Abstract
Adult human brain expresses six isoforms of tau protein as a result of alternative splicing. Alternative splicing of exon 10 (E10) leads to tau isoforms containing either three (3R) or four (4R) microtubule-binding repeats. Imbalance in the 3R-tau/4R-tau ratio causes neurofibrillary degeneration and dementia. Here, we demonstrated that the dual-specificity tyrosine phosphorylation–regulated kinase 1A (Dyrk1A) interacted with the splicing factor 9G8 and phosphorylated it at several serine residues. Dyrk1A itself promoted tau E10 inclusion, whereas 9G8 inhibited E10 inclusion, and these actions were variable depending on the cell types. Co-expression of Dyrk1A and 9G8 led to their translocation from the nucleus to the cytoplasm and suppressed their ability to regulate tau exon 10 splicing. This action is probably due to their interaction-induced translocation from the nucleus, where the regulation of tau E10 splicing occurs, to the cytoplasm. These findings provide novel insights into the molecular mechanism of the regulation of tau E10 splicing and further our understanding of the neurodegeneration caused by dysregulation of tau E10 splicing.
Keywords: Tau, alternative splicing, 9G8, Dyrk1A, neurodegeneration
1. Background
Tau is a major neuronal microtubule-associated protein and plays an important role in the assembly and stabilization of microtubules. It is enriched in the axons of mature and growing neurons. Abnormal hyperphosphorylation and accumulation of this protein into neurofibrillary tangles (NFTs) in neurons, first discovered in Alzheimer disease (AD) brain (Grundke-Iqbal et al., 1986a; Grundke-Iqbal et al., 1986b), is now known to be a characteristic change of several related neurodegenerative disorders that are collectively called tauopathies (Ballatore et al., 2007).
Adult human brain expresses six isoforms of tau generated by alternative splicing of its pre-mRNA from a single gene, which contains 16 exons (Andreadis et al., 1992; Goedert et al., 1989). Exclusion or inclusion of tau exon 10 (E10), which encodes the second microtubule-binding repeat, gives rise to tau isoforms with three (3R) or four (4R) microtubule-binding repeats, respectively. In normal adult human brain, approximately equal levels of 3R-tau and 4R-tau are expressed (Goedert et al., 1989; Kosik et al., 1989). Several specific mutations in the tau gene associated with frontotemporal dementia with Parkinsonism linked to chromosome 17 (FTDP-17) lead to dysregulation of tau E10 splicing and result in a selective increase in either 3R-tau or 4R-tau, suggesting that a disruption of the normal ratio of 3R-tau/4R-tau can cause neurodegeneration and dementia (Goedert and Jakes, 2005; Liu and Gong, 2008; Wolfe, 2009). An altered 3R-tau/4R-tau ratio in the brain is also seen in several other neurodegenerative disorders, such as Pick’s disease, progressive supranuclear palsy, and corticobasal degeneration (Liu and Gong, 2008).
Alternative splicing is controlled by exonic and intronic enhancers and silencers, which in turn are regulated by protein regulators called splicing factors. There are two superfamilies of splicing factors, the SR/SR-like and hnRNP proteins, in mammalian cells (Dreyfuss et al., 2002; Graveley, 2000). Some of the splicing factors are dominant in or restricted to neurons. The SR proteins have a modular structure consisting of one or two copies of an N-terminal RNA-recognition motif (RRM) followed by a C-terminal Arg-Ser–rich (RS) domain, in which the serine residues are targets of phosphorylation by multiple protein kinases.
The function of SR proteins is tightly regulated by phosphorylation. To date, several kinases, including SR-protein kinases 1 and 2 (SRPK1 and SRPK2), the cell cycle-dependent dual specificity kinase (Clk/Sty), Akt/protein kinase B and DNA topoisomerase I (Topo I), have been reported to phosphorylate and regulate SR proteins (Blaustein et al., 2005; Colwill et al., 1996a; 1996b; Gui et al., 1994; Rossi et al., 1996; Wang et al., 1998). We recently found that Dyrk1A (dual-specificity tyrosine phosphorylation-regulated kinase 1A) phosphorylates splicing factor ASF and inhibits its ability to promote tau E10 inclusion (Shi et al., 2008).
One of the SR proteins, 9G8, is believed to be involved in both constitutive splicing and alternative splicing of many pre-mRNAs. In addition, it helps mRNA export to the cytoplasm and enhances the expression of unspliced mRNA (Swartz et al., 2007). A recent study has demonstrated that 9G8 may directly interact with the proximal downstream intron of E10, a clustering region of FTDP mutations, and inhibit tau E10 inclusion (Gao et al., 2007). Like the localization and functions of other SR proteins, the localization and functions of 9G8 are tightly regulated by phosphorylation.
Here, we show that Dyrk1A interacted with and phosphorylated 9G8 in vitro and in live cells. The phosphorylation regulated 9G8’s activity to regulate tau E10 splicing. Dyrk1A also regulated tau E10 splicing in a mechanism that is independent of 9G8 in HEK293 cells. Over-expression of both Dyrk1A and 9G8 led to their translocation from the nucleus to the cytoplasm and thus diminished their effects on tau E10 splicing.
2. Materials and methods
2.1. Plasmids, proteins and antibodies
Recombinant rat Dyrk1A and mammalian expression vector pcDNA3 containing either rat Dyrk1A or kinase-dead Dyrk1AK188R were kindly provided by Dr. Y-W Hwang of the New York State Institute for Basic Research in Developmental Disabilities and prepared as described previously (Chen-Hwang et al., 2002). pCEP4/9G8 tagged with HA (hemagglutinin tag) at C-terminal was a gift from Dr. Tarn of the Institute of Biomedical Sciences, Academia Sinica, Taiwan. pCI-SI9/LI10 or pCI-SI9/SI10, containing a tau minigene SI9/LI10 or SI9/SI10, comprising tau exons 9, 10 and 11 and part of intron 9 and the full length of intron 10 or part of intron 10 was described previously (Yu et al., 2004). Monoclonal antibody 8D9 was raised against a histidine-tagged protein containing the first 160 amino acid residues of rat Dyrk1A (Wegiel et al., 2004). The monoclonal anti-HA, anti-α-tubulin, and anti-β-actin antibodies were bought from Sigma (St. Louis, MO, USA). Peroxidase-conjugated anti-mouse and anti-rabbit IgG were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA, USA); anti-9G8 antibodies, tetramethylrhodamine isothiocyanate (TRITC)–conjugated goat anti-rabbit IgG, and Fluorescein isothiocyanate (FITC)–conjugated goat anti-mouse IgG were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The ECL Kit was from Amersham Pharmacia (Amersham Bioscience, Piscataway, NJ, USA), and [γ-32P]ATP was from MP Biomedicals (Irvine, CA, USA).
