Abstract
The mammalian cryptome consists of bioactive peptides generated by the proteolysis of precursor proteins. It is speculated that the cryptide repertoire increases the complexity of the proteome by an order of magnitude. Cryptides have been found to function in a wide range of processes including neuronal signaling, antigen presentation, and the inflammatory response. Due to their potential as therapeutic agents, there has been an increasing interest in studying cryptides. In this review, we discuss different approaches for discovering these hidden peptides and how proteomic tools can be utilized to aid in their identification and characterization.
Key words: bioactive peptides, cryptides, cryptome, cryptomics, mass spectrometry-based proteomics
INTRODUCTION: WHAT IS CRYPTOMICS AND WHY IS IT IMPORTANT?
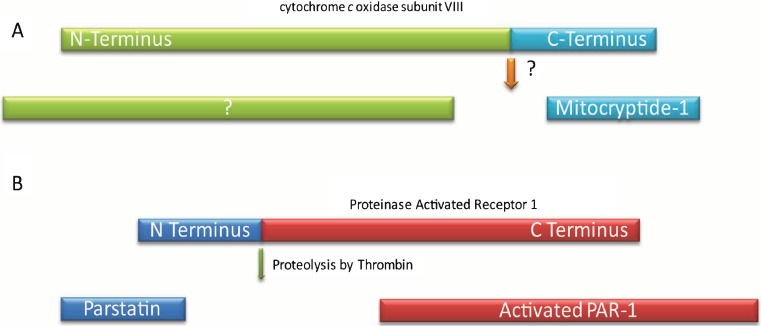
A typical mammalian proteome has tens of thousands of unique proteins. It is staggering to imagine understanding the detailed function of each one. Yet a simple count of the proteins actually underestimates the true complexity of a proteome. Proteins have numerous isoforms. They are modified with a variety of transcriptional and posttranslational modifications. Each variant has a potentially important biological function (1). An added layer of complexity is introduced by proteolytic enzymes that cleave precursor proteins to generate bioactive peptides. These peptides have been termed cryptides, and their study en mass is known as cryptomics (2–4). It is important to note that all peptides generated from proteins are not considered cryptides; a cryptide has a biological activity distinct from its precursor. For example, upon activation of the Notch receptor protein, its cytoplasmic domain is proteolytically cleaved and migrates to the nucleus to regulate the Notch target genes. The cytosolic fragment of Notch is not considered a cryptide since it carries out the main function of the Notch protein (5,6). In contrast, for example, a cryptide is generated when cytochrome c oxidase subunit VIII is cleaved to create mitocryptide-1. The peptide has a distinct function as an activator of neutrophils (Fig. 1a) (7). Another example of a cryptide is parstatin, the N-terminal extracellular domain fragment of proteinase activated receptor 1 (PAR1), which is formed upon cleavage of PAR1 by thrombin. The N-terminal domain has an unexpected function as an inhibitor of angiogenesis and thus has the capability to antagonize the pro-angiogenesis function of its precursor PAR1 (Fig. 1b) (8).
Fig. 1.
a Mitocryptide-1 is the 23-amino acid C-terminal fragment of cytochrome c oxidase subunit VIII originally identified from porcine heart. The protease that cleaves the precursor protein has not been identified yet. Mitocryptide-1 has been found to activate neutrophils. The fate of N-terminal fragment is not known. b Parstatin is the N-terminal fragment of proteinase activated receptor 1 (PAR1). Parstatin is generated when thrombin cleaves PAR1 for its activation. The cryptide has anti-angiogenic activity
The first proof that a bioactive peptide can be the proteolytic product of a larger precursor was provided by the study of proopiomelanocortin, which gives rise to multiple cryptides, including ACTH, β-endorphin, lipotrophins, and melanocyte-stimulating hormones (9–13). Since then a large number of cryptides and their corresponding precursors have been identified along with various proteases that are involved. Recently discovered cryptides are involved in the regulation of a diverse range of cellular processes including neuronal signaling, the inflammatory response, the adaptive immune response, and angiogenesis (Table 1). They are derived from many well-studied proteins, including hemoglobin, cytochromes, laminins, and collagen (2–4,18,19). However, the occurrence and function of cryptides cannot currently be accurately predicted. As such, cryptides are a hidden aspect of the proteome with important yet unknown biological functions. As an evidence of the biodiversity of cryptides, a glycine/leucine-rich bioactive peptide with antimicrobial activity, leptoglycin, has been discovered in the South American frog, although the precursor is still to be identified (14).
Table 1.
