Abstract
A versatile microfluidic platform allowing co-culture of multiple cell populations in close proximity with separate control of their microenvironments would be extremely valuable for many biological applications. Here, we report a simple and compact microfluidic platform that has these desirable features and allows for real-time, live-cell imaging of cell-cell interactions. Using a pneumatically/hydraulically controlled poly(dimethylsiloxane) (PDMS) valve barrier, distinct cell types can be cultured in side-by-side microfluidic chambers with their optimum culture media and treated separately without affecting the other cell population. The platform is capable of both two-dimensional and three-dimensional cell co-culture and through variations of the valve barrier design, the platform allows for cell-cell interactions through either direct cell contact or soluble factors alone. The platform has been used to perform dynamic imaging of synapse formation in hippocampal neurons by separate transfection of two groups of neurons with fluorescent pre- and post-synaptic protein markers. In addition, cross-migration of 4T1 tumor cells and endothelial cells has been studied under normoxic and hypoxic conditions, which revealed different migration patterns, suggesting the importance of the microenvironments in cell-cell interactions and biological activities.
Keywords: Cell culture, Cell-cell interaction, Synapse formation, Cell migration
1 Introduction
Cell co-culture is an important biotechnology required for studying cellular activities and inter-cell communications in diverse cellular populations. Microfluidic cell co-culture platforms have been identified as one particularly promising area where microfluidics can truly make a revolutionary contribution and thereby transform the technology for biological research (Squires and Quake 2005; Dittrich and Manz 2006; El-Ali et al. 2006; Gross et al. 2007; Yeon and Park 2007; Meyvantsson and Beebe 2008). The ability of precise, dynamic control of cellular microenvironments, together with their low cost, high-throughput production and rapid prototyping capacity, offers microfluidic devices unique functions and advantages over traditional in vitro systems.
Microfluidic cell co-culture platforms could have many unique features to address specific needs. A versatile microfluidic cell co-culture platform should be able to (1) load distinct cell types into specified regimes, (2) culture cells with their optimal culture media before the cells reach confluence and could be self-sustained, (3) manipulate the microenvironment of selected cell populations without affecting other cell types, (4) allow for cell-cell interactions in a controlled manner, and (5) facilitate high-resolution real-time, live-cell imaging to study cell-cell interactions.
The advantages and great potential of microfluidic cell co-culture platforms have attracted significant attention and quite a few platforms have been developed for different biological applications, especially in neurobiology and cancer biology, because of the importance of cell-cell interactions in these fields. However, to date, no reported platform has all the desirable features for a versatile microfluidic co-culture platform as discussed above.
One popular microfluidic co-culture technique is surface patterning (Bhatia et al. 1997; Kane et al. 2006; Khetani and Bhatia 2008), which relies on modification of the substrate surface by attaching desired molecules in predetermined patterns. Taking advantage of the selective adhesion to the attached molecules, one type of cells can be loaded to a specified area. Other cells can then be loaded to the remaining surface and co-cultured with the previously loaded cells. For example, Bhatia et al. (1997) performed the pioneering work of patterning molecules on a glass substrate to attach hepatocytes, which were co-cultured with 3T3 fibroblasts that were loaded to the remaining unmodified area. In addition to the surface patterning technique, fluid flow has been used to load different cells to their respective desired regions (Takayama et al. 1999; Khademhosseini et al. 2005; Skelley et al. 2009). These techniques allow for co-culture of different cell populations and examination of cellular activities, which provide the possibility for many interesting biological studies. However, once the cells are loaded, it is difficult for these devices to perform separate treatment on a selected cell population without affecting the entire culture. With syringe pumps to carefully control the pressure of different streams, treatment of selected cell populations can be done. However, there exist several limitations: (1) it takes time to build up the side by side laminar flow; (2) there is a diffusion region between the two streams, which might be undesirable in some studies; and (3) for cell migration studies, cells can migrate across the two streams during long-term treatment, which can lead to failure of the balanced laminar flow approach.
Another popular microfluidic cell co-culture technique is compartmentalization, which uses spatially separated compartments to maintain distinct populations of cells. Micro-grooves (Taylor et al. 2005), collagen tracks (Ravula et al. 2007), semi-permeable membranes (Kimura et al. 2008), or parallel capillaries (Groisman et al. 2005) have been used as barriers to separate different cell populations. The limitation of these devices is that cell-cell interactions through soluble factors always exist. Therefore, it is difficult to completely isolate the two chambers and perform separate treatments on selected cell populations without affecting others. Recently, Hui and Bhatia (2007) reported a reconfigurable co-culture platform, in which two comb-fingers like parts were used to control the separation distance between two cell populations. This development is important in the sense that the platform allows two cell populations to interact with each other in a controlled manner. However, the comb-fingers were made of silicon, leading to imaging and cost issues. In addition, even when the comb-fingers were in the contact mode, there still existed a gap of ~6 μm between the fingers, which might be undesirable for some applications.
Here we report on a simple microfluidic cell co-culture platform, which uses a pressure-controlled PDMS valve barrier to completely separate or connect two adjacent cell populations. It is worth noting that pneumatic/hydraulic valves have been used in microfluidics to control fluid flow (Unger et al. 2000; Thorsen et al. 2002; Studer et al. 2004a; Chen et al. 2008; Kim et al. 2008) or help to sort or trap cells (Studer et al. 2004b; Irimia and Toner 2006); however, we have not seen their usage for separation of distinct cell populations in adjacent chambers. Simple passive pumping method (Walker and Beebe 2002) is chosen to drive media flow inside the microfluidic devices, which eliminates the requirement of complicated connections and expensive external equipments such as syringe pumps. The platform allows for both two-dimensional (2D) and three-dimensional (3D) cell co-culture and for cell-cell interactions through soluble factors alone with a design variation of the valve barrier. The potential applications of the cell co-culture platform have been demonstrated with the dynamic observation of synapse formation in central nervous system (CNS) neurons and tumor cell-endothelial cell cross-migration.
