexin A6 | 78.22 | 43.23 |
| PKM2 | P14618 | Pyruvate kinase isozymes M1/M2 | 72.22 | 90.29 |
| MVP | Q14764 | Major vault protein | 77.37 | 11.93 |
| ALB | P02768 | Serum albumin | 77.78 | 107.67 |
| LGALS3BP | Q08380 | Galectin-3-binding protein | 79 | 48.18 |
| ANXA5 | P08758 | Annexin A5 | 70.18 | 53.52 |
| FLNA | P21333 | Filamin-A | 68.79 | 1.16 |
| ANXA1 | P04083 | Annexin A1 | 63.82 | 75.94 |
| EDIL3 | O43854 | EGF-like repeat and discoidin I-like domain- containing protein 3 | 63.97 | 0.33 |
| MYOC | Q99972 | Myocilin | 67.6 | 0.33 |
| MYH9 | P35579 | Myosin-9 | 58.92 | 38 |
| GAPDH | P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 54.58 | 110.8 |
| MYOF | Q9NZM1 | Myoferlin | 51.48 | 18.71 |
| THBS1 | P07996 | Thrombospondin-1 | 56.9 | 11.96 |
| LAMA4 | Q16363 | Laminin subunit alpha-4 | 56.17 | 0 |
| NT5E | P21589 | 5′-nucleotidase | 50.81 | 1.98 |
| CLTC | Q00610 | Clathrin heavy chain 1 | 47.1 | 30.14 |
| ENO1 | P06733 | Alpha-enolase | 45.46 | 178.5 |
| LTBP2 | Q14767 | Latent-transforming growth factor beta-binding protein 2 | 55.15 | 1.34 |
| GSN | P06396 | Gelsolin | 51.28 | 35.33 |
| DYNC1H1 | Q14204 | Cytoplasmic dynein 1 heavy chain 1 | 38.79 | 24.07 |
| ACTB | P60709 | Actin, cytoplasmic 1 | 40.53 | 199.77 |
| ACTN1 | P12814 | Alpha-actinin-1 | 38.62 | 22.95 |
| TLN1 | Q9Y490 | Talin-1 | 38.98 | 16.51 |
| LAMB1 | P07942 | Laminin subunit beta-1 | 41.24 | 1 |
| HRNR | Q86YZ3 | Hornerin | 40.49 | 6.04 |
| TGFBI | Q15582 | Transforming growth factor-beta-induced protein ig-h3 | 39.46 | 0.34 |
| ACTN4 | O43707 | Alpha-actinin-4 | 33.22 | 40.47 |
| PCOLCE | Q15113 | Procollagen C-endopeptidase enhancer 1 | 42.53 | 0 |
| SDCBP | O00560 | Syntenin-1 | 34.83 | 194.57 |
| MYO1C | O00159 | Myosin-Ic | 31.36 | 100.42 |
| C3 | P01024 | Complement C3 | 31.51 | 60.21 |
| PZP | P20742 | Pregnancy zone protein | 38.39 | 5.28 |
| MSN | P26038 | Moesin | 30.81 | 192.15 |
| ACTC1 | P68032 | Actin, alpha cardiac muscle 1 | 30.31 | 101.39 |
| LAMC1 | P11047 | Laminin subunit gamma-1 | 34.16 | 4.18 |
| TUBB | P07437 | Tubulin beta chain | 29.97 | 32.3 |
| VIM | P08670 | Vimentin | 31.28 | 0 |
| DPP4 | P27487 | Dipeptidyl peptidase 4 | 27.4 | 293.23 |
| HSPA8 | P11142 | Heat shock cognate 71 kDa protein | 30.03 | 92.71 |
| ACTA1 | P68133 | Actin, alpha skeletal muscle | 27.68 | 101.39 |
| STOM | P27105 | Erythrocyte band 7 integral membrane protein | 27.4 | 45.19 |
Figure 2. Analysis of exosomal marker proteins in extracellular membranes prepared from conditioned media taken from human trabecular meshwork cell monolayers.