2.2. Cell culture and transfection
COS-7, N2a, HEK-293 and HEK-293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) at 37 °C. All transfections were performed in triplicates with Lipofactamine 2000 (Invitrogen, Carlsbad, CA, USA) or FuGENE HD (Roche Diagnotics, Indianapolis, In, USA) for N2a cells in 12-well plates. The cells were transfected according to the manufacturer’s protocols. For co-expression experiments, 2.4 μg of total plasmid was used, containing 0.8 μg E10 splicing vector, 0.8 μg 9G8 vectors and 0.8 μg Dyrk1A vector or their control vectors. For N2a cells, after transfection, the cells were differentiated with 20 mM retinoic acid in DMEM supplemented with 2% fetal bovine serum. All transfection experiments were repeated at least three times.
2.3. Knockdown of 9G8 with RNA interference
For inhibition of 9G8 expression, N2a cells cultured in 12-well plates were transfected with short interfering RNA (siRNA) (Santa Cruz, CA, USA) using FuGENE HD. After transfection, the cells were differentiated for 3 days and lysed. The protein and RNA were extracted as described above. The same amount of scramble siRNA was used for controls.
2.4. GST Pull-down
pGEX-4T/9G8 was constructed by PCR amplification from pCEP4/9G8 and subcloned into pGEX-4T to express GST-9G8 fusion protein. GST or GST-9G8 was purified by affinity purification with glutathione-Sepharose, but without elution from the beads. These beads coupled GST or GST-9G8 was incubated with crude extract from rat brain homogenate in buffer (50 mM Tris-HCl, pH 7.4, 8.5% sucrose, 50 mM NaF, 1 mM Na3VO4, 50 nM okadaic acid [OA], 0.1% Triton X-100, 2 mM EDTA, 1 mM PMSF, 10 μg/ml aprotinin, 10 μg/ml leupeptin and 10 μg/ml pepstatin). After 4 h incubation at 4 °C, the beads were washed with washing buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Triton-X 100, 1 mM PMSF, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 1 mM DTT) 6 times, the bound proteins were eluted by boiling in Laemmli sample buffer and the samples were subjected to Western blot analysis.
2.5. In vitro phosphorylation of 9G8 by Dyrk1A
For in vitro 9G8 phosphorylation by Dyrk1A, GST-9G8 or GST (0.2 mg/ml) was incubated with various concentrations of Dyrk1A in a reaction buffer consisting of 50 mM Tris-HCl (pH 7.4), 10 mM β-mercaptoethanol, 0.1 mM EGTA, 10 mM MgCl2 and 0.2 mM [γ-32P]ATP (500 cpm/pmol). After incubation at 30 °C for 30 min, the reaction was stopped by adding equal volume of 2 × Laemmli sample buffer and boiling. The reaction products were separated by SDS-PAGE. Incorporation of 32P was detected by exposure of the dried gel to phosphor-image system.
2.6. Phosphorylation of 9G8 by Dyrk1A in cultured cells
HEK293T cells were transfected with pCEP4-9G8-HA and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. After a 45-h transfection, the medium was replaced with [32P]orthophosphate (10 mCi) in Dulbecco’s modified Eagle’s medium (-phosphate) supplemented with 10% fetal bovine serum plus 20 μM of either Tg003 or harmine, or both. After a 3-h incubation, the cells were harvested in lysate buffer (50mM Tris-HCl, pH 7.4, 150 mM NaCl, 50 NaF, 1 mM Na3VO4, 50 nM okadaic acid, 0.1% Triton X-100, 0.1% Nonidet P-40, 0.25% sodium deoxychalate, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride and 10 μg/ml each of aprotinin, leupeptin and pepstatin). Insoluble materials were removed by centrifugation, and the supernatant was used for immunoprecipitation by using anti-HA, as described below. Immunoprecipitated ASF-HA was resolved in SDS-PAGE and analyzed by immunoblots and autoradiography.
2.7. Co-immunoprecipitation
COS7 cells were co-transfected with pCEP4-9G8-HA and pcDNA3-Dyrk1A for 48 h as described above. The cells were washed twice with PBS, and lysed by sonication in lysate buffer (50 mM Tris-HCl, pH7.4, 8.5% sucrose, 50 mM NaF, 0.1% Triton X-100, 0.1% NP-40, 0.25% sodium deoxychalate, 2 mM EDTA, 1 mM PMSF, 50 nM OA and 10 μg/ml of aprotinin, leupeptin and pepstatin). Insoluble materials were removed by centrifugation; the supernatants were pre-absorbed with protein-G–conjugated agarose beads and incubated with anti-HA or anti-Dyrk1A antibody 8D9 overnight at 4 °C, and then protein G beads were added. After 4 h incubation at 4 °C, the beads were washed with lysate buffer twice and with TBS twice, and bound proteins were eluted by boiling in Laemmli sample buffer. The samples were subjected to Western blot analysis with the indicated primary antibodies.
2.8. Co-localization study
COS7 cells were plated in 12-well plates onto coverslips one day prior to transfection at 50–60 % confluence. HA-tagged 9G8 constructs were singly transfected or co-transfected with Dyrk1A or Dyrk1Ak188R constructs as described above. Two days after transfection, the cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. After being washed with PBS, the cells were blocked with 10% goat serum in 0.2% Triton X-100-PBS for 2 h at 37 °C, and incubated with rabbit anti-HA antibody (1:200) and mouse anti-Dyrk1A (8D9, 1:10000) overnight at 4 °C. After washing and incubation with secondary antibodies (TRITC- conjugated goat anti-rabbit IgG and FITC-conjugated goat anti-mouse IgG, 1:200), the cells were washed extensively in PBS and incubated with 5 μg/ml Hoechst 33342 for 15 min at room temperature. The cells were washed with PBS, mounted with Fluoromount-G, and revealed with a Leica TCS-SP2 laser-scanning confocal microscope.
2.9. Quantitation of tau E10 splicing by reverse transcription-PCR (RT-PCR)
Total cellular RNA was isolated from cultured cells by using the RNeasy Mini Kit (Qiagen, GmbH, Germany). One microgram of total RNA was used for first-strand cDNA synthesis with Oligo-(dT)15–18 by using Omniscript Reverse Transcription Kit (Qiagen, GmbH, Germany). PCR was performed by using the PrimeSTAR™ HS DNA Polymerase (Takara Bio Inc., Otsu, Shiga, Japan) with primers (forward 5′-GGTGTCCACTCCCAGTTCAA-3′ and reverse 5′-CCCTGGTTTATGATGGATGTTGCCTAATGAG-3′) to measure alternative splicing of tau E10 under conditions: 98 °C for 3 min, 98 °C for 10 sec and at 68 °C for 40 sec for 30 cycles and then 68 °C 10 min for extension. The PCR products were resolved on 1.5% agarose gels and quantitated using the Molecular Imager system (Bio-Rad, Hercules, CA, USA). All PCRs were carried out in triplicates.