Examples of Recently Discovered Cryptides with Their Precursors
| Cryptide | Number of amino acids | Precursor | Cellular role |
|---|---|---|---|
| Leptoglycin | 22 | Unknown | Antimicrobial activity (14) |
| Parstatin | 41 | PAR1 | Inhibition of angiogenesis (8) |
| Prolyl-hydroxyproline | 2 | Collagen | Inhibition of chondrocyte differentiation (15) |
| Mytocryptide-1 | 23 | Cytochrome c oxidase subunit VIII | Activation of neutrophils (7) |
| Mytocryptide-2 | 15 | Cytochrome b | Activation of neutrophils (16) |
| NPNA | 15 | Rat neuropeptide FF precursor | Opioid signaling (17) |
PAR1 proteinase activated receptor 1
Cryptides have been divided into three types by Dominic Autelitano and co-workers (2). A type I cryptide is a bioactive peptide detected in vivo with a function entirely different from that of the precursor. A type II cryptide is a peptide found in vivo with activity related but not necessarily identical to that of its precursor. Finally, a type III cryptide is a bioactive peptide produced in vitro by proteolytic digestion of proteins that may or may not exist in vivo (2). This classification scheme organizes the rapidly evolving area of bioactive peptide research. In this review, we discuss recent progress made since the concept of cryptome was first proposed in 2006. We will also discuss different approaches for the discovery of novel cryptides and how mass spectrometry-based proteomics has been and can be utilized in cryptomic research.
Thus far, cryptides have been identified and characterized individually. They are varied in their origins and functions. Type I cryptides are derived from plasma proteins, extracellular matrix proteins, cytosolic, and mitochondrial proteins, among others. For example, plasminogen, a clotting factor, is cleaved to yield angiostatin, a 38-kDa cryptide that is a potent inhibitor of angiogenesis (20). Collagen VIII is cleaved by metalloproteases to generate endostatin, a 20-kDa cryptide with tumor repression activity.(21). Cleavage of hemoglobin results in a variety of cryptides, including hemopressins and hemorphins. Some of these are involved in neuronal signaling (reviewed in (18,19)) and others have antimicrobial activity (22,23). Type II and type III cryptides have also been found from a range of sources and have equally complex biological functions. An example of a type II cryptide is NPNA. The rat neuropeptide FF is cleaved to form a novel neuropeptide NPNA. NPNA influences opioid receptor signaling by reducing the mRNA expression of G-protein α subunits associated with opioid receptors (17,24). As an example of type III cryptides, cytochrome c can be cleaved to generate cell penetrating peptides that have potent apoptogenic activity (25). The apoptogenicity of one of the peptides, Cyt c77–101, was increased by constructing a chimeric synthetic cryptide with a nonapeptide N-terminal extension derived from the C-terminal region of nucleoporin 153. The nucleoporin 153 is a component of nuclear pore complex containing FG repeat at its C terminus. The modified cryptide has apoptogenic activity at sub-micromolar levels, the range of concentration readily achievable for therapeutics.
Although cryptomics is in its infancy, cryptides are already known to originate from a variety of precursors and to possess a vast range of bioactivities. It is not difficult to imagine many more cryptides being generated from unexpected candidate proteins. There is an increasing interest in developing new methodologies to identify and characterize cryptides because of their diverse roles and their potential for therapeutic use (2). The identification of novel cryptides combined with quantitative studies to measure the concentration of individual or sets of cryptides in cells and tissues is expected to answer fundamental questions about their function and regulation.
DISCOVERY OF CRYPTIDES
Varied approaches have been employed to discover novel cryptides. One method involves using a diagnostic assay to screen peptides for a particular biological activity. For example, Daniel Pimenta and co-workers isolated low molecular weight components of dog pancreas and analyzed fractions for bradykinin potentiating activity (26). In this way, they identified cryptides with properties similar to hemorphins and hemorphin-like cryptides, which are derived from hemoglobin (26). A number of other screening approaches, including screening proteolytic digests of purified proteins and screening synthetic random peptide pools, have been successfully applied to identify novel cryptides (3).
Computational biology techniques can be used to predict possible cryptides. For example, Hidehito Mukai and co-workers used the previously identified neutrophil-activating factors mastoparan and mitocryptide-1 as the basis for identifying novel neutrophil-activating factors (4). Mitocryptide-1 is derived from cytochrome c oxidase subunit VIII. Their first step was to generate a list of 15–36 residue peptide fragments predicted to result from the action of various mitochondrial peptidases and cellular proteases on the 441 human mitochondrial proteins (4). They compared the physicochemical properties and three-dimensional structures of the peptides to mastoparan and mitocryptide-1 to generate a list of putative neutrophil-activating peptides. Finally, they synthesized the selected peptides and assayed their ability to activate neutrophils. They identified eight novel peptides that activate neutrophils, including several peptide fragments from cytochrome c (4). To identify cryptides involved in cell adhesion, Yoshihiko Yamada and co-workers synthesized a series of predicted proteolytic fragments of laminin and assayed their biological activity (27–30). Their systematic screen yielded an extensive list of different laminin proteolytic fragments that may affect cell adhesion (27–30).