Membranes isolated by differential centrifugation were floated into linear sucrose gradients, fractions were collected and proteins were analyzed by SDS-PAGE/Western blotting (panel A). Shown are representative blots from one TM cell strain of three that were tested. Protein content of fractions was determined by probing with antibodies specific to Annexin-2, Annexin-5, GAPDH, Emilin-1, Myocilin and Neuropilin-1. Relative protein distributions across fractions were quantified by densitometry and graphed as percent of total abundance for each protein across gradient (panel B). Linearity of gradients was verified by refractometry (inset, panel B).
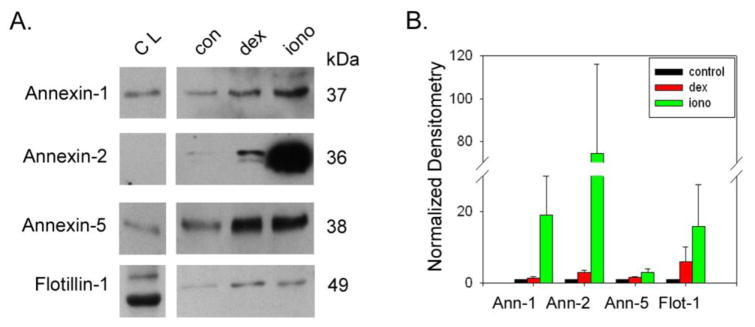
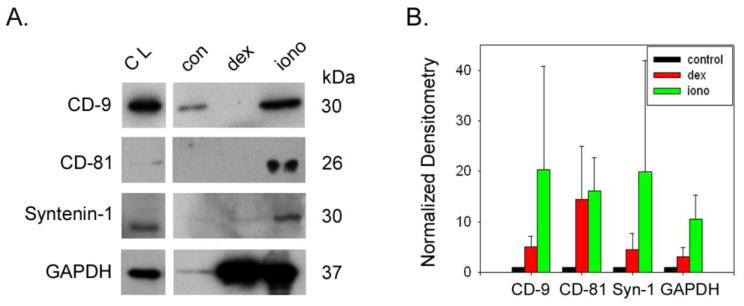
Previously we showed that myocilin release from TM cells and appearance in the exosomal fraction is stimulated by dexamethasone in the long term and ionomycin in the short term [11]. In order to test whether the appearance of the newly identified proteins also are affected by these treatments, we subjected TM cells to dexamethasone (100 nM) for 5 days, collecting conditioned media from the last 48 hours of treatment or dexamethsone for 5 days plus ionomycin (100 μM) for the last 3 hours prior to collection of conditioned media. In response to dexamethasone alone, results show a modest increase in the lipid organizing proteins (Figure 3: annexins and flotilin-1), GAPDH (figure 4) and neuropilin-1 (Figure 5). In contrast, the tetraspanins and myocilin levels were all elevated 5–20 fold following dexamethasone treatment. With the exception of annexin-5 and emilin-1, the entire panel of exosome proteins robustly increased (10–75 fold) in the exosome fraction of conditioned media upon stimulation with ionomycin, similar to exosomal proteins from other cell types [24].
Figure 3. Effect of dexamethasone or dexamethasone plus ionomycin on release of exosomes from trabecular meshwork cell monolayers.
Exosome release was monitored by abundance of exosomal marker proteins in the lipid organizing class (annexin-1, -2, -5 and flotilin-1) in exosomal fraction by SDS-PAGE/Western blotting (panel A). Marker protein content was compared to samples of total cell lysate (CL) prepared from human trabecular meshwork cell monolayers. Effects of drug treatments on protein abundance in exosome fraction were determined using densitometry (panel B). Thus, dexamethasone (dex; 100 nM for 5 days) and dex plus ionomycin (iono; 100 μM for 3 hours) treatments were compared to untreated controls (con). Shown are combined data from a total of 5 independent experiments testing 5 different TM cell strains.
Figure 4. Effect of treatments on abundance of marker proteins in exosomes prepared from trabecular meshwork cell monolayers.
