Abstract
Ornithine decarboxylase (ODC) is the rate-limiting enzyme in polyamine biosynthesis and a target for chemoprevention. Hydroxydibenzoylmethane (HDB), a derivative of dibenzoylmethane of licorice, is a promising chemopreventive agent. In this paper, we investigated whether HDB would inhibit the ODC pathway to enhance apoptosis in human promyelocytic leukemia HL-60 cells. We found ODC enzyme activity was reduced during HDB treatment. Overexpression of ODC in HL-60 parental cells could reduce HDB-induced apoptosis, which leads to loss of mitochondrial membrane potential (Δψm), through lessening intracellular ROS. Furthermore, ODC overexpression protected cytochrome c release and the activation of caspase-3 following HDB treatment. The results demonstrated HDB-induced apoptosis was through a mechanism of down-regulation of ODC and occurred along a ROS-dependent mitochondria-mediated pathway.
Keywords: apoptosis, hydroxydibenzoylmethane, mitochondrial membrane potential, ornithine decarboxylase, reactive oxygen species
Introduction
Hydroxydibenzoylmethane (HDB) is similar to curcumin and identical in structure to dibenzoylmethane (DB) that it possesses a hydroxyl group on one of the aromatic rings. HDB is an inducer of phase 2 detoxification enzymes (Dinkova-Kostova and Talalay, 1999). The induction of phase 2 detoxification enzymes is a sufficient state for obtaining chemoprevention (Jana and Mandlekar, 2009). Chemopreventative strategies include preventing the initial development of cancer and the progression of an already established cancer. Potential targets for chemopreventative agents consist of all steps in the development and progression of carcinogenesis. The induction of apoptosis process is one of the important chemopreventive effects (Sun et al., 2004). Apoptosis is triggered in a cell through two main signaling pathways, which are extrinsic and intrinsic pathways (Circu and Aw, 2010). The extrinsic pathway is activated from outside the cell by specific apoptotic ligands that interact with death receptors on the surface of cells. The intrinsic apoptotic pathway initiates in mitochondria. The caspase family of cysteine proteases plays an essential role in both pathways. A member of this family, caspase-3 is the most extensively studied apoptotic protein (Lavrik et al., 2005). Recent report indicates that HDB is more potent than DB in repressing tumor proliferation and in the induction of caspase-3 and apoptotic cell death (Pan et al., 2003).
Ornithine decarboxylase (ODC) has been reported as a target for cancer chemoprevention and chemotherapy (Gerner, 2010). It is the rate-limiting enzyme in the biosynthesis of polyamines (putrescine, spermidine and spermine), which plays an important role in several biological functions including embryonic development, cell cycle and proliferation. ODC protein expression and enzyme activity increase in malignant cells and tumors compared with normal cells and tissues (Manni et al., 1996; Giardiello et al., 1997; Mohan et al., 1999). ODC overexpression enhances tumorigenesis in vivo and neoplastic transformation in vitro (Auvinen et al., 1992; O'Brien et al., 1997; Smith et al., 1997; Jana and Mandlekar, 2009). A specific and irreversible inhibitor of ODC, difluoromethylornithine (DFMO), could induce apoptosis in cell and animal models (Ploszaj et al., 2000; Fong et al., 2001). Previous our studies and others have reported that overexpression of ODC support survival of cancer cells under TNF-α, H2O2 and curcumin (Park et al., 2002; Liu et al., 2005; Liao et al., 2008). The purpose of the present study was to examine whether HDB-induced apoptosis takes place through an ODC-dependent pathway. In addition, we aimed to determine the mechanism by which ODC mediates HDB-induced apoptosis.