2 The design and characterization of 2D prototype platforms
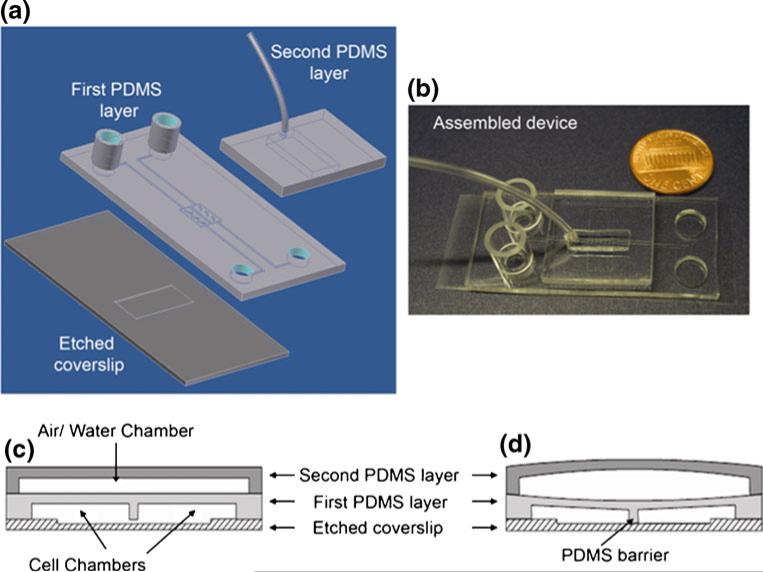
The microfluidic cell co-culture platform is fabricated using standard soft-lithography techniques (Whitesides et al. 2001) with replica molding PDMS (Ellsworth Adhesives, Germantown, WI). To increase the flexibility of the PDMS, the ratio of the polymer base to the curing agent was chosen as 12:1. Two layers of PDMS were bonded to a microscope coverslip to form the functional device (Fig. 1(a), (b)). The first PDMS layer, comprising of a network of microchannels and chambers connecting to a set of wells, was aligned and bonded to a glass coverslip (VWR Vista Vision, Suwanee, GA) with a 1–16 μm deep cell culture region defined by etching using buffered hydrofluoric acid (HF) (Fig. 1(a)). Two adjacent microchambers (6×0.7×0.06 mm, length × width × height), separated by a narrow wall of PDMS valve barrier of 100 μm wide, were designed to confine the culture regions of two cell populations. The second PDMS layer, defining a pressure chamber (16×5×1 mm, length × width × height), was bonded on top of the first layer (Fig. 1(a)). Before bonding, all surfaces except those of the valve barrier and the etched culture region on the coverslip were treated with oxygen plasma to form strong irreversible bonding (Duffy et al. 1998).
Fig. 1.
Design of the microfluidic cell co-culture platforms to study cell-cell interactions. (a) A schematic of the main components. (b) A picture of the fabricated device with a penny for size comparison. Note that the entire device can be placed inside a traditional 75 mm diameter Petri dish. (c,d) Schematic diagrams showing the barrier valve working mechanism. When the valve is inactive, a gap under the PDMS barrier allows interaction and communication between two groups of cells (c). Upon activation of the valve, the two cell populations are isolated completely and cells in the two chambers can be treated separately (d)
Two cloning cylinders (Fisher Scientific, Pittsburgh, PA) were attached to the loading wells as reservoirs for culture media and a microbore tube (Cole-Parmer, Vermon Hills, IL) was attached to the pressure chamber by gluing with liquid PDMS. The entire device was placed in an oven for 30 min at 75°C to cure. During curing, approximately 200 μl of sterilized deionized water was loaded into each reservoir to keep the microchannels filled, maintaining a constant flow in the channels to retain the hydrophilic nature of the PDMS channels (Duffy et al. 1998). Before bioassays, the microfluidic device was sterilized under UV light for 1–2 h.
Depending on the pressure in the pressure chamber, the valve barrier could be either pushed down or released in an ‘up’ position. In the ‘up’ position, the gap between the bottom of the PDMS valve barrier and the surface of the etched coverslip allows for interaction and communication between the two cell populations (Fig. 1(c)). In the ‘down’ position, the valve barrier completely isolates the two chambers for separate culture or treatment of each cell population (Fig. 1(d)). Pressure changes in the pressure chambers were accomplished by injecting air (pneumatic valve) or water (hydraulic valve) with a syringe through the microbore tube. Characterization with Fluorescein isothiocyanate (FITC) dyes (Thermo Fisher Scientific, Rockford, IL) showed that pneumatic valves could successfully separate the two adjacent chambers for 4–6 h while hydraulic valves could work for more than one week. This is because PDMS is a gas permeable material (Merker et al. 2000), which allows air to leak through overtime. It is worth noting that in our characterization, we did not find fluorescent dyes diffuse into the PDMS, unlike that shown by Toepke and Beebe (2006) using Nile red dyes. We noticed that the possibility for soluble factors to diffuse through PDMS walls varies for different molecules (Lee et al. 2003). One more factor in favor of a complete seal between the two chambers is that under pneumatic or hydraulic pressure, the PDMS barrier becomes denser, which helps to prevent small molecules from diffusing through.
Pressure difference between the loading reservoirs and the waste wells generated a continuous fluid flow. This usage of the passive pumping method (Walker and Beebe 2002) allowed the entire co-culture platform to be placed into a Petri-dish and avoided possible contamination from additional instruments, such as syringe pumps. The upstream cylinders could hold enough culture media to maintain continuous flow for more than 24 h before more culture media needed to be added. The waste in the downstream wells was removed by either natural evaporation or manual aspiration. It is worth noting that the media flow rate generated by passive pumping is not constant; and therefore, it might have some effect on certain applications. However, in the assays we performed, the cells were healthy over the entire period of culture up to three weeks as long as fresh media was supplied by filling the loading wells periodically.
Several PDMS semi-lunar shaped supporters were fabricated in the cell chambers to prevent large deformation of the roof of the cell culture chambers when the pressure chamber was pressurized, which could be detrimental to cells. In addition, the shape of these supporters was designed to distribute the coating molecules, cells, and the culture media more evenly, as verified by modeling of the flow field and observation of cell distributions after loading (Majumdar et al. 2011).