Exosome release was monitored by abundance of exosomal marker proteins in the tetraspanin category (CD-9, CD-81 and syntenin-1) plus GAPDH in exosomal fraction by SDS-PAGE/Western blotting (panel A). Marker protein content was also monitored in total cell lysates (CL) prepared from human trabecular meshwork cell monolayers. Effects of drug treatments on protein abundance in exosome fraction were determined using densitometry (panel B). Thus, dexamethasone (iono; 100 nM for 5 days) and dex plus ionomycin (iono; 100 μM for 3 hours) treatments were compared to untreated controls (con). Shown are combined data from a total of 3–5 independent experiments testing 3–5 different TM cell strains.
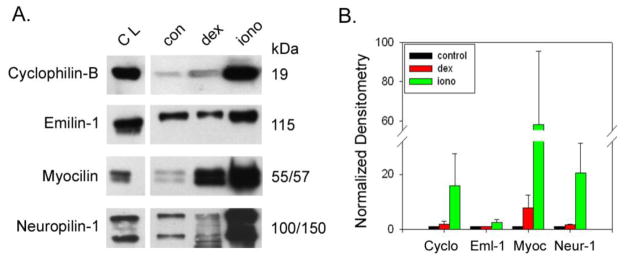
Figure 5. Effect of treatments on appearance of TM-specific markers in exosomes prepared from trabecular meshwork cell monolayers.
In response to treatment with dexamethasone (dex) or dexamethasone plus ionomycin (iono), abundance of TM-specific proteins (cyclophilin-B, emilin-1, myocilin and neuropilin-1) in exosomes was monitored by SDS-PAGE/Western blotting (panel A). Marker protein content was also monitored in total cell lysates (CL) prepared from human trabecular meshwork cell monolayers. Effects of drug treatments on protein abundance in exosome fraction were determined using densitometry (panel B). Thus, dexamethasone (100 nM for 5 days) and dex plus ionomycin (100 μM for 3 hours) treatments were compared to untreated controls (con). Shown are combined data from a total of 5 independent experiments testing 5 different TM cell strains.
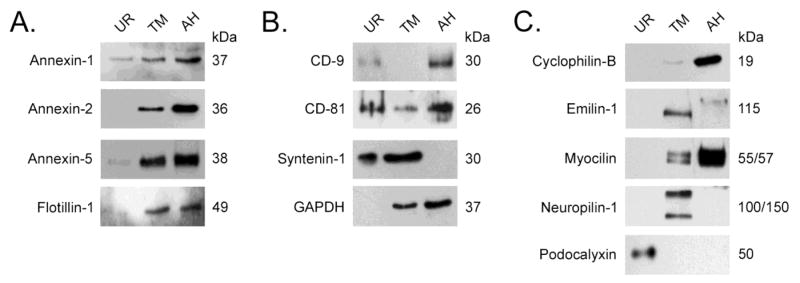
To compare our findings with cultured human ocular cells to human biological fluids, we next tested the expression of candidate proteins in exosome fraction prepared from TM cells to exosomes prepared from aqueous humor and urine. In figure 6 results show that all candidate proteins tested appeared in both TM conditioned media and aqueous humor except for emilin-1 and syntenin-1. We did see evidence for neuropilin-1 in aqueous humor when blots were exposed to film longer (data not shown). In urine, we detected all common exosomal proteins that were screened. Annexin-2 and GAPDH were only seen in urine samples concentrated 10X over data shown in figure 6 (data not shown). The TM specific proteins, those not found in common between TM and urine with proteomic analysis, were also not found in urine, even in 10X concentrated samples. As a positive control for urine exosomes, we probed blots with antibodies that specifically recognize podocalyxin and observed that this podocyte-specific protein was not present in either TM or aqueous humor samples, but was found abundantly in urine.
Figure 6. Comparison of exosomal proteins present in human TM exosomes to those prepared from human urine and aqueous humor.