Results
Hydroxydibenzoylmethane (HDB) induced HL-60 cell apoptosis
Treatment with HDB (Figure 1A) at a concentration of 10 to 100 µM for 12 h resulted in a dose-dependent decrease in cell viability of HL-60 cells (Figure 1B) using trypan blue exclusion assay. The data were presented as proportional viability (%) by comparing the HDB treated group with the control group, the viability of which was assumed to be 100%. Cells undergoing apoptosis reveals a characteristic cleavage of DNA into oligonucleosome fragments manifesting as DNA laddering, a hallmark of apoptosis. HDB-treated cells induced significantly DNA fragmentation in a dose-dependent and time-dependent manner (Figure 1C).
Figure 1.
Hydroxydibenzoylmethane (HDB) promoted HL-60 cell apoptosis. (A) Chemical structure of HDB. (B) The cells were treated with different concentrations of HDB at 12 h. Cell viability was determined by the trypan blue exclusion assay. (C) DNA fragmentation was detected by gel electrophoresis following 0, 10, 50 and 100 µM HDB stimulation for 12 h, and 50 µM at 0, 6, 12 and 24 h. M, DNA ladder maker. Data were representative of at least three experiments.
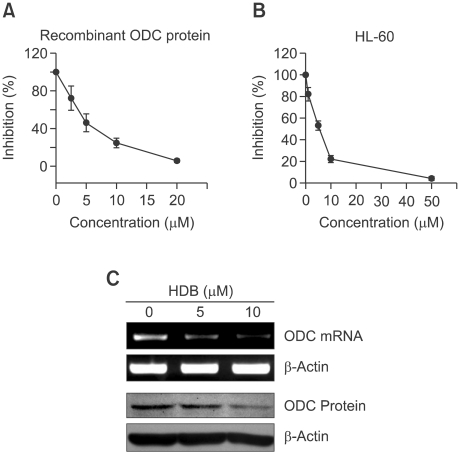
HDB inhibited ODC enzyme activity and expression
The ODC enzyme activity has been found to be associated with increasing malignancy grade for many tumors. Here, purified human ODC recombinant protein was incubated with different concentrations of HDB for 1 h and then the enzyme activity was determined by a luminescent assay. ODC activity was decreased in a dose-dependent manner (Figure 2A). HL-60 cells were treated with HDB and then harvested to measure the enzyme activity of ODC. There was the dose-dependent effect of HDB on reducing ODC enzyme activity (Figure 2B). Furthermore, HDB inhibited the expression of ODC mRNA and protein (Figure 2C). These results showed HDB could significantly decrease ODC enzyme activity and expression.
Figure 2.
HDB inhibited ODC activity and expression. (A) Recombinant ODC protein was added with different concentrations of HDB to analyze ODC enzyme activity. (B) HL-60 cells were treated with different concentrations of HDB for 6 h to analyze ODC enzyme activity. (C) ODC protein and mRNA were detected by immunoblotting and RT-PCR following 0, 5 and 10 µM HDB stimulation for 12 h. Data were representative of at least three experiments.
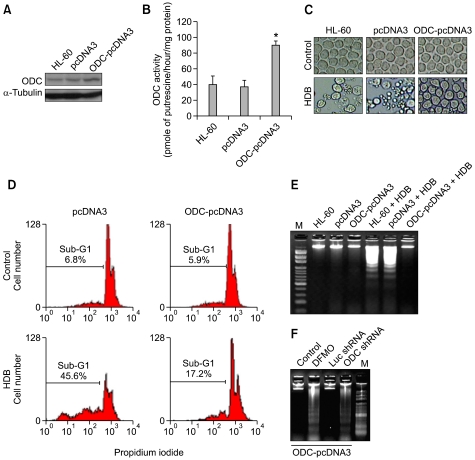
ODC resisted HDB-induced apoptosis
To determine whether the HDB-induced apoptotic pathway was correlated with ODC status, we introduced ODC cDNA into the system of mammalian expression plasmid, pcDNA3 and produced the empty vector (pcDNA3) and overexpressing ODC (ODC-pcDNA3) in parental HL-60 cells. ODC enzyme activity and protein expression were greater in ODC-pcDNA3 cells than in HL-60 and pcDNA3 cells (Figures 3A and 3B). In typical apoptotic morphologic study, HDB-treated HL-60 and pcDNA3 cells charged significantly into the round and lobulate appearances of apoptotic cells, while HDB-treated ODC-pcDNA3 cells maintained normal cell morphology as well as untreated cells (Figure 3C). Furthermore, ODC overexpression could repress HDB-induced sub-G1 fraction and DNA fragmentation (Figures 3D and 3E). Finally, HDB induced DNA fragmentation was recovered by DFMO and ODC shRNA in cells overexpressing ODC (Figure 3F). These results showed that ODC-overexpressing human promyelocytic leukemia HL-60 cells survived and escaped HDB-induced apoptosis.