MASS SPECTROMETRY-BASED PROTEOMIC APPROACHES FOR THE DISCOVERY OF CRYPTIDES
Mass spectrometry-based approaches are powerful and comprehensive tools for analyzing the cryptome. These approaches have been extensively used to qualitatively and quantitatively characterize predicted proteomes in multitudes of organisms, tissues, and cell types. Mass spectrometry-based cryptomics can be conceptually utilized in two ways: (a) to search for cryptides in homogenized tissue and (b) to identify and determine the tissue distribution of cryptides in situ (Figs. 2 and 3). In the following sections, we will discuss the mass spectrometry-based technologies that can be exploited to identify novel cryptides from biological samples, borrowing mainly from the technologies utilized for peptidome profiling. These methods are expected to reveal a variety of peptides present in a sample, which can be tested subsequently for their bioactivity. Moreover, quantitative proteomic tools now allow the simultaneous quantitation of multiple cryptides with high sensitivity and accuracy in a single experiment.
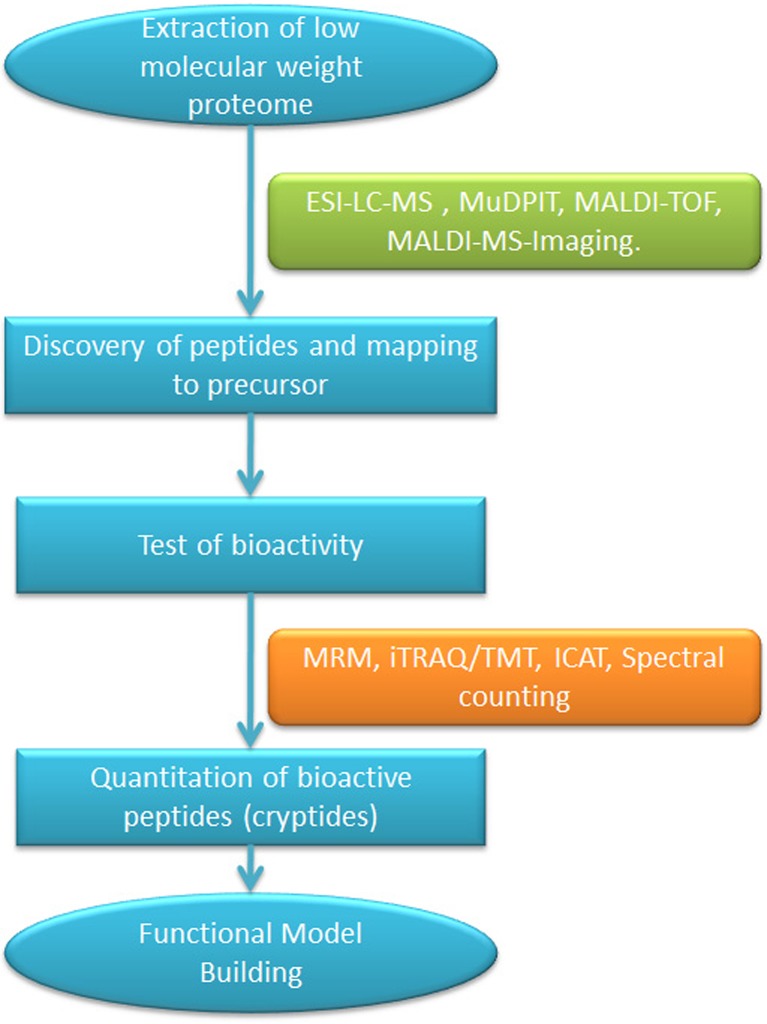
Fig. 2.
Mass spectrometry-based cryptide discovery and quantitation work flow: Cryptomics work flow for the high throughput discovery of cryptides would start with fractionation of tissue to enrich for low molecular weight proteome. A variety of mass spectrometry approaches can be used to identify the enriched cryptides. In most cases, the identified peptides can be mapped to the precursor proteins. Putative cryptides are tested for their bioactivities using either biochemical or cell-based assays. After the validation of bioactivity, the cryptides can be quantitated using a variety of approaches. The advantage of using mass spectrometry-based proteomic tools for quantitation is that large numbers of cryptides can be accurately quantitated in a single experiment. This information can be utilized for building higher order models of the functions of the cryptides. LC liquid chromatography, MS mass spectrometry, MALDI matrix-assisted laser desorption ionization, ESI electrospray ionization, iTRAQ isobaric tag for absolute and relative quantitation, TMT tandem mass tag, ICAT isotope coded affinity tags, MuDPIT multi-dimensional protein identification technology, TOF time of flight, MRM multiple reaction monitoring
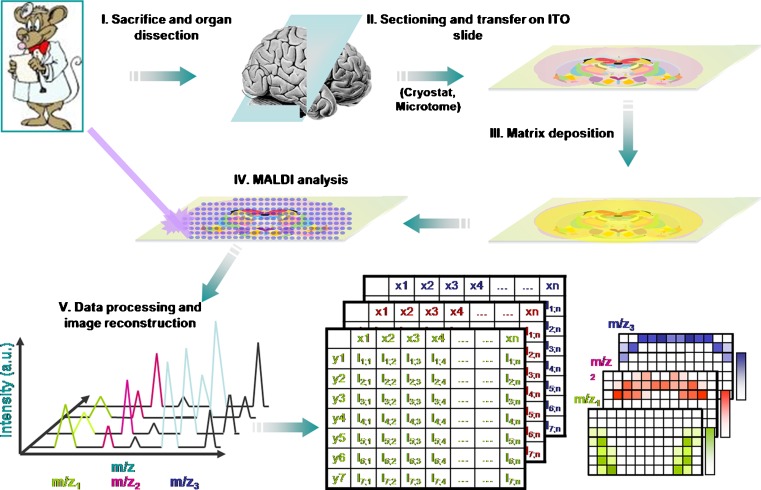
Fig. 3.