Exosomes from TM conditioned media, aqueous humor (AH) and urine (UR) were prepared by differential centrifugation and analyzed by SDS-PAGE/Western blotting. Panel A shows comparison of lipid organizing protein content (annexin-1, -2 and -5 and flotillin-1); panel B shows comparison of tetraspanin proteins (CD-9, CD-81 and syntenin-1) and GAPDH content; while panel C shows TM-specific (cyclophilin-B, emilin-1, myocilin, neuropilin-1) versus urine-specific (podocalyxin) protein profile. Data shown are representative examples of exosomal samples that were prepared from 5 different TM cell strains. Urine samples and aqueous humor samples were pooled from several different donors before exosomal preparation and analysis. During preparation of samples, purified exosomes from each cell strain/bodily fluid were concentrated to 100X original volume before loading onto gels.
Discussions
Using a proteomic approach, the present study defines for the first time the protein profile of exosomes from TM cells. We observed that exosomes produced by TM cells have an expression pattern typical of exosomes prepared from other sources. According to the ExoCarta database, exosomes from TM cells contain 108 of the 143 most represented exosomal proteins produced by other cell types or found in bodily fluids such as blood or urine [23]. Of these, eight exosomal marker proteins were confirmed using western blot analysis. A number of exosomal proteins present in TM samples, but not urine, were also found by mass spectrometry and confirmed using western blot, including cyclophilin-B, emilin-1, neuropilin-1 and the glaucoma-causing protein, myocilin.
While treatment with dexamethasone alone had relatively little effect on the majority of the exosomal marker proteins in TM, in every case the addition of the calcium ionophore triggered increased levels of all marker proteins. The magnitude of increase varied considerably for a few from 2-fold to 80-fold, but the majority of proteins increased 10–20 fold. This robust and immediate increase in exosomal proteins in the extracellular space is consistent with exosomes that are contained in the multivesticular body awaiting a calcium-based signal for fusion with the plasma membrane and subsequent release of their cargo of exosomes.
The total number of spectral counts (an indirect measure of protein abundance [25]) and consequently the total number of proteins identified was significantly lower in one of the TM samples. This was most likely due to lower total protein harvested for analysis. Despite this variation, 69% of TM proteins were observed in all three exosome samples and 97.4% of TM proteins were observed in two of three samples, providing confidence in our reported proteome profile.
A primary function of the TM is to clear cell debris from the aqueous humor before reaching the downstream resistance generating region of the conventional outflow pathway. As such, TM cells, like macrophages must endocytose, process and dispose of cellular components, particularly lipids. The use of the endocytic pathway and MVB by TM cells to transform large pieces of cellular material into manageable exosomal vesicles seems a practical and efficient way to maintain homeostasis in the region of the eye that controls IOP. Consistent with this idea, others have suggested that exosomal release from cells located in drainage systems such as gut and kidney may be used for similar disposal functions (in addition to signaling function) [17]. This is the first study to our knowledge to suggest such a mechanism for the TM.
Myocilin was a prominent component of aqueous humor and TM exosomes but not detectible in urine exosomes. Compared to the cellular responsibilities of non-ocular tissues that express robust amounts of myocilin (i.e.: skeletal muscle and heart), the filtering function and associated lipid processing appears uniquely important for the conventional outflow tissues. Such a distinction may explain why mutations in myocilin, a ubiquitously expressed protein, result in focal disease; limited to the development of ocular hypertension and glaucoma [8, 26]. For instance, myocilin mutations may interfere with its role in the exosomal pathway, limiting the ability of TM cells to effectively process cell debris, resulting in increased outflow resistance and elevated IOP. The mutation-specific patient phenotypes that associate with significantly different age of POAG diagnosis and maximum IOP [8] may relate to the degree with which a specific mutation interferes with myocilin function in the exosome pathway and the capacity of the outflow system over time.