Figure 3.
Overexpression of ODC prevented HDB-induced apoptosis. HL-60 cells were transfected with pcDNA3 and ODC-pcDNA3 plasmids, and then cells were harvested to measure ODC protein (A) and enzyme activity (B). HL-60, pcDNA3 and ODC-pcDNA3 cells were treated with HDB for 12 h. The cells were harvested to measure morphology (C), the percentage of sub-G1 (D) and DNA fragmentation (E) by light microscope, flow cytometry using PI staining and DNA gel electrophoresis. (F) Prior to HDB treatment, ODC-pcDNA3 cells were pretreated with vehicle (control) or 1 mM DFMO for 1 h, or transfected with Luc shRNA or ODC shRNA for 12 h. Genomic DNA was collected and analyzed for DNA fragmentation. Data were representative of at least three experiments. *P < 0.05 as compared with H6-60 cells.
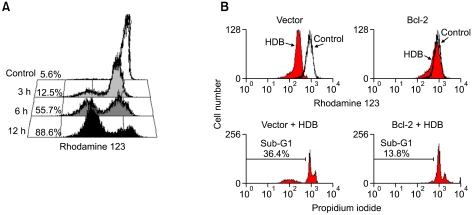
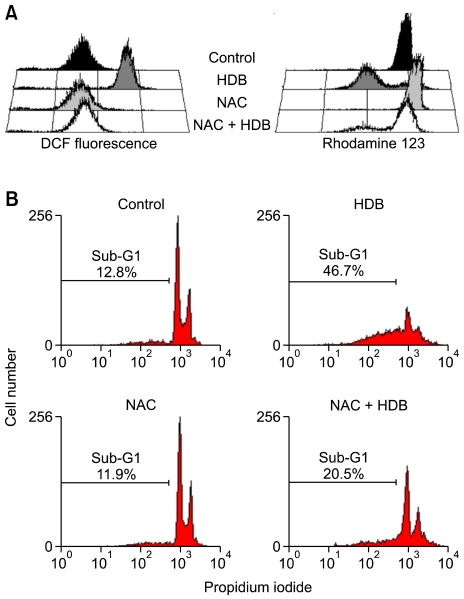
HDB induced apoptosis, the loss of Δψm and intracellular ROS generation
To observe the effect of HDB on mitochondria, HL-60 cells were treated with HDB for different lengths of time and measured their membrane potential by staining with rhodamine 123. Figure 4A indicated mitochondrial membrane depolarization to be observed. HDB induced a loss of Δψm after 3 h, 6 h and 12 h of treatment, which decreased to 7%, 50% and 83% of total cell counts compared with the Δψm in the control groups. The Bcl-2 protein could abrogate cytochrome c release from mitochondria to repress Δψm loss. The transient overexpression of Bcl-2 in HDB-treated HL-60 cells prevented the loss of Δψm and reduced sub-G1 fraction (Figure 4B). To clarify the role of ROS in the mechanism of HDB-induced apoptosis, we pretreated with NAC, a ROS scavenger. The intracellular production of ROS was monitored by flow cytometry using the flurorescent probe DCFH-DA, which was permeable into the HDB-treated with HL-60 cells (Figure 5A left). The results of flow cytometric analysis showed that HDB induced the intracellular ROS generation. NAC was shown to be capable of significantly maintaining Δψm (Figure 5A right) and reducing sub-G1 fraction (Figure 5B).