3 Applications of 2D cell co-culture platforms
3.1 Dynamic observation of synapse formation among CNS neurons
Synapses are highly specialized cell-cell junctions that allow for communication between neurons. CNS synapses are structurally composed of a pre-synaptic axonal terminal and a post-synaptic region, which in the case of excitatory synapses is composed of dendritic spines. Spines and axons are dynamic structures and their plasticity is thought to be the basis of cognitive functions such as learning and memory (Levy and Steward 1979; Dunaevsky et al. 1999; Gray et al. 2006). Not surprisingly, abnormalities in these structures are associated with a number of neurological disorders, including autism, mental retardation, schizophrenia, epilepsy, and Alzheimer's disease (Fiala et al. 2002; Selkoe 2002).
It is currently very difficult to image synapse formation in real-time, which has limited the advancement toward identifying key molecular signals that regulate synaptic assembly. Synapse formation is a fast, dynamic process involving the assembly of hundreds of proteins into highly specific structures (McAllister 2007). Studies of the molecular mechanisms of synapse formation heavily rely on imaging fluorescent-tagged proteins at synaptic contacts in cultured neurons. This approach faces challenges particularly for molecules such as actins and cadherins, which are found at synapses in both pre- and post-synaptic terminals. It would be of great advantage to be able to separately tag the proteins in the pre- and post-synaptic terminals with different fluorescent markers and observe the protein dynamics, which has not been achieved (Taylor et al. 2010). Recently, Taylor et al. (2010) demonstrated that they could infect neurons in pre- and post-synaptic chambers separately with green fluorescent protein (GFP) and red fluorescent protein (RFP) using Sindbis virus and visualize synapses in long (900 μm) and slim micro-grooves. However, they did not image synapses as they formed. The reported microfluidic cell co-culture platform, with its ability to treat two cell populations separately, could be readily adapted for dynamic observation of synapse formation among CNS neurons.
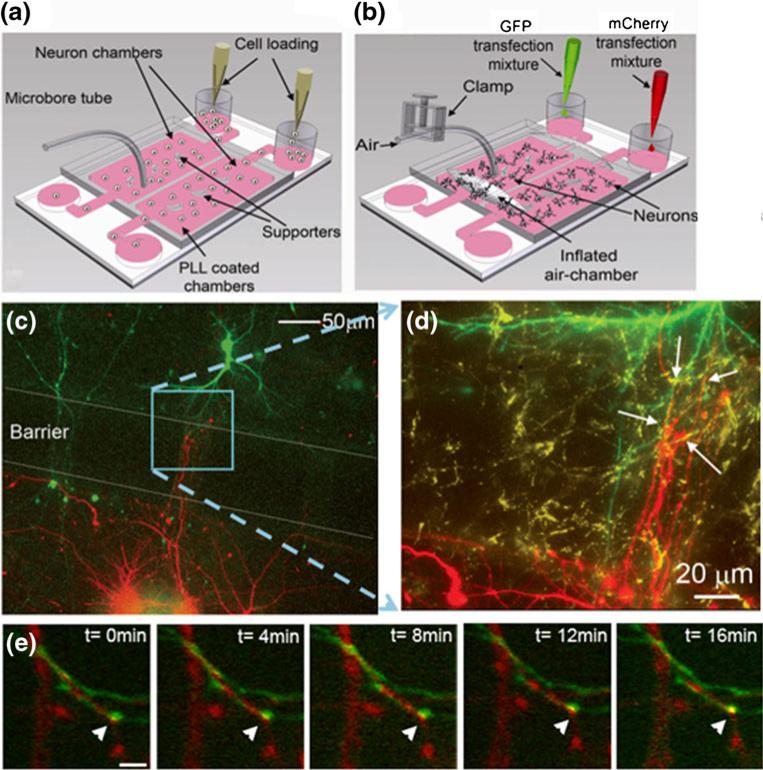
Towards this goal, the microfluidic platforms were UV sterilized and coated with poly-L-lysine (PLL) by flowing 1 mg/ml PLL through the chambers for 12 h at 37°C to coat the glass coverslips. To remove excess uncoated PLL, the chambers were washed with sterilized water for at least 1 h at 37°C. The chambers were then equilibrated with B27 supplemented Neurobasal™ media (GIBCO™ Invitrogen, Carlsbad, CA) containing L-glutamine. Following coating, dissociated hippocampal neurons, which were isolated from E19 rat embryos, were re-suspended in B27 Neurobasal™ media at a density of 5,000 cells per microliter of media and loaded into each of the reservoir cylinders by adding 15–20 μl of the neuron suspension to the loading wells (Fig. 2(a)). The microfluidic platforms were then placed in a cell culture incubator with 5% CO2 for 2–3 h at 37°C to allow neurons to attach to the PLL-coated glass coverslips. When cells had visibly attached to the surface, approximately 300 μl of culture media was added to one reservoir and half of this volume was added to the other reservoir. This allows for sufficient flow rate through the gap between the two chambers. Every 36–48 h, neurons were supplied with fresh B27 Neurobasal™ media, which was conditioned for at least 24 h over a confluent monolayer of glial cells. Glia were isolated from brains of rats post-natal day 2 and cultured as previously described (Goslin et al. 1998).
Fig. 2.