In addition to the presence of myocilin in TM exosomes, we identified other proteins that were richly present in TM but not urine exosomes or in the ExoCarta database, including emilin-1 and neuropilin-1. Elastin microfibril interface-located protein-1 (EMILIN-1) is an extracellular matrix protein, localized at the interface between microfibrils and elastin and secreted by endothelial cells. Deficiency in emilin-1 results in functional defects in the vascular system [27, 28]. Interestingly, one hallmark of ocular hypertension in glaucoma is accumulation of elastin plaque-like material in the TM, near Schlemm’s canal [29]. Neuropilin-1 (NRP-1) is present on the cell surface of endothelial cells, or as a soluble truncated variant. Membrane NRP-1 is proposed to enhance angiogenesis by promoting the formation of a VEGF-heparin sulfate signaling complex, whereas the soluble NRP-1 is thought to act as an antagonist of signaling complex formation [30]. Both VEGF and heparin sulfate are present in the conventional outflow pathway, likely functioning in mediating tissue homeostasis with respect to ECM turnover; a function integral to IOP regulation [31]. Interestingly, while myocilin was found abundantly in aqueous humor exosomes, neither emilin-1 nor neuropilin-1 was present. Since TM cells contribute little to exosome content in aqueous humor, it is likely that emilin-1 and neuropilin-1 are TM cell-specific.
Conclusions
This is the first study to define the proteome of exosomes produced from TM cells and for that matter any ocular cell type/tissue to our knowledge. Significantly, we validate proteins found in cultured TM cells in human samples of aqueous humor, a bodily fluid that may be used in the future as a source of ocular disease biomarkers. Moreover, the prominence of myocilin, a protein that when mutated causes glaucoma, in exosomes takes a step forward to resolve the controversy as to how myocilin exits TM cells and enters the extracellular space [6]. The present study provides strong evidence that myocilin exits cells via exosomes and not using traditional secretory machinery. Since the trabecular meshwork is the diseased tissue responsible for ocular hypertension and that myocilin is found uniquely and in abundance on exosomes, we hypothesize that a functional exosomal pathway is critical for proper intraocular pressure regulation over time. Future research directed at understanding how the exosomal pathway functions in IOP control will likely reveal novel therapeutic targets for the treatment of ocular hypertension; an endpoint that when controlled effectively halts vision loss in those with glaucoma.
Supplementary Material
Acknowledgments
Grant support from NIH: HL100016 (KLS) and EY12797 (WDS). The authors thank Zhen Wang and Salisha Hill as well as from the Vanderbilt Mass Spectrometry Research Center for technical support. Support for sample collection of the clinical samples was provided through the Vanderbilt CTSA (1 UL1 RR024975 from NCRR/NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.AGIS-Investigators. The Advanced Glaucoma Intervention Study (AGIS): The relationship between control of intraocular pressure and visual field deterioration. Am J Ophthalmol. 2000;130:429–440. doi: 10.1016/s0002-9394(00)00538-9. [DOI] [PubMed] [Google Scholar]
- 2.Grant WM. Clinical tonography. Trans Am Acad Ophthalmol Otolaryngol. 1951;55:774–81. [PubMed] [Google Scholar]
- 3.Grant WM. Open-angle glaucoma. AMA Arch Ophthalmol. 1953;50:125–6. [PubMed] [Google Scholar]
- 4.Grant WM. Experimental aqueous perfusion in enucleated human eyes. Arch Ophthalmol. 1963;69:783. doi: 10.1001/archopht.1963.00960040789022. [DOI] [PubMed] [Google Scholar]
- 5.Allingham RR, Liu Y, Rhee DJ. The genetics of primary open-angle glaucoma: a review. Exp Eye Res. 2009;88:837–44. doi: 10.1016/j.exer.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Resch ZT, Fautsch MP. Glaucoma-associated myocilin: a better understanding but much more to learn. Exp Eye Res. 2009;88:704–12. doi: 10.1016/j.exer.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone EM, Fingert JH, Alward WLM, Nguyen TD, Polansky JR, Sunden SLF, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Shefield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–670. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 8.Alward W, Fingert J, Coote M, Johnson A, Lerner S, Junqua D, Durcan F, McCarnty P, Mackey D, Sheffield V, Stone E. Clinical features associated with mutations in the chromosome 1 open-angle glaucoma gene (GLC1A) N Engl J Med. 1998;338:1022–1027. doi: 10.1056/NEJM199804093381503. [DOI] [PubMed] [Google Scholar]