Figure 4.
Overexpression of Bcl-2 maintained Δψm and reduced apoptosis after HDB treatment. (A) HL-60 cells were treated with 50 µM HDB for the indicated time. The cells were harvested for measuring Δψm by flow cytometry with rhodamine 123. (B) Parental HL-60 and overexpression of Bcl-2 cells were treated with HDB. Cells were harvested for measuring Δψm and sub-G1 peak ratio by flow cytometry with rhodamine 123 and PI fluorescence respectively. Data were representative of at least three experiments.
Figure 5.
NAC, a ROS scavenger, decreased intracelluar ROS generation, the loss of Δψm and sub-G1 peak ratio after HDB treatment. (A) Parental HL-60 cells were pretreated with 10 mM NAC for 1 h and treated with or without HDB for 12 h. Intracellular ROS was detected by flow cytometry measuring fluorescence of DCF. Δψm was detected by flow cytometry with measuring fluorescence of rhodamine 123. (B) The percentage of sub-G1 was measured by flow cytometry using PI staining. Data were representative of at least three experiments.
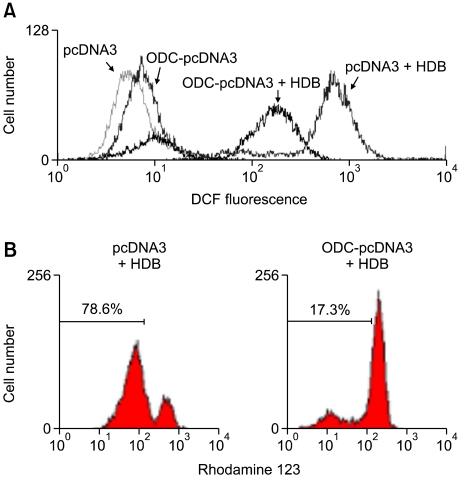
Overexpression of ODC protected HDB-induced intracellular ROS production, the loss of Δψm, cytochrome c release, and the activation of caspase-3
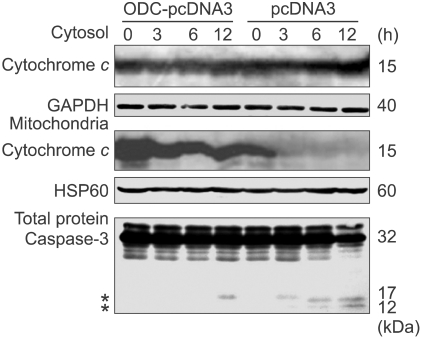
According to above data, HDB-induced apoptosis could be through increasing ROS production. To examine the effect of ODC in HDB-mediated ROS production during the cell death process, pcDNA3 and ODC-pcDNA3 cells were treated with HDB for 1 h. The intracellular ROS generation of ODC-pcDNA3 cells was decreased as compared with pcDNA3 cells (Figure 6A). Furthermore, the Δψm of ODC-pcDNA3 cells was increased as compared with pcDNA3 cells (Figure 6B). Besides, cytosol and mitochondrial proteins of HDB-treated cells were detected by immunoblotting with anti-cytochrome c antibody. Cytochrome c release from mitochondria into cytosol decreased on ODC-pcDNA3 to compare with pcDNA3 cells (Figure 7). The release of cytochrome c is considered to trigger a series of events leading to the activation of effector caspases such as caspase-3. In the ODC-pcDNA3 cells, the cleavage of caspase-3 was diminished to compare with the pcDNA3 cells following HDB treatment (Figure 7).
Figure 6.
Overexpression of ODC decreased intracellular ROS generation and the loss of Δψm. pcDNA3 and ODC-pcDNA3 cells were treated with HDB and then intracellular ROS (A) and Δψm (B) were detected by flow cytometry with fluorescence of DCF and rhodamine 123. Data were representative of at least three experiments.