2.10. Mass spectrometry
GST-9G8 fusion protein was phosphorylated by Dyrk1A as described above. In order to maximize the yield of the phosphorylated protein, the reaction was carried out for 1 h with high amounts of the kinase (6:1 molar ratio of GST-9G8 and GST-Dyrk1A). The phosphorylated products were separated in SDS-PAGE and stained by Coomassie blue. The GST-9G8–containing gel piece was in-gel tryptic–digested. Proteolytic peptides were extracted from the gel, followed by TiO2 IMAC enrichment for the phosphopeptides. The resulting fraction was concentrated and reconstituted in 10 μl of 5% formic acid for LC-MS/MS analysis.
2.11. Statistical analysis
When appropriate, the data are presented as the means ± S.D. Data points were compared by the unpaired two-tailed Student’s t test, and the calculated p-values are indicated in the figures.
3. Results
3.1. 9G8 suppresses tau exon 10 inclusion
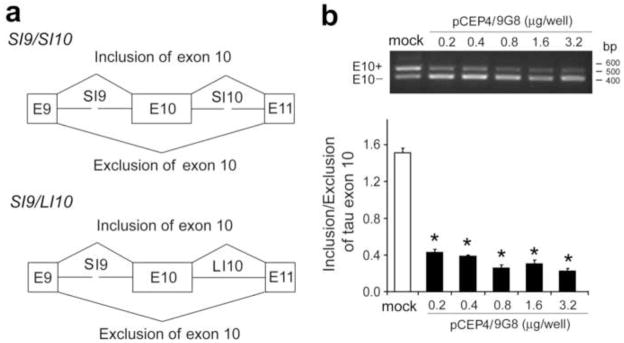
To confirm 9G8’s regulating role in tau E10 splicing in our experimental system, we co-transfected mini-tau gene pCI-SI9/SI10, consisting of tau exons 9–11, part of intron 9 (SI9) and intron 10 (SI10) (Fig. 1a) (Yu et al., 2004), together with various amounts of pCEP4-9G8 into COS7 cells and detected the inclusion and exclusion of E10 by reverse-transcription (RT)-PCR. We found that 9G8 inhibits E10 inclusion dose-dependently (Fig. 1b). This effect was not cell-type–specific, as over-expression of 9G8 in other types of cells, including HEK293T, HeLa and SH-SY5Y, also down-regulated E10 inclusion (data not shown).
Fig. 1.
Over-expression of 9G8 suppresses tau exon 10 inclusion. (a) Diagrams of mini-tau genes, pCI-SI9/SI10, comprising tau exons 9, 10 and 11 and part of intron 9 and intron 10, and pCI-SI9/LI10, containing a full length of intron 10. (b) Over-expression of 9G8 inhibited tau E10 inclusion concentration dependently. COS7 cells were co-transfected with pCI-SI9/SI10 and various amounts of pCEP4/9G8 for 48 h. Total RNA was subjected to RT-PCR for measurement of tau E10 splicing. The relative ratios of E10+/E10− were calculated and are shown in the graph. “Con” indicates cells transfected with pCI-SI9/SI10 alone. “pCEP4” indicates co-transfection with pCI-SI9/SI10 and the empty vector pCEP4.
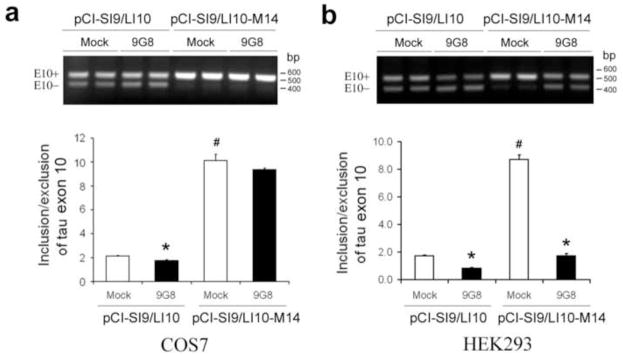
A previous study suggests that 9G8 may inhibit tau E10 inclusion by binding to the proximal downstream intron of E10, a clustering region for mutations seen in FDDP-17 (Gao et al., 2007) and that a “c” at +14 is required for the effect. Thus, we overexpressed 9G8 with wild type (WT) mini-tau gene, pCI-SI9/LI10, or M14-mini-tau gene (pCI-SI9/LI10-M14) (in the latter of which “c” at +14 was mutated to “t”) into COS7 cells, and then measured the alternative splicing of E10 by RT-PCR. We found that, consistent with the previous study (Gao et al., 2007), E10 inclusion in the M14-mini-tau–transfected cells was dramatically increased (~5-fold) as compared to the WT-mini-tau–transfected cells and that 9G8 co-expression did not inhibit the E10 inclusion significantly in the M14-mini-tau–transfected cells as it did in the WT-mini-tau–transfected cells (Fig. 2a). Surprisingly, in contrast to COS7 cells, 9G8 over-expression suppressed dramatically the E10 inclusion caused by the mutation at +14 of the intron 10 in HEK293 cells (Fig. 2b), suggesting that the “c” at this position is not required for the regulation of tau E10 splicing by 9G8 in HEK293 cells.
Fig. 2.
Effects of 9G8 on tau exon 10 inclusion when intron 10 at +14 was mutated. PCI-SI9/LI10 and pCI-SI9/LI10-M14 were cotransfected with pCEP4/9G8 into COS7 cells (a) and HEK293 cells (b), and the total RNA was subjected to RT-PCR for measurement of tau E10 splicing after 48 h transfection.
3.2. Dyrk1A phosphorylates 9G8 in vitro and in cultured cells
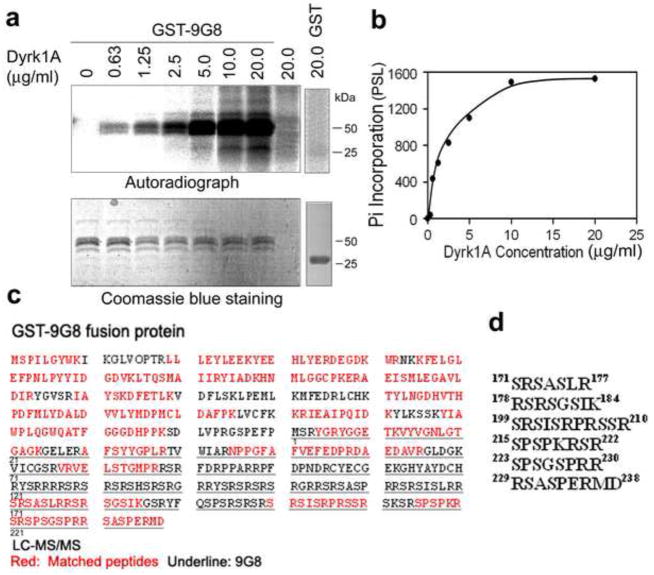
The biological activity of 9G8 is tightly regulated by its phosphorylation. To study whether Dyrk1A phosphorylates 9G8, we incubated GST-9G8 with Dyrk1A in vitro and found that GST-9G8, but not GST, was phosphorylated in an enzyme concentration–dependent manner (Fig. 3a, b).