- 9.Wilkinson C, van der Straaten D, Craig J, Coote M, McCartney P, Stankovich J, Stone E, Mackey D. Tonography demonstrates reduced facility of outflow of aqueous humor in myocilin mutation carriers. J Glaucoma. 2003;12:237–242. doi: 10.1097/00061198-200306000-00010. [DOI] [PubMed] [Google Scholar]
- 10.Hardy K, Hoffman E, McKay B, Gonzalez P, Stamer W. Extracellular trafficking of myocilin in human trabecular meshwork cells. J Biol Chem. 2005;280:28917–28926. doi: 10.1074/jbc.M504803200. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman EA, Perkumas KM, Highstrom LM, Stamer WD. Regulation of myocilin-associated exosome release from human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2009;50:1313–8. doi: 10.1167/iovs.08-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Resch ZT, Hann CR, Cook KA, Fautsch MP. Aqueous humor rapidly stimulates myocilin secretion from human trabecular meshwork cells. Exp Eye Res. doi: 10.1016/j.exer.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perkumas K, Hoffman E, McKay BS, Stamer W. Myocilin-associated exosomes in human samples. Exp Eye Res. 2006;84:209–212. doi: 10.1016/j.exer.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel-Bakky MS, Gutwein P, Altevogt P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007;72:1095–102. doi: 10.1038/sj.ki.5002486. [DOI] [PubMed] [Google Scholar]
- 15.Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760–71. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–83. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 17.Johnstone RM. Exosomes biological significance: A concise review. Blood Cells Mol Dis. 2006;36:315–21. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 73:1907–20. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 20.Stamer W, Seftor R, Williams S, Samaha H, Snyder R. Isolation and culture of human trabecular meshwork cells by extracellular matrix digestion. Curr Eye Res. 1995;14:611–617. doi: 10.3109/02713689508998409. [DOI] [PubMed] [Google Scholar]
- 21.Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr Opin Mol Ther. 2002;4:242–50. [PubMed] [Google Scholar]
- 22.Stamer WD, Roberts BC, Howell DN, Epstein DL. Isolation, culture and characterization of endothelial cells from Schlemm’s canal. Invest Ophthalmol Vis Sci. 1998;39:1804–1812. [PubMed] [Google Scholar]
- 23.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 24.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. Journal of Biological Chemistry. 2003;278:20083–90. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 25.Hendrickson EL, Xia Q, Wang T, Leigh JA, Hackett M. Comparison of spectral counting and metabolic stable isotope labeling for use with quantitative microbial proteomics. Analyst. 2006;131:1335–41. doi: 10.1039/b610957h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamm ER. Myocilin and glaucoma: facts and ideas. Prog Retin Eye Res. 2002;21:395–428. doi: 10.1016/s1350-9462(02)00010-1. [DOI] [PubMed] [Google Scholar]
- 27.Danussi C, Spessotto P, Petrucco A, Wassermann B, Sabatelli P, Montesi M, Doliana R, Bressan GM, Colombatti A. Emilin1 deficiency causes structural and functional defects of lymphatic vasculature. Mol Cell Biol. 2008;28:4026–39. doi: 10.1128/MCB.02062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zacchigna L, Vecchione C, Notte A, Cordenonsi M, Dupont S, Maretto S, Cifelli G, Ferrari A, Maffei A, Fabbro C, Braghetta P, Marino G, Selvetella G, Aretini A, Colonnese C, Bettarini U, Russo G, Soligo S, Adorno M, Bonaldo P, Volpin D, Piccolo S, Lembo G, Bressan GM. Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell. 2006;124:929–42. doi: 10.1016/j.cell.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Lutjen-Drecoll E, Futa R, Rohen J. Ultrahistochemical studies on tangential sections of the trabecular meshwork in normal and glaucomatous eyes. Invest Ophthalmol Vis Sci. 1981;21:563–573. [PubMed] [Google Scholar]
- 30.Uniewicz KA, Cross MJ, Fernig DG. Exogenous recombinant dimeric neuropilin-1 is sufficient to drive angiogenesis. J Biol Chem. doi: 10.1074/jbc.M110.190801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keller KE, Aga M, Bradley JM, Kelley MJ, Acott TS. Extracellular matrix turnover and outflow resistance. Exp Eye Res. 2009;88:676–82. doi: 10.1016/j.exer.2008.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.