Figure 7.
Overexpression of ODC protected cytochrome c release and the cleavage of caspase-3. pcDNA3 and ODC-pcDNA3 cells were treated with HDB for the indicated time. Cytosol and mitochondrial proteins were separated and detected by immunoblotting with anti-cytochrome c, heat shock protein 60 (HSP60, as a mitochondrial marker) and Glyceraldehyde-3-phosphate dehydrogenase (GADPH, as a cytosolic marker) antibodies. The all of the proteins were extracted for immunoblotting with specific antibody of caspase-3. *Indicated the activated and cleaved forms of caspase-3. Data were representative of at least three experiments.
Discussion
The results of the present study demonstrated for the first time that hydroxydibenzoylmethane (HDB) promoted human hematopoietic cell apoptosis. HDB induces programmed cell death through increasing the production reactive oxygen species (ROS) in human epidermoid carcinoma cells (Pan et al., 2005). Here, we demonstrated the mechanism of HDB-induced apoptosis in human promyelocytic leukemia cells. That was, intracellular ROS accumulation in the initial one hour and the disruption of Δψm was in twelve hour after HDB treatment. NAC, a ROS scavenge, could reduce the accumulation of intracellular ROS, prevent loss of Δψm and protect at least part of the cells from apoptosis. These data provided evidence that HDB caused Δψm disruption and apoptosis in human promyelocytic leukemia HL-60 cells through ROS generation. Overexpression of Bcl-2 also prevented HDB-induced Δψm disruption and apoptosis. Furthermore, cytochrome c release from mitochondria and activation of caspase-3 are found after HDB treatment. There is an evidence to suggest that the mitochondria-mediated pathway participates in HDB-induced apoptosis.
ROS including oxygen radicals (superoxide and hydroxyl radial) and nonradical derivatives of O2 (hydrogen peroxide), have been shown to mediate and control chemopreventive agent-induced apoptosis (Rigas and Sun, 2008). Herein, our results showed overexpression of ODC could prevent HDB-induced apoptosis in HL-60 cells. Further, overexpressed ODC could decrease the accumulation of ROS following HDB treatment as the performance of ROS scavengers. ODC can promote the ability of the anti-oxidant defense system to decrease the inherent ROS level. There are several possible mechanisms to explain how ODC can reduce ROS. First, overexpression of ODC augments the concentration of polyamines and the polyamine spermine can function directly as a free radical scavenger (Ha et al., 1998). Second, inhibition of ODC by DFMO leads to an imbalance in polyamine pools and raises intracellular ROS (Ploszaj et al., 2000; Liu et al., 2005). Third, the key regulatory enzyme of polyamine catabolism, polyamine oxidase, could continue and result in the production of ROS (Casero and Pegg, 2009). Therefore, ODC might be through the polyamine biosynthetic pathway to interfere in HDB-stimulated ROS generation and apoptosis.
ODC exhibits the high enzymatic activity in a variety of tumor cells. The elevated ODC activation promotes cell transformation and tumor proliferation (Auvinen et al., 1992; O'Brien et al., 1997). Therefore, ODC is an important target for cancer prevention and antineoplastic therapy. The irreversible inhibitor of ODC, DFMO, has been found as a cancer chemopreventive agent (Meyskens and Gerner, 1999). Our findings indicated that HDB inhibited ODC activation (Figure 2). Phase 2 detoxification enzymes are recognized to play vital roles in cancer chemoprevention because they augment cytoprotection through detoxification and elimination of activated carcinogens at tumor initiation. Phase 2 enzymes are including glutathione transferases, NAD(P)H:quinone oxidoreductase, glucuronosyltransferases and epoxide hydrolase. NAD(P)H:quinone oxidoreductase plays a role in regulating ubiquitin-independent degradation of ODC by the 20S proteasomes (Asher et al., 2005). HDB can elevate the activities of phase 2 detoxification enzymes of xenobiotic metabolism (Dinkova-Kostova and Talalay, 1999). Therefore, the characteristic of HDB-inhibited ODC might be a potential approach in the interference of tumor processing.