Dynamic imaging of synapse formation among hippocampal neurons. (a,b) Schematic diagrams showing cell loading and separate transfection. (c) Neurons in one chamber expressed mCherry and neurons in the other chamber expressed GFP. The expression of each protein was confined to the individual chamber. The neuronal processes extended to the opposite chamber and formed synapses. (d) A high magnification image of the boxed region of (c). To demonstrate that neurons formed synapses, cells in the device were immunostained for the synaptic marker SV2. (e) Time lapse images show a synapse forming (yellow puncta, arrowhead) between a neuron expressing mCherry-synaptophysin (red puncta) and a neuron expressing GFP-PSD-95 (green puncta)
Three days later the valve barrier was pushed down pneumatically to separate the two chambers. Neurons in each chamber were subsequently transfected with different cDNA solutions for approximately 1.5 h using a modified calcium phosphate method (Zhang et al. 2003). After the two chambers were isolated, the transfection solution containing different cDNAs, one encoding GFP (green) and the other m-Cherry fluorescent protein (red), were loaded into the two reservoir cylinders (Fig. 2(b)). The cDNA-calcium phosphate precipitates were introduced into the respective chambers by continuous flow and some complexes were observed to attach onto the cell body surface, which is expected for a successful transfection. Since a low, compared to a high flow-rate led to an improvement in transfection efficiency, we added a drop of culture media into each waste well 10–15 min after loading the transfection solution to reduce the flow-rate. The procedure consistently led to an approximately 40% transfection efficiency, which is significantly higher than the efficiency that we achieved on traditional culture plates under identical experimental conditions (Majumdar et al. 2011). When transfected neurons in each chamber were visualized by fluorescent microscopy, expression of the fluorescent proteins, GFP (green) and mCherry (red), was observed. Importantly, expression of mCherry was confined to the culture chamber that received the mCherry cDNA transfection solution and was not seen in the adjacent chamber. Similar results were obtained with GFP, which indicates the pneumatic valve successfully isolated the chambers and prevented cross flow between the two chambers.
After transfection, the DNA-calcium phosphate complexes were washed off and fresh glia-conditioned culture media were filled into the loading reservoirs. The valve barrier was then released, allowing for neurons in one chamber to interact and contact neurons in the adjacent chamber. Neuronal processes (axons and dendrites) appeared to interact with each other across the two chambers (Fig. 2(c)). To show that neurons formed synapses, cells in the device were immunostained for the synaptic marker SV2. Immunostaining began by fixing the neurons in the chambers with 4% paraformaldehyde/ 4% sucrose in phosphate-buffered saline (PBS). The fixative was flowed through the chambers for 30 min at room temperature. After three 15 min washes with PBS, the neurons were blocked by flowing 20% goat serum for 30 min at room temperature. The neurons were immunostained for SV2 (Development studies hybridoma bank, The University of Iowa, Iowa City, IA) by allowing the primary antibody solution (1:500 in 5% goat serum in PBS) to flow through the devices for 1 h at room temperature or overnight at 4°C. After three 15 min washes with PBS, a secondary antibody solution (Alexa Fluor® 647 from Molecular probes, Eugene, OR) of a dilution of 1:500 in 5% goat serum in PBS, was flowed through the chambers for 45 min at room temperature. Three 15 min washes with PBS were performed before the device was ready for microscopy. A high magnification image shows that the neuronal contacts did form synapses as indicated by the presence of SV2 clusters (Fig. 2(d), yellow).
To demonstrate the capability of dynamic observation of synapse formation, neurons in the two chambers were transfected with a fluorescently tagged pre-synaptic protein, mCherry-synaptophysin and a post-synaptic marker, GFP-tagged postsynaptic density protein-95 (PSD-95). We then performed live-cell imaging between the chambers to determine if we could dynamically capture contact between the pre- and post-synaptic markers. Live-cell imaging was performed using the Quorum WaveFX confocal microscope for 12 h with 4 min intervals. The microfluidic chambers were maintained at 37°C under humidified conditions during the entire duration of imaging. As shown in Fig. 2(e), we imaged a neuron expressing mCherry-synaptophysin forming a synapse with a GFP-PSD-95 expressing neuron, suggesting that we can dynamically image synapse formation using these devices. This ability will allow us to study the function of various molecules that are considered important to CNS synapse formation by labeling the molecules of interest and imaging them at synapses.
3.2 Cross-migration of 4T1 tumor cells and endothelial cells in normoxic and hypoxic microenvironments
Cross-talk between tumor cells and host vasculature endothelium are critical in tumor growth, progression and metastasis (Kaelin 2008; Coleman and Ratcliffe 2009). Migration of both cell types is a key component of vascular recruitment through the process of angiogenesis, as well as transendothelial cell migration in the processes of intravasation and metastasis (Li et al. 2000). A hurdle to dissect the contribution of these two processes in tumor migration is the lack of appropriate co-culture platforms. Most existing co-culture platforms either mix the two cell types or seed one type of cells on the confluence layer of another. The lack of initial spatial separation makes it difficult to interpret the nature of the interactions. Additionally, it is of great interest to understand the effects of microenvironments on tumor-endothelial cross-migration, which requires separate treatment of one cell type without affecting the other.
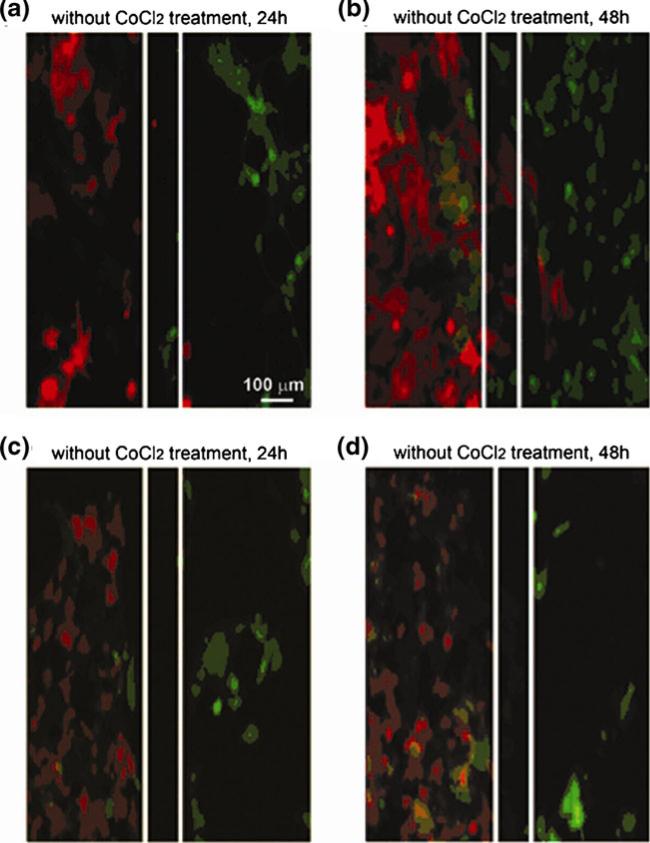
Here, we applied the microfluidic platform to study the dynamic interaction between tumor cells and endothelial cells in vitro. In control experiments, while the barrier valve was closed by hydraulic pressure, murine 4T1 mammary tumor cells, which stably express RFP (4T1-RFP), were seeded in one chamber, and human dermal microvascular endothelial cells stably labeled with GFP (HDVEC-GFP) in the other chamber. Culture media (high glucose Dulbecco's Modified Eagle's Medium, (Mediatech, Inc., Herndon, VA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin) were continuously flowed through the chamber to provide a physiological environment. When cell density reached ~75% confluence (about 24 h), the valve barrier was lifted and the kinetics of cell migration was imaged for two additional days. Under these conditions, we found that cell migration was bidirectional: tumor cells migrated toward endothelial cells and endothelial cells also migrated toward the tumor cells (Fig. 3(a), (b)), indicative of tumor angiogenesis and intravasation/extravasation.