Overview of MALDI-MS imaging for tissue-based cryptomics. In MALDI-MS imaging, 10–20-μm-thick frozen tissue sections are cut using a microtome and transferred to a conductive surface. After matrix is deposited onto the section, MALDI ionization is carried out in the defined regions of the tissue section and spectral data acquired. The mass spectrometry data are computationally processed to generate a pseudoimage, showing abundances of individual peptides and proteins. a.u. arbitrary units, ITO iridium tin oxide (reprinted with permission from (31))
The development of efficient peptide fractionation using liquid chromatography coupled to electrospray ionization has led to a tremendous capability for peptide identification and quantification. Multi-dimensional protein identification technology (MuDPIT) has been utilized in a number of large-scale proteome profiling studies (32–34), and variations have been specifically used to profile endogenous small peptides. For example, MuDPIT-based strategies have been utilized to profile the peptidome of urine and to compare the self-antigen peptidomes of plasma and lymphatic fluid (35,36). A number of studies have analyzed the blood peptidome, many with the goal of discovering cancer biomarkers (37). These approaches have identified many low molecular weight components of the blood proteome, many of which are expected to be cryptides (37).
Electrospray ionization-Fourier transform mass spectrometry (ESI-FTMS) has been used to identify proteolytic cleavage products of the plasma proteome. Yufeng Shen and co-workers first depleted the 12 most abundant plasma proteins and used size-exclusion chromatography to enrich for peptides with a molecular weight less than 20 kDa (38). Then, using ultra-high-pressure liquid chromatography (UHPLC) ESI-FTMS on a LTQ-Orbitrap mass spectrometer, they identified more than 200 peptides from 29 precursors (38). Using this UHPLC-ESI-FTMS strategy, they also performed a peptidomic profiling of yeast whole cell lysate and have identified about 1,100 peptides from approximately 200 precursor proteins (39).
Similar strategies have also been used to identify novel candidate cryptides from tissue homogenates. Per Andrén and co-workers used nanoLC electrospray ionization quadrupole time-of-flight mass spectrometry to identify peptides from brain tissue (40). Their innovative enrichment for possible cryptides employed a microfilter device to remove proteins larger than 10 kDa from the mixture. They analyzed the filtered sample using ESI-MS to measure the masses of the peptides. They were able to detect approximately 1,500 endogenous brain peptides. Using tandem mass spectrometry, they were able to identify the sequences of 10% of the detected peptides, including novel peptides (40). In another study, Hanfa Zou and co-workers used ultrafiltration followed by size-exclusion chromatography to fractionate the low molecular weight proteome of liver (41). They directly analyzed the fractions with peptides smaller than 3 kDa and digested the fractions with larger molecules with trypsin before analysis. Using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) and LC-ESI-MS, they were able to identify more than 1,000 peptides from 400 precursor proteins (41). A similar strategy has been used by William Hancock and co-workers to generate a serum peptidome profile (42).
Peptidome profiling from tissue homogenates is a highly sensitive and fast approach for identifying and quantitating a large number of molecules. But often, one is interested in the tissue-specific distribution of peptides. Moreover, the levels of particular peptides may not be constant throughout a given tissue. The resulting dilution in a tissue homogenate makes it harder to identify them. Therefore, peptidome profiling of specific tissue regions or cells not only helps in understanding the tissue-specific distribution but also allows the identification of additional peptides that may be at elevated levels only in the small regions of the tissue. One technique that has been extensively utilized for visualizing tissue-specific distribution of peptides and proteins is MALDI-MS imaging (Fig. 3) (43). The technology and applications of MALDI-MS imaging have been comprehensively reviewed in (31,44). Recently, a number of studies have used MALDI-TOF-MS to analyze single cells (45,46). These studies demonstrate the potential for utilizing the mass spectrometry-based approaches for the discovery of novel cryptides.
MASS SPECTROMETRY-BASED PROTEOMIC APPROACHES FOR QUANTITATION OF CRYPTIDES
Proteomic tools have grown beyond their application for discovery of novel targets and can be used to generate quantitative profiles of cryptides. A number of techniques have been developed in the past several years that can determine relative levels of peptides and proteins using mass spectrometry (47). For quantitation, the power of mass spectrometry-based proteomics lies in its ability to detect a wide dynamic range of peptide concentrations. These proteomic approaches include label-free methods, metabolic labeling with stable isotopes, and post-lysis labeling of peptides with isotopic tags (48). Because levels of cryptides are likely to change in response to external stimuli or signaling (2,3), quantitative proteomic tools will be important for characterizing the biological roles of cryptides.