In conclusion, the study described here proved that inhibiting ODC activation launched the responsible mechanism for HDB-promoted apoptotic cell death. Overexpression of ODC maintained Δψm and prevented apoptosis after HDB treatment through decreasing intracellular ROS. In addition, ODC overexpression could prevent cytochrome c release from the mitochondria, and inactivate caspase-3. Therefore, HDB-induced apoptosis was a ROS-dependent mitochondria-mediated pathway via suppressing the activation of ODC. Further, the effect of HDB-inhibited ODC-resulted in apoptosis seems to exist in the convenient application of cancer chemoprevention.
Methods
Cell culture and chemicals
HL-60 cells were grown in RPMI 1640 medium supplemented with 10% of fetal bovine serum (FBS) at a temperature of 37℃ under a humidified and 5% CO2. Hydroxydibenzoylmethane, NAC, 2'-7'-dichlorofluoroscein diacetate (DCFH-DA), rhodamine 123 and DFMO were purchased from Sigma. ODC shRNA and Luc shRNA were purchased from National RNAi Core Facility (Taiwan).
Cell viability
Cell numbers were counted using trypan blue exclusion assay. The extent of cell viability was calculated by the viable cell numbers from experiment groups in contrast with the untreated control group.
Human odc gene sub-cloning and cell transfection
Parental HL-60 cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated FBS for 3 h, then the harvested cells were gently rinsed in PBS. Purification of mRNA was carried out according to the supplier's instructions (MDBio, Taiwan) and the cDNA was synthesized by reverse transcriptase (RT) (Promega, Madison, WI). Polymerase chain reaction (PCR) amplification of the encoding region of the human odc cDNA was performed with our designed primers derived from the human odc sequence. The PCR product was sub-cloned to a eukaryotic expression vector, pcDNA3 and then sequenced. The plasmid of ODC expression was constructed by inserting the BamHI-EcoRI 1,415 bp coding region fragment. Parental HL-60 cells were transfected with ODC-pcDNA3 (overexpressing ODC) and pcDNA3 (empty vector) plasmids according to calcium phosphate-mediated transfections, respectively. Stably transfected cells were selected with the antibiotic G418 (400 µg/ml). Three weeks later, isolated G418-resistant clones were individually analyzed for expression of ornithine decarboxylase. The ODC expressions of individual clones were examined by immunoblotting and enzyme activity assay.
Enzyme activity assay of ODC
ODC enzyme activity was assayed at 37℃ by measuring its product, putrescine, as we described previously (Liu et al., 2005). Cells were harvested, lysed in ODC buffer (50 µM EDTA, 25 µM pyridoxal phosphate and 2.5 mM DTT in 25 mMTris HCl, pH 7.1), homogenized and centrifuged. The supernatant or recombinant human ODC protein was incubated with ODC buffer containing 2 nmole L-ornithine for 1 h and material was spotted onto p81 phosphocellulose (Whatman, Maidstone, England). Diamines were eluted from the dried papers by shaking at 37℃ for 1 h with 0.5 ml quantities of elution buffer (0.5 M magnesium chloride in 0.2 M boric acid-borax buffer, pH 8.4). After that, samples were supplemented by luminescence reagent (11.7 µg/ml luminal, 30 µg/ml peroxidase type II and 67 mM glycine buffer, pH 8.6 (at 1:1:2.5, v/v/v)) to each cuvette. Keeping the cuvettes in the dark for 30 min, the background was measured in the TR 717 microplate luminometer (Perkin-Elmer, Foster, CA). Then 5 µl of diamine oxidase (4.61 µg/µl) (Sigma) were injected into each cuvette, luminescence was recorded for 40 s at 37℃ and results were calculated according to the standard curve using putrescine.