Fig. 3.
Dyrk1A phosphorylates 9G8 in vitro. (a and b) Phosphorylation of 9G8 by Dyrk1A. GST-9G8 or GST (0.2 mg/ml) was incubated with the indicated concentrations of Dyrk1A at 30 °C for 30 min, and the reaction products were separated by SDS-PAGE. Incorporation of 32P into GST-9G8 or GST was detected by autoradiogram (a), and the quantifications are shown in (b). (c) The amino acid sequence of GST-9G8. The red-colored sequences represent those detected by LC-MS/MS. The underlined sequence indicates the 9G8 sequence, which is numbered underneath. (d) Six phospho-peptides from the Dyrk1A-phosphorylated 9G8 samples detected by LC-MS/MS.
To map the phosphorylation sites of 9G8 by Dyrk1A, we phosphorylated GST-9G8 with high-concentration Dyrk1A (130 μg/ml, an enzyme/substrate molar ratio of ~1:6) for 60 min. The non-phosphorylated and the phosphorylated GST-9G8 were resolved by SDS-PAGE, followed by in-gel trypsin digestion and LC-MS/MS. We identified six phospho-peptides from the phosphorylated GST-9G8 digest, which were all located at the C-terminal half of 9G8 molecule (Fig. 3c, d). Because no threonine residue was present in these six phosphor-peptides, our results indicate that Dyrk1A phosphorylates 9G8 at at least six serine residues of the C-terminal half of the molecule in vitro.
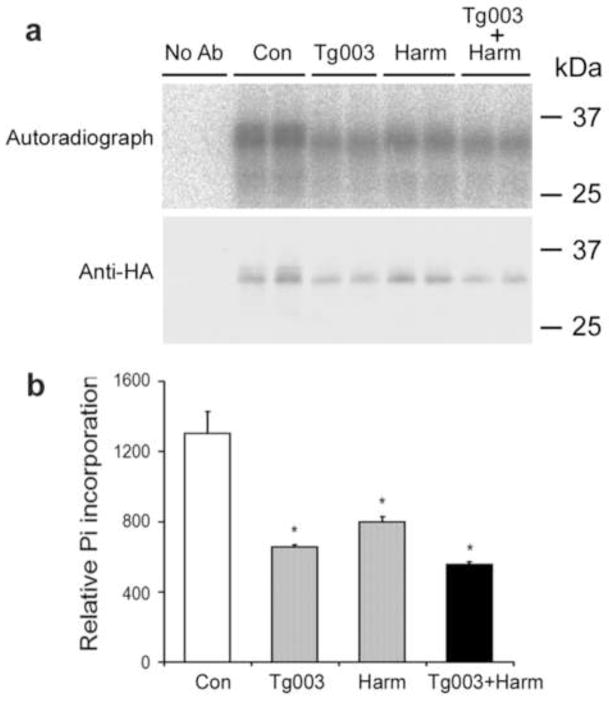
To study if Dyrk1A also phosphorylates 9G8 in live cells, we over-expressed pCEP4-9G8 in HEK293T cells and then labeled the cells with [32P]orthophosphate for 48 h. Autoradiograph of the affinity-purified 9G8 showed strong radioactive signals of double bands, suggesting phosphorylation of 9G8 to different extents in the cells (Fig. 4). When we treated the transfected HEK293T cells with Tg003, an inhibitor of Clk/Sty that is a known 9G8 kinase (Colwill et al., 1996b), we observed a ~50% reduction of the radioactive signals, as expected. Approximately 40% of inhibition was also achieved when the cells were treated with harmine, a specific Dyrk1A inhibitor (Bain et al., 2007). A combination of both inhibitors induced further inhibition of 9G8 phosphorylation (Fig. 4b). These results indicated that 9G8 was phosphorylated by Dyrk1A in live cells.
Fig. 4.
Dyrk1A phosphorylates 9G8 in cultured cells. HEK293T cells were transfected with pCEP4-9G8-HA for 45 h and then treated with 20 μM of either Tg003 or harmine (harm), or both. At the same time, [32P]orthophosphate was added to label the phosphoproteins. After a 3-h treatment and phospholabeling, the cells were harvested, and the cell lysates were subjected to immunoprecipitation with anti-HA. The immunoprecipitated 9G8-HA was resolved in SDS-PAGE and analyzed by autoradiography and Western blot with anti-HA (a). In quantitation of 9G8 phosphorylation (b), the 32P incorporated into 9G8-HA was normalized by the 9G8-HA level detected with anti-HA. *, p<0.05 vs. control group.
3.3. 9G8 interacts with Dyrk1A
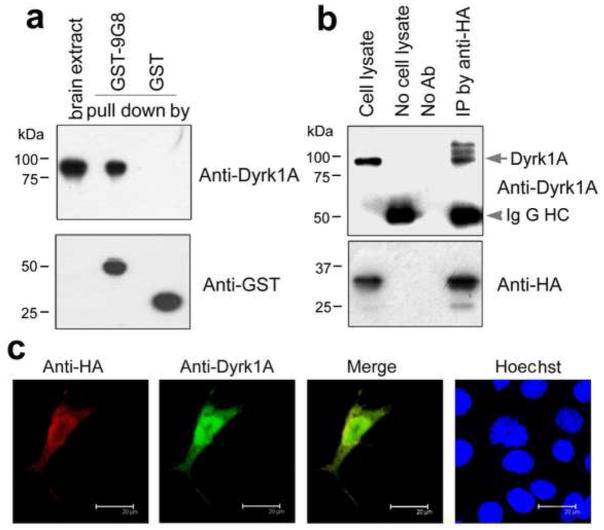
We investigated whether 9G8 and Dyrk1A interact with each other. By employing a GST-pulldown assay, we found that GST-9G8 pulled down Dyrk1A from rat brain extract (Fig. 5a). As a negative control, GST itself did not pull down any Dyrk1A. These results suggest an interaction between 9G8 and Dyrk1A. We also performed co-immunoprecipitation studies of the cell lysates of the COS7 cells that over-expressed both HA-tagged 9G8 and Dyrk1A. We found that Dyrk1A was co-immunoprecipitated by antibody to HA (Fig. 5b). These results confirm the interaction between 9G8 and Dyrk1A in vitro.
Fig. 5.