Fig. 3.
The interaction of tumor cells and endothelial cells. To investigate the cross-migration of tumor and endothelial cells under normoxic and hypoxic conditions, HDVEC-GFP cells were seeded on the right side of the chamber, and 4T1-CRHG (RFP) were seeded on the left side of the chamber and cultured for 24–48 h with (c,d) or without (a,b) CoCl2 (300 μM). After flowing fresh media to remove the residual CoCl2, the chambers were reconnected and cell migration was imaged under fluorescence microscopy for an additional 48 h. Images in panels (a,b) show bidirectional cell migration under normoxic condition at 24 and 48 h. Images in panels (c,d) show migration of endothelial cells only with hypoxic treatment of tumor cells at 24 and 48 h
One major advantage of this device is to permit separate treatments of cells in each chamber, which we use to observe the effects of hypoxia on tumor angiogenesis. Hypoxia is a key regulator of angiogenesis. When tissue growth outpaces the growth of blood vessels, the tissue becomes hypoxic. Hypoxia upregulates a variety of genes that promote angiogenesis toward tissues and restore homeostasis (Ikeda et al. 1995; Carmeliet et al. 1998; Mandriota and Pepper 1998; Pugh and Ratcliffe 2003). For experiments studying the effect of hypoxia, 4T1-RFP tumor cells and endothelial cells were seeded in separated chambers as previous described. After cells reached ~75% confluence, 300 μM cobalt chloride (CoCl2) (Sigma-Aldrich, Atlanta, GA) was added to the tumor cell chamber for another 24 h to induce hypoxia. CoCl2 mimics hypoxia by binding to the oxygen-dependent degradation domain (ODDD) in HIF-1α (Liu et al. 1999; Yuan et al. 2003). This binding prevents ubiquitination and degradation of HIF-1α and leads to the accumulation/stabilization of HIF in cells. After CoCl2 treatment, fresh media was allowed to flow through to remove the CoCl2 from the system and then the two chambers were connected by releasing the valve barrier. We continuously examined the kinetics of cell-cell interactions for an additional two days. Intriguingly, under this condition, it became only a one-way traffic. Only migration of endothelial cells toward tumor cells was observed, but not vice versa (Fig. 3(c), (d)), which was quite different from the observations we made under normoxic conditions. These experiments suggest that under normoxic condition, both tumor cells and endothelial cells recruit each other. However, the hypoxic treatment of tumor cells suppresses their migration ability, and prompts release of angiogenic growth factors to attract endothelial cells and the process is angiogenesis only.
4 Demonstration of 3D cell co-culture platforms
To better mimic the in vivo microenvironment, it is desirable to co-culture cells and study cell-cell interactions in 3D. The cell-matrix interaction in the 3D setting mediates physiological responses that are important for cell growth, differentiation, and survival (Bissell and Radisky 2001; Jacks and Weinberg 2002). For example, fibroblasts cultured within an extracellular matrix (ECM) showed different morphology and migration patterns from those cultured in 2D (Cukierman et al. 2001).
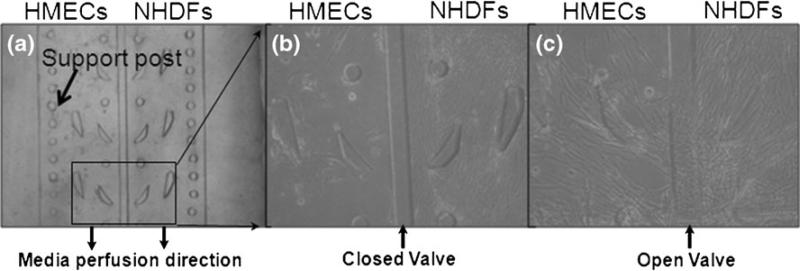
By loading cells together with biogels, the reported microfluidic cell co-culture platform can be adapted for 3D cell co-culture. To demonstrate this, human mammary epithelial cells (HMECs) and normal human dermal fibroblasts (NHDFs) were prepared and loaded into different chambers with the valve barrier in the ‘down’ position. Because the cell-biogel mixtures are viscous, a vacuum-driven loading method was employed to load HMECs and NHDFs with their distinct 3D matrix as follows. The harvested HMECs and NHDFs were suspended in their respective growth media at a density of 1×106 cells/ml. The suspended media and the microfluidic platform with hydraulic valve closed were placed on ice prior to cell loading. The HMECs were prepared in Matrigel™ (BD Biosciences, San Jose, CA) at the following volumes: 25 μl cells with 75 μl of 100% Matrigel™. After mixing these two components to homogeneity, the 100 μl mixture was added to the loading well of HMECs, upon which vacuum was applied at the corresponding waste well to help the mixture to flow down to the cell culture chamber. For the NHDFs, 40 μl of type I collagen (BD Biosciences, San Jose, CA) and 60 μl of NHDF suspended medium was mixed homogeneously with the help of 0.5 μl of 1 M NaOH. The mixture was then added to the loading well of NHDFs and vacuum was applied in the same manner as for HMECs. The co-culture platform was then placed into a humidified incubator at 37°C for 30 min to polymerize the matrices, after which the loading reservoirs of the HMECs and NHDFs were filled with their respective media (Fig. 4(a)).