Reverse transcription-polymerase chain reaction (RT-PCR)
RNA was isolated from 2×106 cells by Trizol according to the manufacturer's instructions. Synthesis of cDNA was performed using mRNA templates, reverse transcriptase (RT) and 500 ng of dT15 primers. The reaction mixture was incubated for 90 minutes at 37℃. For PCR assay, cDNA was added to 30 µl mixture buffer containing 75 mM Tris-HCl, pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween-20 (v/v), 1 mM MgCl2, 0.2 mM dNTPs, 0.5 µM forward and reverse primers, and 1 U Taq DNA polymerase (MDBio). PCR was set at the condition of 5 minutes at 95℃, 25-36 cycles (30 seconds, 95℃; 30 seconds, 55℃; 90 seconds, 72℃) and 10 minutes at 72℃ by a Mastercycler (Eppendorf, Hamburg, Germany), and products were analyzed on a 2% agarose gel. The following primer pairs were used: β-Actin (309 bps) 5'-AGCGGGAAATCGTGCGTG-3' and 5'-CAG GGTACATGGTGGTGC-3'; ODC (533 bps) 5'-TTACTGC CAAGGACATTCTG-3' and 5'-GCTGACACCAACAACATCG-3'.
DNA fragmentation assay
Cells were harvested and lysed overnight in a digestion buffer (0.5% sarkosyl, 0.5 mg/ml proteinase K, 50 mM Tris-HCl, pH 8.0 and 10 mM EDTA) at 55℃. Subsequently cells were treated with 0.5 µg/ml RNase A for 2 h. The genomic DNA was extracted by phenol/chloroform/isoamyl alcohol extraction and analyzed by gel electrophoresis using 2% agarose.
Sub-G1 fraction analysis
After HDB treatment, 1×106 cells were harvested, washed with PBS, resuspended in 0.2 ml of PBS and fixed in 0.8 ml of ice-cold 100% ethanol at -20℃ overnight. The cell pellets were collected by centrifugation, resuspended in 1 ml of hypotonic buffer (0.5% Triton X-100 in PBS and 0.5 µg/ml RNase A) and incubated at 37℃ for 30 min. Then, 1 ml of PI solution (50 µg/ml) was added and the mixture was allowed to stand on ice for 30 min. The nuclei were analyzed in a FACSCAN laser flow cytometer (Becton Dickenson, USA).
Detection of intracellular ROS
Intracellular oxidative stress was assayed by measuring intracellular oxidation of 2', 7'-dichlorofluorescin (DCFH) (Carter et al., 1994; Amer et al., 2003). The substrate is DCFH-DA, which easily diffuses into the cell and is then deacetylated by cellular esterases to the more hydrophilic, nonfluorescent DCFH. ROS generation in the cell oxidizes DCFH to the fluorescent 2', 7'-dichlorofluorescein (DCF). DCF fluorescence was measured in a flow cytometer using the WinMDI software. In each study, 10,000 events (cells) were counted.
Analysis of the mitochondrial membrane potential (Δψm)
The Δψm was monitored by fluorescence of rhodamine 123 (Liu et al., 2005). Cells were incubated with 10 µM rhodamine 123 for 10 min. Finally, cells were detached and fluorescence was measured in a flow cytometer. In each study, 10,000 events (cells) were counted. Data were acquired and analyzed using WinMDI software.
Immunoblotting
To extract mitochondrial proteins, cells were washed once with PBS, and then resuspended with mitochondrial buffer (25 mM Tris, pH 6.8, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF and 250 mM sucrose). To purify the total proteins, cells were harvested and lysed in cold lysis buffer (10% v/v glycerol, 1% v/v Triton X-100, 1 mM sodium orthovanadate, 1 mM EGTA, 10 mM NaF, 1 mM sodium pyrophosphate, 20 mM Tris, pH 7.9, 100 µM β-glycerophosphate, 137 mM NaCl, 5 mM EDTA, 1 mM PMSF, 10 µg/ml aprotinin and 10 µg/ml leupeptin), homogenized, centrifuged, and then the supernatant was boiled in a loading buffer with an aliquot corresponding to 50 µg of protein being separated by SDS-PAGE. After blotting, PVDF membranes were incubated with the primary antibodies for 6 h and the secondary antibody labeled with horseradish-peroxidase was incubated for 1 h. The antigen-antibody complexes were visualized by the enhanced chemiluminescence.