9G8 interacts with Dyrk1A. (a) Pull-down of Dyrk1A from rat brain extract by GST-9G8. GST-9G8 or GST coupled onto glutathione-Sepharose beads was incubated with rat brain extracts. After washing, the bound proteins were subjected to Western blots by using anti-GST and anti-Dyrk1A, respectively. (b) Co-immunoprecipitation of Dyrk1A with 9G8. HA-tagged 9G8 and Dyrk1A were co-expressed in COS7 cells for 48 h. The cell extract was incubated with anti-HA, and then protein G beads were added into the mixture. The bound proteins were subjected to Western blots by using antibodies indicated at the right of each blot. (c) Co-localization of 9G8 with Dyrk1A. HA-9G8 and Dyrk1A were co-transfected into COS7 cells. After 48 h transfection, the cells were fixed and immunostained by polyclonal anti-HA and monoclonal anti-Dyrk1A, followed by TRITC-anti-rabbit IgG (red) and FITC-anti-mouse IgG (green), respectively. Hoechst (blue) was used for nuclear staining.
To study their interaction in live cells, we co-expressed HA-tagged 9G8 and Dyrk1A in COS7 cells and examined their subcellular localization by using confocal microscopy. We found that 9G8 and Dyrk1A were co-localized in the cytoplasm and nucleus (Fig. 5c). These results provide evidence of their interaction in live cells.
3.4. Interaction between Dyrk1A and 9G8 suppresses their ability to regulate tau exon 10 splicing
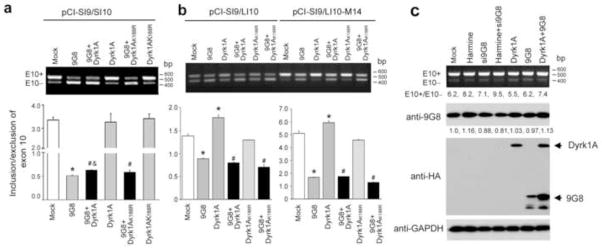
To study the effects of interactions between Dyrk1A and 9G8 on their roles in the regulation of tau E10 splicing, we overexpressed Dyrk1A and 9G8 individually and in combination in the COS7 cells transfected with pCI-SI9/SI10. Splicing products of E10 inclusion and exclusion were then quantitated by RT-PCR. We found that over-expression of 9G8 alone inhibited tau E10 inclusion dramatically, but over-expression of Dyrk1A alone did not have any effect on tau E10 splicing (Fig. 6a). Co-over-expression of Dyrk1A with 9G8 prevented slightly, but significantly, 9G8’s inhibition of tau E10 inclusion. To investigate whether the observed effect on E10 inclusion resulted from 9G8 phosphorylation by Dyrk1A, we co-transfected cells with 9G8 and the kinase-dead Dyrk1A (Dyrk1AK188R). We found that co-transfection with Dyrk1AK188R did not affect tau E10 splicing significantly, as compared to transfection with 9G8 alone. These results suggest that Dyrk1A-induced phosphorylation of 9G8 suppresses slightly its role in inhibiting tau E10 inclusion.
Fig. 6.
Role of Dyrk1A and 9G8 in the regulation of tau exon 10 splicing. Mini-tau gene pCI-SI9/SI10 (a), pCI-SI9/LI10 or pCI-SI9/LI10-M14 (b) was co-transfected with 9G8 and either Dyrk1A or Dyrk1AK188R into COS7 cells (a) or HEK293 cells (b). The total RNA was extracted 48 h later and subjected to measurement of tau E10 splicing by using RT-PCR. * p < 0.05 versus control group; # p < 0.05 versus Dyrk1A or group; and & p < 0.05 versus 9G8 Dyrk1AK188R group. (c) N2a cells were transfected with Si9G8, pCI-Neo/HA-Dyrk1A-Flag or pCEP4-9G8-HA for 48 h and cultured in the presence or absence of 20 μM harmine for additional 24 h. The cells were then harvested for measuring tau E10 splicing by RT-PCR and determination of the expression of 9G8 and Dyrk1A by Western blots. GAPDH blot was included as a loading control. The numbers under the upper panel indicate the ratios of E10+/E10−. The numbers of the anti-9G8 blot indicate the relative intensities of the 9G8 bands.
The mRNA splicing is regulated differently in different cell types. Thus, we further investigated the effects of co-expression of 9G8 and Dyrk1A on tau E10 splicing in SI9/LI10- or SI9/LI10-M14-transfected HEK293 cells. We found that 9G8 suppressed tau E10 inclusion in HEK293 cells, as it did in COS7 cells (Fig. 6b). However, unlike in COS7 cells, overexpression of active Dyrk1A, but not dead Dyrk1AK188R, promoted tau E10 inclusion. When 9G8 and Dyrk1A were co-transfected, tau E10 splicing was similar to that in HEK293 cells transfected with 9G8 alone, suggesting that Dyrk1A, either active or inactive, does not significantly affect 9G8’s inhibition of E10 inclusion in HEK293 cells. However, the active Dyrk1A-induced increase in E10 inclusion was blocked by co-transfection with 9G8. Taken together, these results suggest that in HEK293 cells, Dyrk1A also regulates tau E10 splicing in a mechanism dependent on its kinase activity, but not on 9G8, and that the interaction of 9G8 with Dyrk1A blocks this action of Dyrk1A.
Because tau is mainly expressed in the neuron and because altered tau E10 splicing leads to neurodegeneration, we investigated the role of 9G8 and Dyrk1A in alternative tau E10 splicing in neurons. This was achieved by studying endogenous tau E10 splicing 3 days after up- and down-regulation of 9G8 and Dyrk1A in differentiated N2a cells. We found that the differentiated N2a cells expressed much more 4R-tau mRNA than 3R-tau mRNA (the ratio of E10 inclusion over E10 exclusion was 6.2) (Fig. 6c). Inhibition of Dyrk1A by harmine promoted tau E10 inclusion. In contrast, over-expression of Dyrk1A suppressed E10 inclusion. Knockdown of 9G8 expression also enhanced tau E10 inclusion slightly, but transfection of 9G8 gene did not affect the alternative splicing. Down-regulation of 9G8 and Dyrk1A increased E10 inclusion synergistically. However, over-expression of both Dyrk1A and 9G8 also increased E10 inclusion. These results suggest that Dyrk1A activity suppresses tau E10 inclusion in neuronal cells. However, these results could not explain the role of 9G8 and the interactions between 9G8 and Dyrk1A in regulation of tau E10 splicing because of the apparent inconsistent results.
To further understand the results above, we determined the levels of 9G8 and Dyrk1A and found that siRNA of 9G8 knocked down the 9G8 expression successfully, but transfection with 9G8 vector failed to induce an increase in the 9G8 level (Fig. 6c). These results could explain why transfection with 9G8 vector did not affect tau E10 splicing. Interestingly, we observed that co-transfection of 9G8 and Dyrk1A resulted in a higher level of expression, as compared to transfection with 9G8 or Dyrk1A alone (Fig. 6c). These data suggest that Dyrk1A and 9G8 overexpression promoted each other’s expression, but inhibited their role in suppression of E10 inclusion, resulting in an increase of 4R-tau expression (see below for explanation).