Fig. 4.
The 3D co-culture platform. (a) With the hydraulic valve closed, HMECs in Matrigel™ and NHDFs in type I collagen were vacuum-loaded into each chamber. (b) The cells were cultured for 7 days; and (c) 7 days after the valve was released, fibroblasts entered the epithelial chamber
The HMECs and NHDFs were maintained for 7 days while the valve was closed (Fig. 4(b)). On day 8 the valve was opened. The NHDFs were observed to cross over to the HMEC chamber on day 14 (Fig. 4(c)). Interestingly, the HMECs responded to the NHDFs by forming small rounded structures reminiscent of acinar structures (Fig. 4(c)).
One problem encountered in the 3D cell co-culture is that collagen fibers will contract during cell culture in response to cell-matrix interactions. To overcome this issue, a column of posts were patterned near the outer edge of each chamber as anchors for the gel fibers to adhere, as indicated in Fig. 4(a). Multiple experiments showed that these anchors could effectively prevent the collagen fibers from contracting during cell culture and interaction period.
5 Cell co-culture and interactions through soluble factors alone with an agarose-coupled valve barrier
Cell-cell interactions can occur through direct cell-cell physical contact involving both soluble factors and surface receptors or through soluble factors alone. For example, it was found that the clonogenic growth of human breast cancer cells was dramatically and consistently enhanced when tumor cells were co-cultured in direct contact with fibroblasts (Samoszuk et al. 2005). It would be extremely desirable to have a cell co-culture platform that can differentiate cell-cell interactions through either direct cell contact or soluble factors alone. The reported microfluidic platforms, as discussed above, allow for cell-cell interactions through direct cell contact. However, through inclusion of an agarose-coupled valve barrier, our platform can be used to study cell-cell interactions through soluble factors alone.
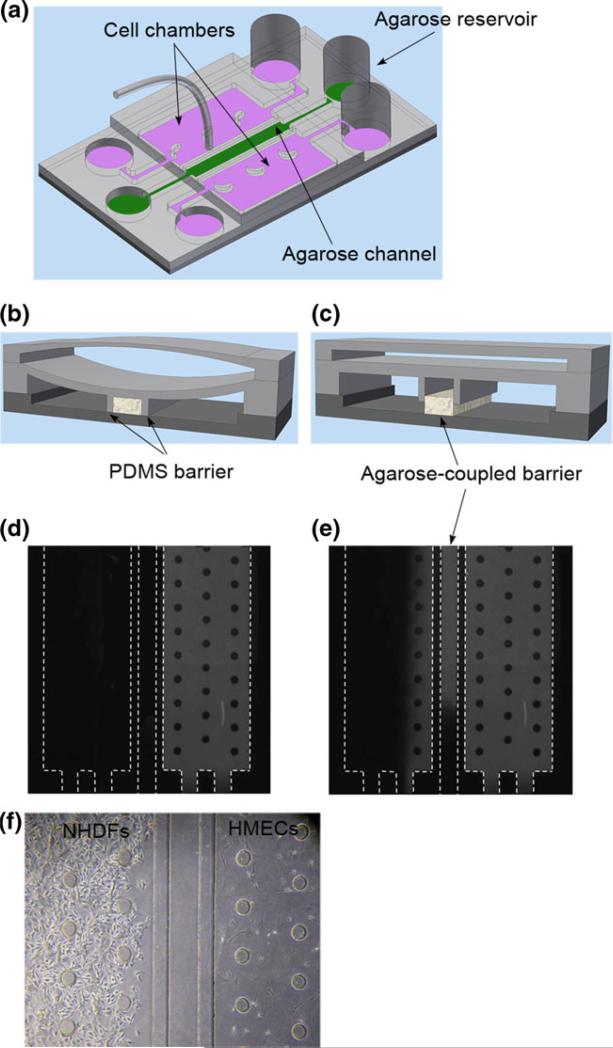
The agarose-coupled valve barrier was fabricated using the two PDMS barriers that formed a channel after being pushed down, into which the pre-gel solution of agarose was filled (Fig. 5(a), (b)). Note that to facilitate the agarose loading, separate loading and waste wells and connecting channels were included into the first PDMS layer (Fig. 5(a)). After polymerization, the PDMS valve barriers were released into the ‘up’ position and the agarose remained attached to the glass substrate to block transport of cells (Fig. 5(c)). In this way, the platform with an agarose-coupled valve barrier permitted cell-cell interactions solely through soluble factors.
Fig. 5.
The cell co-culture platform with an agarose-coupled valve barrier. (a) A schematic of the microfluidic device with an agarose-coupled barrier. (b,c) Schematics of the working mechanisms of the agarose-coupled barrier. (d,e) Fluorescent dyes were added to one cell chamber with the PDMS valve barrier closed (d), after the valve barrier was released into an ‘up’ position (e), fluorescent dyes were observed in the opposite chamber. (f) HMECs in Matrigel™ and NHDFs in type I collagen were loaded and cultured side by side in a platform with the agarose-coupled valve barrier. The two cell populations could communicate only through soluble factors because the nanoporous agarose prevented the cell from migrating over the barrier
To reduce the bonding between the agarose and the PDMS for the agarose to stay attached to the substrate when the valve barriers are released, the PDMS barriers were pretreated with ethylene glycol or Fluoronic 127 following published protocols (Tang et al. 2003; Golden and Tien 2007). In addition, the glass slide was coated with aminopropyl ethylsilane (APES) to form aminosilane on the surface, which enhanced the agarose attachment to the glass coverslip and ensured the proper function of the device. The details of these treatments are described as follows. After PDMS was peeled off from the SU8 mold and the reservoirs were punched, it was adhered to a clean coverslip. This formed a reversible bonding. Ethylene glycol (Sigma-Aldrich, Atlanta, GA) or Fluronic 127 (Sigma-Aldrich, Atlanta, GA) was filled in by capillary force to treat the channel surfaces at 37°C. After 1 h, deionized water was filled into the channel to wash off the excess ethylene glycol or Fluronic 127 (30 min at 37°C). The PDMS was then peeled off from the coverslip and dried overnight. For the glass substrates, the microscopic slides were immersed in a 10% nitric acid solution at 80-90°C with gentle shaking for 1 h. After rinsing with water, the slides were then placed in a 10% solution of APES (Sigma-Aldrich, Atlanta, GA) at a pH of 3.4. The solution was heated to 70°C with stirring for an additional 3 h. The silanated slides were then thoroughly washed with water and dried overnight.