Selected reaction monitoring/multiple reaction monitoring (SRM/MRM) is a targeted mass spectrometry approach to identify and/or quantify peptide(s) and can be utilized for quantitating cryptide(s) that is known a priori (49). This is a very powerful technique, with the highest dynamic range and sensitivity of all mass spectrometry-based proteomic tools (50). In addition, SRM/MRM can be used for absolute quantitation of cryptides. In this method, a cryptide can be chosen based on prior mass spectrometry data or in silico proteolytic digestion of proteins. Fragmentation spectra of the target cryptide can be either generated or extracted from spectral libraries. An internal standard peptide (corresponding to the sequence identical to the cryptide of interest) is synthesized with a stable isotope label, and a known amount is spiked into the sample, a strategy known as stable isotope dilution (51). Reporter fragment ion peaks, called transitions, are selected, and peak areas for the heavy and light cryptides are compared to extract quantitative information. Since the amount of the internal standard is known, MRM allows absolute quantitation of the cryptide in a sample (49,52). SRM/MRM is expected to have a great potential for measuring the changing abundance of cryptides in cells and tissues and also for detecting computationally predicted cryptides.
Isobaric tag for relative and absolute quantitation (iTRAQ) and tandem mass tag (TMT) are approaches that use isobaric mass tags that react with the amino group at the N terminus and the epsilon amino group of lysine residues (53,54). Both iTRAQ and TMT reagents have three functional groups, an amine reactive group that reacts with primary amines, a mass balancer region that allows different tags to have the same mass, and a reporter region whose mass varies between different tags. In this approach, two or more samples are allowed to react with the tag, mixed together, processed, and analyzed using tandem mass spectrometry. Identical cryptides with isobaric mass tags from different samples are expected to migrate together during chromatography, and since they are isobaric, they will be selected simultaneously for fragmentation inside the mass spectrometer. Upon fragmentation, the reporter ions will be released from the peptide in the low mass region of the spectrum. Quantitative information can be gleaned by comparing the intensities of the different reporter ions in the same spectra. The spectrum also provides sequence information for the identification of the cryptide. iTRAQ has been multiplexed up to eight channels, while TMT has been multiplexed up to six channels (55,56). Isobaric mass tagging methods are expected to be useful for quantifying the low molecular weight cryptides in multiple samples without a priori knowledge of the identity of the molecular species.
Stable isotope labeling by amino acids in cell culture (SILAC) yields relative quantitative information (57). In the SILAC approach, one cell culture is grown in unlabeled medium while another is grown in medium with a stable heavy isotope, usually C13 or N15, such that one of the amino acids, such as lysine or arginine, is labeled. The cells are lysed and equal amounts of protein from the two samples are mixed together. The mixed samples are processed and analyzed using liquid chromatography coupled to tandem mass spectrometry. Cryptides can be identified by their fragmentation patterns, and relative quantitative information can be generated by comparing the integrated peak areas at the precursor levels for the labeled and unlabeled cryptides. The biggest advantage of SILAC is that experimental variables in processing of the samples are normalized, as the two samples for comparison are processed together after cell lysis. SILAC has been successfully used in a number of studies involving a wide range of processes (58–63).
SILAC was originally developed for bottom-up proteomics. More recently, it has been used also for top-down proteomics, and, as such, it is conceptually possible to determine the changes in the levels of cryptide precursors. Mathias Mann and co-workers initially demonstrated the applicability of SILAC-based quantitation in top-down proteomics (64), and later, David Muddiman and co-workers used SILAC to identify 11 intact proteins from human embryonic stem cells (65). These studies were designed only to test the possibility of identifying differentially expressed proteins with SILAC-based quantitation, but the results were promising for the future applications of this technology (64,65). It is possible to imagine determining changes in the levels of cryptide precursors using SILAC. Muddiman and co-workers also mathematically modeled the variability introduced by the biochemical processes of incorporation of labeled amino acids and metabolic conversion of arginines to prolines inside the cells during cell culture, information that will be very helpful for future top-down SILAC quantitation experiments (65).
Another isotope tagging-based quantitation method relies on tagging of the amino group by either deuterated (heavy) or protiated (light) acetic anhydride (66). Lloyd Fricker and co-workers have used this labeling approach to determine the changes in the brain peptidome under different condition and to identify differences in the levels of hemoglobin-derived peptides in blood, heart, and brain (18,66,67).
CONCLUSION
There are a multitude of proteomic tools available that can be readily adapted to the study of cryptides. The strategic advantage of proteomic methods is that multiple cryptides can be assayed in a single experiment, which is not possible with any other approach. Discovery-mode cryptomic tools can be used to identify novel cryptides, and quantitative-mode tools can be used to build functional models for the action of the cryptide(s) of interest (Fig. 2). Adaptation of these tools is expected to lead to an explosion of information about the cryptome. An important challenge in utilizing mass spectrometry-based proteomics for cryptomic analysis is to establish the methodological paradigms for their routine use. We envision that the interaction between these two fields will not only benefit cryptomics but will also lead to the development of new analytical technologies that will benefit other areas of research.