3.5. Interaction between Dyrk1A and 9G8 drives them from the nucleus into the cytoplasmic compartment
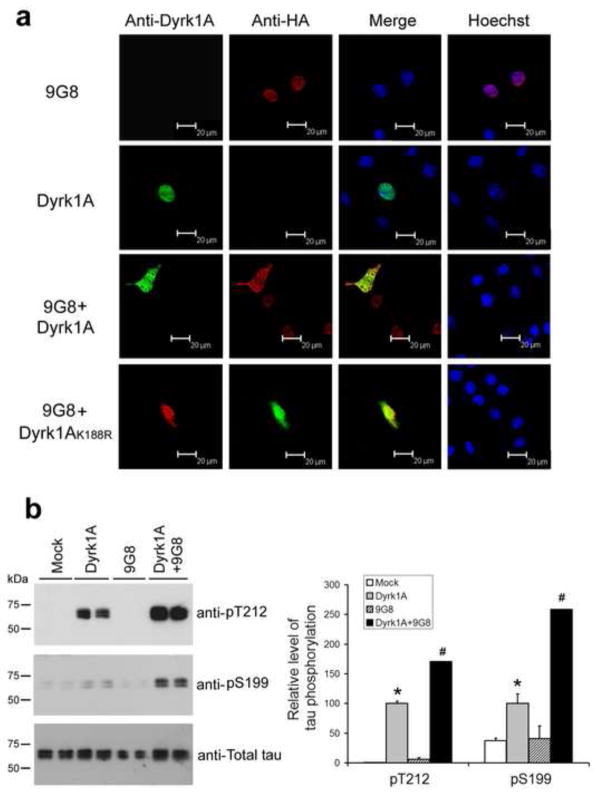
To investigate the possible mechanism by which Dyrk1A and 9G8 suppress their biological activity in regulation of tau E10 splicing, we determined the subcellular localization of 9G8 and Dyrk1A. We over-expressed 9G8, Dyrk1A or kinase-dead Dyrk1AK188R alone or in combination in COS7 cells. By employing laser confocal microscopy, 9G8 and Dyrk1A were found to be localized exclusively in the cell nucleus when they were transfected separately (Fig. 7a). However, when 9G8 and Dyrk1A were co-expressed, both were translocated into the cytoplasm, and these two proteins were localized more in the cytoplasm than in the nucleus (Fig. 7a). To our surprise, co-expression of 9G8 with the inactive Dyrk1AK188R also led to their translocation into the cytoplasm. We also determined the in situ phosphorylation of tau, a cytoplasmic substrate of Dyrk1A, and found more tau phosphorylation after co-transfection with Dyrk1A and 9G8 than after transfection with Dyrk1A alone (Fig. 7b). These results are consistent to the 9G8-induced export of Dyrk1A. Taken together, our results suggest that Dyrk1A interacts with 9G8, drives them from the nucleus into the cytoplasm, and prevents them from regulating tau E10 splicing. This role of Dyrk1A appears to be partially independent on its kinase activity.
Fig. 7.
Co-expression of Dyrk1A or Dyrk1AK188R with 9G8 drives their translocalization from the nucleus into the cytoplasm. (a) COS7 cells were transfected, as indicated at the left of the figure. After 48 h transfection, the cells were fixed and immunostained by monoclonal anti-Dyrk1A and polyclonal anti-HA, followed by FITC-anti-mouse IgG (green) and TRITC-anti-rabbit IgG (red), respectively. Hoechst (blue) was used for nucleus staining. (b) HEK293FT cells were transfected with pCI-Neo/HA-Dyrk1A-Flag, pCEP4-9G8-HA and pCI-Neo/tau 40 for 48 h. The cells were then collected and the cell lysates were subjected to Western blots for measuring tau phosphorylation at T212 and S199. The blots were also quantified, and the data after being normalized with the total tau level are shown in graph at the right side. Data are presented as mean±SD (n=4). * P<0.05 versus mock control; # P<0.05 versus Dyrk1A alone.
4. Discussion
Recent studies have demonstrated an important role of proper regulation of tau E10 alternative splicing, which leads to approximately equal levels of 3R-tau and 4R-tau in the adult human brain (for reviews, see (Andreadis, 2005; D’Souza and Schellenberg, 2005; Goedert and Jakes, 2005)), in neuronal survival and function. The discovery of several mutations in intron 10 of the TAU gene, which alter the alternative splicing of tau E10 but do not change the primary sequence of tau protein, in familial FTDP-17 indicates that alterations of 3R-tau/4R-tau ratio alone can cause neurodegeneration and dementia (D’Souza et al., 1999; Gao et al., 2000; Grover et al., 1999; Hutton et al., 1998; Stanford et al., 2003). Dysregulation of E10 splicing may also contribute to the pathogenesis of several other tauopathies, such as Pick disease, progressive supranuclear palsy, corticobasal degeneration, and Down syndrome, because altered 3R-tau/4R-tau ratio is observed in the brains of individuals with these disorders (for review, see (Liu and Gong, 2008). Altered tau E10 splicing also causes neurodegeneration in experimental animals. For example, overexpression of human 3R-tau, but not 4R-tau, in mice (where 4R-tau is the major tau isoform in adult mouse brain) produces age-dependent tauopathy (Gotz et al., 1995; Ishihara et al., 1999). Thus, investigation of the regulation of tau E10 splicing and of the mechanisms leading to dysregulation of tau E10 splicing, which remains largely elusive, is highly important.
The alternative splicing of tau E10 is regulated by several regulatory sequences (cis-elements) within exon 10 and intron 10 of TAU gene, as well as several trans-acting splicing factors (for review, see (Liu and Gong, 2008)). We recently demonstrated that ASF is the major splicing factor for regulating tau E10 splicing, and its activity to promote E10 inclusion is inhibited by its phosphorylation with Dyrk1A (Shi et al., 2008). How 9G8 regulates tau E10 alternative splicing was less known, and the role of Dyrk1A in 9G8-mediated E10 splicing has not been reported. In the present study, we found that, in contrast to ASF, 9G8 suppressed tau E10 inclusion in several types of cells and that 9G8’s role in regulation of tau E10 splicing was modulated by Dryk1A.
A recent study reported that 9G8 may interact with the proximal downstream intron of tau E10 directly, resulting in inhibition of tau E10 inclusion (Gao et al., 2007). The cytosine (C) at the position +14 downstream of E10 is thought to be critical for interaction between 9G8 and the tau pre-mRNA, and the FTDP-17 mutation at this position (C to T) abolishes 9G8 action in COS7 cells (Gao et al., 2007). Such a dependency of 9G8’s action on tau E10 splicing was also observed in COS7 cells in the present study. However, in HEK293 cells, we found that 9G8 suppressed tau E10 inclusion in a manner independent of the presence of cytosine at the +14 position of intron 10, because 9G8 overexpression also suppressed tau E10 inclusion of the M14-mini-tau. These results suggest that the mechanisms by which 9G8 regulates tau E10 splicing and/or the binding sites of 9G8 to TAU gene are different in different cell types. Our results also suggest that there are different or additional binding sites of tau gene for 9G8 in HEK293 cells, which remain to be identified.