After the platform was assembled and the two PDMS valve barriers were pushed into the ‘down’ position to form a channel (Fig. 5(b)), agarose pre-gel solution was loaded into the channel with the entire platform placed on a hotplate at 60°C. The agarose pre-gel solution was prepared by adding phosphate buffer to a measured amount of agarose powder (Sigma-Aldrich, Atlanta, GA) to a concentration of 1 mg/ml. The mixture slurry was then heated in an oven to 90°C till the agarose powder was completely dissolved. The hot agarose pre-gel solution was then loaded into the middle channel, after which the platform was removed from the hotplate to cool down at room temperature allowing for gelation of the agarose.
Figure 5(d) and (e) show the exchange of fluorescent dyes between the two chambers. While the valve barrier was closed, one chamber (right-hand side in the figure) was filled with aqueous FITC. Figure 5(d) shows that in this case, FITC cannot perfuse into the opposite chamber. However, after the valve barriers were released, FITC was observed to perfuse through the agarose barrier into the opposite chamber (Fig. 5(e)).
We further demonstrated that the agarose-coupled valve barrier could block cells from moving into the opposite chamber by loading the two chambers on each side of the valve barrier with HMECs in Matrigel™ and NHDFs in type I collagen, following the procedure as previously described. The pore size of agarose varies from 50 nm to several hundred nanometers depending on the concentration (Narayanan et al. 2006). After the PDMS valve barriers were released into the ‘up’ position, the nanoporous agarose-coupled valve barrier allowed soluble factors to perfuse through but prevented cells from crossing the barrier and migrating to the opposite chamber (Fig. 5(f)).
It is worth noting that we observed the above phenomena in multiple experiments using multiple devices, which suggests that agarose attaches well to the glass surface. This is because cells can squeeze through small micro-sized channels (Luscinskas et al. 2002; Friedl and Wolf 2003) and if the agarose detaches from the glass surface when the PDMS barrier is released, it is highly likely that some cells will squeeze through from underneath the detached region, which has not been observed in our multiple experiments.
6 Summary
We developed a microfluidic cell co-culture platform that uses a pneumatic/hydraulic valve to separate two cell populations in two chambers that are close to each other, which allows for culture of distinct cell types with their optimum culture media and treatment of each cell population without affecting the other. The platform is capable of both 2D and 3D cell co-culture and real-time, live-cell imaging of cell-cell interactions. Through variations of the valve barrier design, the platform allows for cell-cell interactions through either direct cell contact or soluble factors alone. We believe that the platform would have extensive applications for neurobiology and cancer biology. To demonstrate the potential applications, we applied the platform to culturing hippocampal neurons and achieved dynamic imaging of synapse formation in the CNS, which provides a powerful technique to identify the function of various molecules in synapse formation. In addition, we successfully studied the cross-migration of tumor and endothelial cells under normoxic and hypoxic conditions. The results demonstrate that under normoxic condition, both 4T1 tumor cells and endothelial cells migrate toward each other, indicating that both angiogenesis and intravasation/extravasation are important. However, the hypoxic treatment of the tumor cell suppresses their migration, and only endothelial cells migrate to the tumor chamber, suggesting that in this case, only angiogenesis occurs.
Acknowledgements
We thank Anne Marie Craig, Mark Dewhirst, and Freda Miller for kindly providing us with reagents and Dmitry Markov for helpful discussions. We also thank Lan Hu for technical assistance in preparing neuronal cultures. This work was supported by grants MH071674 and GM092914 from NIH and S10RR025524 from the National Center for Research Resources at NIH to D.J.W. and grant CBET0643583 from NSF and W81XWH-07-1-0507 from DoD to D.L.
Contributor Information
Yandong Gao, Department of Mechanical Engineering, Vanderbilt University, Nashville, TN 37235, USA.
Devi Majumdar, Department of Biological Sciences and Vanderbilt Kennedy Center for Research on Human Development, Vanderbilt University, Nashville, TN 37235, USA.
Bojana Jovanovic, Department of Cancer Biology, Vanderbilt University, Nashville, TN 37235, USA.
Candice Shaifer, Department of Biochemistry and Cancer Biology, Meharry Medical College, Nashville, TN 37208, USA.
P. Charles Lin, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Andries Zijlstra, Department of Pathology, Vanderbilt University, Nashville, TN 37235, USA.
Donna J. Webb, Department of Biological Sciences and Vanderbilt Kennedy Center for Research on Human Development, Vanderbilt University, Nashville, TN 37235, USA Department of Cancer Biology, Vanderbilt University, Nashville, TN 37235, USA.
Deyu Li, Department of Mechanical Engineering, Vanderbilt University, Nashville, TN 37235, USA.