Acknowledgments
We acknowledge Elizabeth M. Link for helpful comments and suggestions in the preparation of this manuscript. P.S. and A.J.L. were supported by NIH grant GM64779.
References
- 1.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422(6928):193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 2.Autelitano DJ, Rajic A, Smith AI, Berndt MC, Ilag LL, Vadas M. The cryptome: a subset of the proteome, comprising cryptic peptides with distinct bioactivities. Drug Discov Today. 2006;11(7–8):306–314. doi: 10.1016/j.drudis.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Pimenta DC, Lebrun I. Cryptides: buried secrets in proteins. Peptides. 2007;28(12):2403–2410. doi: 10.1016/j.peptides.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Ueki N, Someya K, Matsuo Y, Wakamatsu K, Mukai H. Cryptides: functional cryptic peptides hidden in protein structures. Pept Sci. 2007;88(2):190–198. doi: 10.1002/bip.20687. [DOI] [PubMed] [Google Scholar]
- 5.Fortini ME. Notch and presenilin: a proteolytic mechanism emerges. Curr Opin Cell Biol. 2001;13(5):627–634. doi: 10.1016/S0955-0674(00)00261-1. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398(6727):518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 7.Mukai H, Hokari Y, Seki T, Takao T, Kubota M, Matsuo Y, et al. Discovery of mitocryptide-1, a neutrophil-activating cryptide from healthy porcine heart. J Biol Chem. 2008;283(45):30596–30605. doi: 10.1074/jbc.M803913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zania P, Gourni D, Aplin AC, Nicosia RF, Flordellis CS, Maragoudakis ME, et al. Parstatin, the cleaved peptide on proteinase-activated receptor 1 activation, is a potent inhibitor of angiogenesis. J Pharmacol Exp Ther. 2009;328(2):378–389. doi: 10.1124/jpet.108.145664. [DOI] [PubMed] [Google Scholar]
- 9.Mains RE, Eipper BA, Ling N. Common precursor to corticotropins and endorphins. Proc Natl Acad Sci USA. 1977;74(7):3014–3018. doi: 10.1073/pnas.74.7.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakanishi S, Inoue A, Kita T, Nakamura M, Chang ACY, Cohen SN, et al. Nucleotide sequence of cloned cDNA for bovine corticotropin-[beta]-lipotropin precursor. Nature. 1979;278(5703):423–427. doi: 10.1038/278423a0. [DOI] [PubMed] [Google Scholar]
- 11.Roberts JL, Herbert E. Characterization of a common precursor to corticotropin and beta-lipotropin: cell-free synthesis of the precursor and identification of corticotropin peptides in the molecule. Proc Natl Acad Sci USA. 1977;74(11):4826–4830. doi: 10.1073/pnas.74.11.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts JL, Herbert E. Characterization of a common precursor to corticotropin and beta-lipotropin: identification of beta-lipotropin peptides and their arrangement relative to corticotropin in the precursor synthesized in a cell-free system. Proc Natl Acad Sci USA. 1977;74(12):5300–5304. doi: 10.1073/pnas.74.12.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert E. Discovery of pro-opiomelanocortin—a cellular polyprotein. Trends Biochem Sci. 1981;6:184–188. doi: 10.1016/0968-0004(81)90068-2. [DOI] [Google Scholar]
- 14.Sousa JC, Berto RF, Gois EA, Fontenele-Cardi NC, Honorio JE, Jr, Konno K, et al. Leptoglycin: a new glycine/leucine-rich antimicrobial peptide isolated from the skin secretion of the South American frog Leptodactylus pentadactylus (Leptodactylidae) Toxicon. 2009;54(1):23–32. doi: 10.1016/j.toxicon.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Nakatani S, Mano H, Sampei C, Shimizu J, Wada M. Chondroprotective effect of the bioactive peptide prolyl-hydroxyproline in mouse articular cartilage in vitro and in vivo. Osteoarthritis cartilage OARS Osteoarthritis Res Soc. 2009;17(12):1620–1627. doi: 10.1016/j.joca.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Mukai H, Seki T, Nakano H, Hokari Y, Takao T, Kawanami M, et al. Mitocryptide-2: purification, identification, and characterization of a novel cryptide that activates neutrophils. J Immunol. 2009;182(8):5072–5080. doi: 10.4049/jimmunol.0802965. [DOI] [PubMed] [Google Scholar]
- 17.Suder P, Nawrat D, Bielawski A, Zelek-Molik A, Raoof H, Dylag T, et al. Cryptic peptide derived from the rat neuropeptide FF precursor affects G-proteins linked to opioid receptors in the rat brain. Peptides. 2008;29(11):1988–1993. doi: 10.1016/j.peptides.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Gelman JS, Sironi J, Castro LM, Ferro ES, Fricker LD. Hemopressins and other hemoglobin-derived peptides in mouse brain: comparison between brain, blood, and heart peptidome and regulation in Cpefat/fat mice. J Neurochem. 2010;113(4):871–880. doi: 10.1111/j.1471-4159.2010.06653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes I, Dale C, Casten K, Geigner M, Gozzo F, Ferro E, et al. Hemoglobin-derived peptides as novel type of bioactive signaling molecules. AAPS J. 2010;12:658–669. doi: 10.1208/s12248-010-9217-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Moses M, et al. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 1994;79(2):315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 21.O’Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, et al. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88(2):277–285. doi: 10.1016/S0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 22.Deng LX, Pan XL, Wang Y, Wang LL, Zhou XE, Li M, et al. Hemoglobin and its derived peptides may play a role in the antibacterial mechanism of the vagina. Hum Reprod. 2009;24(1):211–218. doi: 10.1093/humrep/den318. [DOI] [PubMed] [Google Scholar]