Similar to splicing of other exons, cis elements within exon 10 or around its nearby 5′ and 3′ splicing sites in the tau gene play crucial roles in the regulation of its splicing (Gao et al. 2000). It was suggested by using a tau mini-gene construct that distal sequences beyond 300 base pairs upstream or downstream of exon 10 may have little effects on the splicing of exon 10 or that the effects are counteracted among these elements (Gao et al. 2000). In the present study, we used two mini-tau genes, pCI/SI9-SI10 (Gao et al., 2000, Yu et al., 2004) and pCI/SI9-LI10 (Yu et al 2004) and found that SI9/LI10 produced mRNAs that excluded exon 10 more than LI9/SI10 (Figs. 6a, 6b), which is consistent with the previous studies. However, over-expression of 9G8 suppressed exon 10 inclusion in both mini-genes (Figs. 6a, 6b), and down-regulation of 9G8 promoted the 4R-tau expression in endogenous TAU gene. Many splicing factors are known to be regulated by phosphorylation, especially at the SR domains (for review, see (Liu and Gong, 2008)). However, neither the phosphorylation sites nor the kinases for 9G8 phosphorylation have been elucidated. Here, we found, for the first time, that 9G8 is phosphorylated by Dyrk1A at at least six serine residues at the SR domain in the C-terminal half of the molecule. Phosphorylation of 9G8 by Dyrk1A was also seen in cultured cells. Dyrk1A is a Ser/Thr protein kinase that phosphorylates substrate proteins preferably at the R-X-(X)-S/T-P motif. Within the six phosphorylated peptides derived from 9G8 after phosphorylation by Dyrk1A, there are three serine residues (Ser217, Ser223 and Ser232) that match with the R-X-(X)-S/T-P motif, as determined by mass spectrometry (Fig. 3). Thus, these three serine residues are most likely phosphorylated by Dyrk1A. In addition, Dyrk1A also phosphorylates 9G8 at sites other than the R-X-(X)-S/T-P motif.
It appears that phosphorylation of 9G8 affects its activity of regulating tau E10 splicing only slightly and variably in different types of cells. We observed that in COS7 cells, co-transfection of 9G8 with wild type Dyrk1A, but not with inactive Dryk1AK188R, prevented 9G8-induced inhibition of tau E10 inclusion only very slightly, whereas no such prevention was observed in HEK293 cells. Interestingly, we found that Dyrk1A itself, without being co-transfected with 9G8, promoted tau E10 inclusion, and this action was dependent on the kinase activity because no promotion of E10 inclusion was observed with the inactive Dyrk1AK188R. When 9G8 was co-transfected with Dyrk1A, the Dyrk1A’s action in promoting tau E10 inclusion was blocked. Further studies of the subcellular localizations indicated that co-transfection led to interaction between these two proteins, which was independent of the kinase activity, and to their translocation from the nucleus to the cytoplasmic compartment, where they lost their role in regulating tau E10 splicing.
In neuronal N2a cells, we found that over-expression of Dyrk1A suppressed tau E10 inclusion, whereas inhibition of Dyrk1A led to increased E10 inclusion. This phenomenon was different from what was seen in HEK293 and COS7 cells. These observations suggest that the regulation of tau E10 splicing by Dyrk1A is cell type-specific and that in neurons, Dyrk1A over-expression suppresses tau E10 inclusion. We previously found that Dyrk1A suppresses tau E10 inclusion by inhibiting ASF-induced promotion of tau E10 inclusion (Shi et al. 2008). It is thus possible that ASF is the major splicing factor that regulates tau E10 splicing and is regulated by Dyrk1A in the neuron.
Dyrk1A is over-expressed in the brain of individuals with Down syndrome due to trisomy 21, because its gene is localized in the Down syndrome critical region of chromosome 21 (Guimera et al., 1999). Our recent studies indicate an approximately 50% increase in both the protein level and the kinase activity of Dyrk1A in Down syndrome brain, as compared to control brains (Liu et al., 2008; Shi et al., 2008). We have further demonstrated that Dyrk1A phosphorylates ASF and inhibits its activity in the promotion of tau E10 inclusion, leading to an increase in the ratio of 3R-tau/4R-tau and neurodegeneration in Down syndrome brain (Shi et al., 2008). The present study showed that while 9G8 suppressed tau E10 inclusion in all of the cell types that were tested, Dyrk1A’s role in the regulation of tau E10 splicing was cell type-specific. In the neuronal cells, Dyrk1A suppressed tau E10 inclusion. It is clear now that Dyrk1A can regulate tau E10 splicing through multiple mechanisms by interacting and/or phosphorylating various splicing factors. Different mechanisms might dominate in different cell types, leading to different net regulation of tau E10 splicing. In Down syndrome brain, overexpression of Dyrk1A correlates to the increase in 3R-tau/4R-tau ratio (Shi et al., 2008), suggesting that ASF, rather than 8G9, is the major splicing factor that regulates tau E10 splicing in human brain. Consistent with this finding, we observed that ASF had a much stronger effect than 9G8 on tau E10 splicing in culture cells (unpublished observations).
In conclusion, we have demonstrated that Dyrk1A interacted with 9G8 and phosphorylated 9G8 at several serine residues at the SR domain both in vitro and in live cells, and that these two proteins colocalized and co-transported from the nucleus to the cytoplasm. Dyrk1A promoted tau E10 inclusion, whereas 9G8 inhibited E10 inclusion, and these actions were variable, depending on the cell types. Co-expression of Dyrk1A and 9G8 suppresses their ability to regulate tau E10 splicing, probably because their interaction drives their translocation from the nucleus, where the regulation of tau E10 splicing occurs, into the cytoplasm. These findings provide novel insight into the molecular mechanism of the regulation of tau E10 splicing and further our understanding of neurodegeneration caused by dysregulation of tau E10 splicing.
Acknowledgments
We thank Ms. J. Murphy for secretarial assistance and Ms. M. Marlow for editorial assistance. This work was supported in part by Nantong University, by the New York State Office of Mental Retardation and Developmental Disabilities, and by grants from the National Natural Science Foundation of China (30770468 to FL and 30801202 to JS), the Natural Science Foundation of Jiangsu Province, China (BK2009159 to FL), and the U.S. National Institutes of Health (AG027429 to CXG, and AG019158 to KI).
Footnotes
Disclosure statement
We state that there are no potential conflicts of interest, including any financial, personal or other relationships with people or organizations that could inappropriately influence the current study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Shaohong Ding, Email: dingshaohong_2000@yahoo.com.cn.
Jianhua Shi, Email: ntshijianhua@hotmail.com.
Wei Qian, Email: weiqianhuang@hotmail.com.