References
- Bhatia SN, Yarmush ML, et al. J. Biomed. Mater. Res. 1997;34:189–199. doi: 10.1002/(sici)1097-4636(199702)34:2<189::aid-jbm8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Bissell MJ, Radisky D. Nat. Rev. Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, et al. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Chen H, Gu W, et al. Anal. Chem. 2008;80:6110–6113. doi: 10.1021/ac800506n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, Ratcliffe PJ. Nat. Med. 2009;15:491–493. doi: 10.1038/nm0509-491. [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, et al. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Dittrich PS, Manz A. Nat. Rev. Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- Duffy DC, McDonald JC, et al. Anal. Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Dunaevsky A, Tashiro A, et al. Proc. Natl. Acad. Sci. USA. 1999;96:13438–13443. doi: 10.1073/pnas.96.23.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ali J, Sorger PK, et al. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, et al. Brain Res. Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Nat. Rev. Cancer. 2003;3:362–374. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- Golden AP, Tien J. Lab Chip. 2007;7:720–725. doi: 10.1039/b618409j. [DOI] [PubMed] [Google Scholar]
- Goslin K, Asmussen H, et al. Rat hippocampal neurons in low-density culture. MIT; Cambridge: 1998. [Google Scholar]
- Gray NW, Weimer RM, et al. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman A, Lobo C, et al. Nat. Methods. 2005;2:685–689. doi: 10.1038/nmeth784. [DOI] [PubMed] [Google Scholar]
- Gross PG, Kartalov EP, et al. J. Neurosci. 2007;252:135–143. doi: 10.1016/j.jns.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Hui EE, Bhatia SN. Proc. Natl. Acad. Sci. USA. 2007;104:5722–5726. doi: 10.1073/pnas.0608660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda E, Achen MG, et al. J. Biol. Chem. 1995;270:19761–19766. doi: 10.1074/jbc.270.34.19761. [DOI] [PubMed] [Google Scholar]
- Irimia D, Toner M. Lab Chip. 2006;6:345–352. doi: 10.1039/b515983k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Weinberg RA. Cell. 2002;111:923–925. doi: 10.1016/s0092-8674(02)01229-1. [DOI] [PubMed] [Google Scholar]
- Kaelin Jr WG. Nat. Rev. Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- Kane BJ, Zinner MJ, et al. Anal. Chem. 2006;78:4291–4298. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A, Yeh J, et al. Lab Chip. 2005;5:1380–1386. doi: 10.1039/b508096g. [DOI] [PubMed] [Google Scholar]
- Khetani SR, Bhatia SN. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- Kim JY, Park H, et al. Biomed. Microdevices. 2008;10:11–20. doi: 10.1007/s10544-007-9105-z. [DOI] [PubMed] [Google Scholar]
- Kimura H, Yamamoto T, et al. Lab Chip. 2008;8:741–746. doi: 10.1039/b717091b. [DOI] [PubMed] [Google Scholar]
- Lee JN, Park C, Whitesides G. Anal. Chem. 2003;75:6544–6554. doi: 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- Levy WB, Steward O. Brain Res. 1979;175:233–245. doi: 10.1016/0006-8993(79)91003-5. [DOI] [PubMed] [Google Scholar]
- Li CY, Shan S, et al. J. Natl. Cancer Inst. 2000;92:143–147. doi: 10.1093/jnci/92.2.143. [DOI] [PubMed] [Google Scholar]
- Liu XH, Kirschenbaum A, et al. Clin. Exp. Metastas. 1999;17:687–694. doi: 10.1023/a:1006728119549. [DOI] [PubMed] [Google Scholar]
- Luscinskas FW, Ma S, et al. Immunol. Rev. 2002;186:57–67. doi: 10.1034/j.1600-065x.2002.18606.x. [DOI] [PubMed] [Google Scholar]
- Majumdar D, Gao Y, et al. J. Neurosci. Meth. 2011;196:38–44. doi: 10.1016/j.jneumeth.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandriota SJ, Pepper MS. Circ. Res. 1998;83:852–859. doi: 10.1161/01.res.83.8.852. [DOI] [PubMed] [Google Scholar]
- McAllister AM. Annu. Rev. Neurosci. 2007;30:425–450. doi: 10.1146/annurev.neuro.29.051605.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker TC, Bondar VI, et al. J. Polym. Sci. 2000;38:415–434. [Google Scholar]
- Meyvantsson I, Beebe DJ. Annu. Rev. Anal. Chem. 2008;1:423–449. doi: 10.1146/annurev.anchem.1.031207.113042. [DOI] [PubMed] [Google Scholar]
- Narayanan J, Xiong J-Y, et al. J. Phys. Conf. Ser. 2006;28:4. [Google Scholar]
- Pugh CW, Ratcliffe PJ. Nat. Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- Ravula SK, Wang MS, et al. J. Neurosci. Meth. 2007;159:78–85. doi: 10.1016/j.jneumeth.2006.06.022. [DOI] [PubMed] [Google Scholar]
- Samoszuk M, Tan J, et al. Breast Cancer Res. 2005;7:R274–283. doi: 10.1186/bcr995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Skelley AM, Kirak O, et al. Nat. Methods. 2009;6:147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires TM, Quake SR. Rev. Mod. Phys. 2005;77:977–1026. [Google Scholar]
- Studer V, Hang G, et al. J. Appl. Phys. 2004a;95:393–398. [Google Scholar]
- Studer V, Jameson R, et al. Microelectron. Eng. 2004b;73–74:852–857. [Google Scholar]
- Takayama S, McDonald JC, et al. Proc. Natl. Acad. Sci. USA. 1999;96:5545–5548. doi: 10.1073/pnas.96.10.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang MD, Golden AP, et al. J. Am. Chem. Soc. 2003;125:12988–12989. doi: 10.1021/ja037677h. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, et al. Nat. Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AM, Dieterich DC, et al. Neuron. 2010;66:57–68. doi: 10.1016/j.neuron.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen T, Maerkl SJ, et al. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- Toepke MW, Beebe DJ. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- Unger MA, Chou HP, et al. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Walker GM, Beebe DJ. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- Whitesides GM, Ostuni E, et al. Annu. Rev. Biomed. Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- Yeon JH, Park JK. Biochip J. 2007;1:17–27. [Google Scholar]
- Yuan Y, Hilliard G, et al. J. Biol. Chem. 2003;278:15911–15916. doi: 10.1074/jbc.M300463200. [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, et al. J. Cell Biol. 2003;161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]