- 23.Liepke C, Baxmann S, Heine C, Breithaupt N, Standker L, Forssmann WG. Human hemoglobin-derived peptides exhibit antimicrobial activity: a class of host defense peptides. J Chromatogr B. 2003;791(1–2):345–356. doi: 10.1016/S1570-0232(03)00245-9. [DOI] [PubMed] [Google Scholar]
- 24.Dylag T, Pachuta A, Raoof H, Kotlinska J, Silberring J. A novel cryptic peptide derived from the rat neuropeptide FF precursor reverses antinociception and conditioned place preference induced by morphine. Peptides. 2008;29(3):473–478. doi: 10.1016/j.peptides.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 25.Jones S, Holm T, Mager I, Langel U, Howl J. Characterization of bioactive cell penetrating peptides from human cytochrome c: protein mimicry and the development of a novel apoptogenic agent. Chem Biol. 2010;17(7):735–744. doi: 10.1016/j.chembiol.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Ianzer D, Konno K, Xavier CH, Stocklin R, Santos RA, de Camargo AC, et al. Hemorphin and hemorphin-like peptides isolated from dog pancreas and sheep brain are able to potentiate bradykinin activity in vivo. Peptides. 2006;27(11):2957–2966. doi: 10.1016/j.peptides.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Nomizu M, Kim WH, Yamamura K, Utani A, Song S-Y, Otaka A, et al. Identification of cell binding sites in the laminin 1 chain carboxyl-terminal globular domain by systematic screening of synthetic peptides. J Biol Chem. 1995;270(35):20583–20590. doi: 10.1074/jbc.270.35.20583. [DOI] [PubMed] [Google Scholar]
- 28.Nomizu M, Kuratomi Y, Malinda KM, Song S-Y, Miyoshi K, Otaka A, et al. Cell binding sequences in mouse laminin α1 chain. J Biol Chem. 1998;273(49):32491–32499. doi: 10.1074/jbc.273.49.32491. [DOI] [PubMed] [Google Scholar]
- 29.Nomizu M, Kuratomi Y, Ponce ML, Song S-Y, Miyoshi K, Otaka A, et al. Cell adhesive sequences in mouse laminin [beta]1 chain. Arch Biochem Biophys. 2000;378(2):311–320. doi: 10.1006/abbi.2000.1828. [DOI] [PubMed] [Google Scholar]
- 30.Nomizu M, Kuratomi Y, Song S-Y, Ponce ML, Hoffman MP, Powell SK, et al. Identification of cell binding sequences in mouse laminin γ1 chain by systematic peptide screening. J Biol Chem. 1997;272(51):32198–32205. doi: 10.1074/jbc.272.51.32198. [DOI] [PubMed] [Google Scholar]
- 31.Franck J, Arafah K, Elayed M, Bonnel D, Vergara D, Jacquet A, et al. MALDI imaging mass spectrometry. Mol Cell Proteomics. 2009;8(9):2023–2033. doi: 10.1074/mcp.R800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20(10):1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Link AJ. Multidimensional peptide separations in proteomics. Trends Biotechnol. 2002;20(12 Suppl):S8–S13. doi: 10.1016/S1471-1931(02)00202-1. [DOI] [PubMed] [Google Scholar]
- 34.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17(7):676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 35.Machtejevas E, Marko-Varga G, Lindberg C, Lubda D, Hendriks R, Unger KK. Profiling of endogenous peptides by multidimensional liquid chromatography: on-line automated sample cleanup for biomarker discovery in human urine. J Sep Sci. 2009;32(13):2223–2232. doi: 10.1002/jssc.200900058. [DOI] [PubMed] [Google Scholar]
- 36.Clement CC, Cannizzo ES, Nastke MD, Sahu R, Olszewski W, Miller NE, et al. An expanded self-antigen peptidome is carried by the human lymph as compared to the plasma. PLoS One [Article] 2010;5(3):e9863. doi: 10.1371/journal.pone.0009863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petricoin EF, Belluco C, Araujo RP, Liotta LA. The blood peptidome: a higher dimension of information content for cancer biomarker discovery. Nat Rev Cancer. 2006;6(12):961–967. doi: 10.1038/nrc2011. [DOI] [PubMed] [Google Scholar]