Khalid Iqbal, Email: khalid.iqbal.ibr@gmail.com.
Inge Grundke-Iqbal, Email: i_g_iqbal@yahoo.com.
Cheng-Xin Gong, Email: chengxin.gong@csi.cuny.edu.
Fei Liu, Email: feiliu63@hotmail.com.
References
- Andreadis A. Tau gene alternative splicing: expression patterns, regulation and modulation of function in normal brain and neurodegenerative diseases. Biochim Biophys Acta. 2005;1739:91–103. doi: 10.1016/j.bbadis.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Andreadis A, Brown WM, Kosik KS. Structure and novel exons of the human tau gene. Biochemistry. 1992;31:10626–10633. doi: 10.1021/bi00158a027. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Caceres JF, Coso OA, Srebrow A. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Hwang MC, Chen HR, Elzinga M, Hwang YW. Dynamin is a minibrain kinase/dual specificity Yak1-related kinase 1A substrate. J Biol Chem. 2002;277:17597–17604. doi: 10.1074/jbc.M111101200. [DOI] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996a;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. Embo J. 1996b;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- D’Souza I, Poorkaj P, Hong M, Nochlin D, Lee VM, Bird TD, Schellenberg GD. Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements. Proc Natl Acad Sci U S A. 1999;96:5598–5603. doi: 10.1073/pnas.96.10.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza I, Schellenberg GD. Regulation of tau isoform expression and dementia. Biochim Biophys Acta. 2005;1739:104–115. doi: 10.1016/j.bbadis.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Gao L, Wang J, Wang Y, Andreadis A. SR protein 9G8 modulates splicing of tau exon 10 via its proximal downstream intron, a clustering region for frontotemporal dementia mutations. Mol Cell Neurosci. 2007;34:48–58. doi: 10.1016/j.mcn.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao QS, Memmott J, Lafyatis R, Stamm S, Screaton G, Andreadis A. Complex regulation of tau exon 10, whose missplicing causes frontotemporal dementia. J Neurochem. 2000;74:490–500. doi: 10.1046/j.1471-4159.2000.740490.x. [DOI] [PubMed] [Google Scholar]
- Goedert M, Jakes R. Mutations causing neurodegenerative tauopathies. Biochim Biophys Acta. 2005;1739:240–250. doi: 10.1016/j.bbadis.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer’s disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- Gotz J, Probst A, Spillantini MG, Schafer T, Jakes R, Burki K, Goedert M. Somatodendritic localization and hyperphosphorylation of tau protein in transgenic mice expressing the longest human brain tau isoform. Embo J. 1995;14:1304–1313. doi: 10.1002/j.1460-2075.1995.tb07116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. Rna. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A, Houlden H, Baker M, Adamson J, Lewis J, Prihar G, Pickering-Brown S, Duff K, Hutton M. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J Biol Chem. 1999;274:15134–15143. doi: 10.1074/jbc.274.21.15134. [DOI] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem. 1986a;261:6084–6089. [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci USA. 1986b;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci U S A. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimera J, Casas C, Estivill X, Pritchard M. Human minibrain homologue (MNBH/DYRK1): characterization, alternative splicing, differential tissue expression, and overexpression in Down syndrome. Genomics. 1999;57:407–418. doi: 10.1006/geno.1999.5775. [DOI] [PubMed] [Google Scholar]
- Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Ishihara T, Hong M, Zhang B, Nakagawa Y, Lee MK, Trojanowski JQ, Lee VM. Age-dependent emergence and progression of a tauopathy in transgenic mice overexpressing the shortest human tau isoform. Neuron. 1999;24:751–762. doi: 10.1016/s0896-6273(00)81127-7. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Orecchio LD, Bakalis S, Neve RL. Developmentally regulated expression of specific tau sequences. Neuron. 1989;2:1389–1397. doi: 10.1016/0896-6273(89)90077-9. [DOI] [PubMed] [Google Scholar]
- Liu F, Gong CX. Tau exon 10 alternative splicing and tauopathies. Mol Neurodegener. 2008;3:8. doi: 10.1186/1750-1326-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Liang Z, Wegiel J, Hwang YW, Iqbal K, Grundke-Iqbal I, Ramakrishna N, Gong CX. Overexpression of Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. Faseb J. 2008;22:3224–3233. doi: 10.1096/fj.07-104539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer G, King A, Rappsilber J, Calvio C, Watson M, Ajuh P, Sleeman J, Lamond A, Mann M. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat Genet. 1998;20:46–50. doi: 10.1038/1700. [DOI] [PubMed] [Google Scholar]
- Rossi F, Labourier E, Forne T, Divita G, Derancourt J, Riou JF, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature. 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhang T, Zhou C, Chohan MO, Gu X, Wegiel J, Zhou J, Hwang YW, Iqbal K, Grundke-Iqbal I, Gong CX, Liu F. Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J Biol Chem. 2008;283:28660–28669. doi: 10.1074/jbc.M802645200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford PM, Shepherd CE, Halliday GM, Brooks WS, Schofield PW, Brodaty H, Martins RN, Kwok JB, Schofield PR. Mutations in the tau gene that cause an increase in three repeat tau and frontotemporal dementia. Brain. 2003;126:814–826. doi: 10.1093/brain/awg090. [DOI] [PubMed] [Google Scholar]
- Stoilov P, Meshorer E, Gencheva M, Glick D, Soreq H, Stamm S. Defects in pre-mRNA processing as causes of and predisposition to diseases. DNA Cell Biol. 2002;21:803–818. doi: 10.1089/104454902320908450. [DOI] [PubMed] [Google Scholar]
- Swartz JE, Bor YC, Misawa Y, Rekosh D, Hammarskjold ML. The shuttling SR protein 9G8 plays a role in translation of unspliced mRNA containing a constitutive transport element. J Biol Chem. 2007;282:19844–19853. doi: 10.1074/jbc.M701660200. [DOI] [PubMed] [Google Scholar]
- Wang J, Xiao SH, Manley JL. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 1998;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegiel J, Imaki H, Wang KC, Wegiel J, Rubenstein R. Cells of monocyte/microglial lineage are involved in both microvessel amyloidosis and fibrillar plaque formation in APPsw tg mice. Brain Res. 2004;1022:19–29. doi: 10.1016/j.brainres.2004.06.058. [DOI] [PubMed] [Google Scholar]
- Wolfe MS. Tau mutations in neurodegenerative diseases. Journal of Biological Chemistry. 2009;284:6021–6025. doi: 10.1074/jbc.R800013200. [DOI] [PubMed] [Google Scholar]
- Yu Q, Guo J, Zhou J. A minimal length between tau exon 10 and 11 is required for correct splicing of exon 10. J Neurochem. 2004;90:164–172. doi: 10.1111/j.1471-4159.2004.02477.x. [DOI] [PubMed] [Google Scholar]
- Zheng ZM. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J Biomed Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]