- 38.Shen Y, Liu T, Toli N, Petritis BO, Zhao R, Moore RJ, et al. Strategy for degradomic–peptidomic analysis of human blood plasma. J Proteome Res. 2010;9(5):2339–2346. doi: 10.1021/pr901083m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Y, Hixson KK, Toli N, Camp DG, Purvine SO, Moore RJ, et al. Mass spectrometry analysis of proteome-wide proteolytic post-translational degradation of proteins. Anal Chem. 2008;80(15):5819–5828. doi: 10.1021/ac800077w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skold K, Svensson M, Kaplan A, Bjorkesten L, Astrom J, Andren PE. A neuroproteomic approach to targeting neuropeptides in the brain. Proteomics. 2002;2(4):447–454. doi: 10.1002/1615-9861(200204)2:4<447::AID-PROT447>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 41.Hu L, Li X, Jiang X, Zhou H, Jiang X, Kong L, et al. Comprehensive peptidome analysis of mouse livers by size exclusion chromatography prefractionation and NanoLC-MS/MS identification. J Proteome Res. 2007;6(2):801–808. doi: 10.1021/pr060469e. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X, Baker H, Hancock WS. Analysis of the low molecular weight serum peptidome using ultrafiltration and a hybrid ion trap-Fourier transform mass spectrometer. J Chromatogr A. 2006;1120(1–2):173–184. doi: 10.1016/j.chroma.2006.01.098. [DOI] [PubMed] [Google Scholar]
- 43.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69(23):4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 44.Burnum KE, Frappier SL, Caprioli RM. Matrix-assisted laser desorption/ionization imaging mass spectrometry for the investigation of proteins and peptides. Annu Rev Anal Chem. 2008;1(1):689–705. doi: 10.1146/annurev.anchem.1.031207.112841. [DOI] [PubMed] [Google Scholar]
- 45.Millet LJ, Bora A, Sweedler JV, Gillette MU. Direct cellular peptidomics of supraoptic magnocellular and hippocampal neurons in low-density cocultures. ACS Chem Neurosci. 2009;1(1):36–48. doi: 10.1021/cn9000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubakhin SS, Churchill JD, Greenough WT, Sweedler JV. Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal Chem. 2006;78(20):7267–7272. doi: 10.1021/ac0607010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaudel M, Sickmann A, Martens L. Peptide and protein quantification: a map of the minefield. Proteomics. 2010;10(4):650–670. doi: 10.1002/pmic.200900481. [DOI] [PubMed] [Google Scholar]
- 48.Wilm M. Quantitative proteomics in biological research. Proteomics. 2009;9(20):4590–4605. doi: 10.1002/pmic.200900299. [DOI] [PubMed] [Google Scholar]
- 49.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100(12):6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: a general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35(3):265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Multiple reaction monitoring for quantitative biomarker analysis in proteomics and metabolomics. J Chromatogr B. 2009;877(13):1229–1239. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 53.Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 55.Choe L, D’Ascenzo M, Relkin NR, Pappin D, Ross P, Williamson B, et al. 8-Plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer’s disease. Proteomics. 2007;7(20):3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80(8):2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 57.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.M200025-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Mittler G, Butter F, Mann M. A SILAC-based DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19(2):284–293. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8(3):443–450. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ong S-E, Schenone M, Margolin AA, Li X, Do K, Doud MK, et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc Natl Acad Sci. 2009;106(12):4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teckchandani A, Toida N, Goodchild J, Henderson C, Watts J, Wollscheid B, et al. Quantitative proteomics identifies a Dab2/integrin module regulating cell migration. J Cell Biol. 2009;186(1):99–111. doi: 10.1083/jcb.200812160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 63.Jorgensen C, Sherman A, Chen GI, Pasculescu A, Poliakov A, Hsiung M, et al. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 2009;326(5959):1502–1509. doi: 10.1126/science.1176615. [DOI] [PubMed] [Google Scholar]
- 64.Waanders LF, Hanke S, Mann M. Top-down quantitation and characterization of SILAC-labeled proteins. J Am Soc Mass Spectrom. 2007;18(11):2058–2064. doi: 10.1016/j.jasms.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Collier TS, Sarkar P, Rao B, Muddiman DC. Quantitative top-down proteomics of SILAC labeled human embryonic stem cells. J Am Soc Mass Spectrom. 2010;21(6):879–889. doi: 10.1016/j.jasms.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 66.Che F-y, Fricker LD. Quantitation of neuropeptides in Cpefat/Cpefat mice using differential isotopic tags and mass spectrometry. Anal Chem. 2002;74(13):3190–3198. doi: 10.1021/ac015681a. [DOI] [PubMed] [Google Scholar]
- 67.Che FY, Biswas R, Fricker LD. Relative quantitation of peptides in wild-type and Cpe(fat/fat) mouse pituitary using stable isotopic tags and mass spectrometry. J Mass Spectrom. 2005;40(2):227–237. doi: 10.1002/jms.742. [DOI] [PubMed] [Google Scholar]