Statistical analysis
Data shown represent means ± standard deviation (S.D.). Statistical analyses for detection of significant differences between the control and experimental groups were carried out using Student's t test.
Acknowledgements
This study was financially supported by the grants from the Chung Shan Medical University and the Tung's Taichung MetroHarbor hospital (CSMU-TTM-098-005).
Abbreviations
- DB
dibenzoylmethane
- HDB
hydroxydibenzoylmethane
- ODC
ornithine decarboxylase
References
- 1.Amer J, Goldfarb A, Fibach E. Flow cytometric measurement of reactive oxygen species production by normal and thalassaemic red blood cells. Eur J Haematol. 2003;70:84–90. doi: 10.1034/j.1600-0609.2003.00011.x. [DOI] [PubMed] [Google Scholar]
- 2.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auvinen M, Paasinen A, Andersson LC, Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 4.Carter WO, Narayanan PK, Robinson JP. Intracellular hydrogen peroxide and superoxide anion detection in endothelial cells. J Leukoc Biol. 1994;55:253–258. doi: 10.1002/jlb.55.2.253. [DOI] [PubMed] [Google Scholar]
- 5.Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J. 2009;421:323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinkova-Kostova AT, Talalay P. Relation of structure of curcumin analogs to their potencies as inducers of Phase 2 detoxification enzymes. Carcinogenesis. 1999;20:911–914. doi: 10.1093/carcin/20.5.911. [DOI] [PubMed] [Google Scholar]
- 8.Fong LY, Nguyen VT, Pegg AE, Magee PN. Alpha-difluoromethylornithine induction of apoptosis: a mechanism which reverses pre-established cell proliferation and cancer initiation in esophageal carcinogenesis in zincdeficient rats. Cancer Epidemiol Biomarkers Prev. 2001;10:191–199. [PubMed] [Google Scholar]
- 9.Gerner EW. Cancer chemoprevention locks onto a new polyamine metabolic target. Cancer Prev Res (Phila) 2010;3:125–127. doi: 10.1158/1940-6207.CAPR-09-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giardiello FM, Hamilton SR, Hylind LM, Yang VW, Tamez P, Casero RA., Jr Ornithine decarboxylase and polyamines in familial adenomatous polyposis. Cancer Res. 1997;57:199–201. [PubMed] [Google Scholar]
- 11.Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA., Jr The natural polyamine spermine functions directly as a free radical scavenger. Proc Natl Acad Sci USA. 1998;95:11140–11145. doi: 10.1073/pnas.95.19.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jana S, Mandlekar S. Role of phase II drug metabolizing enzymes in cancer chemoprevention. Curr Drug Metab. 2009;10:595–616. doi: 10.2174/138920009789375379. [DOI] [PubMed] [Google Scholar]
- 13.Lavrik IN, Golks A, Krammer PH. Caspases: pharmacological manipulation of cell death. J Clin Invest. 2005;115:2665–2672. doi: 10.1172/JCI26252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao YF, Hung HC, Hour TC, Hsu PC, Kao MC, Tsay GJ, Liu GY. Curcumin induces apoptosis through an ornithine decarboxylase-dependent pathway in human promyelocytic leukemia HL-60 cells. Life Sci. 2008;82:367–375. doi: 10.1016/j.lfs.2007.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Liu GY, Hung YC, Hsu PC, Liao YF, Chang WH, Tsay GJ, Hung HC. Ornithine decarboxylase prevents tumor necrosis factor alpha-induced apoptosis by decreasing intracellular reactive oxygen species. Apoptosis. 2005;10:569–581. doi: 10.1007/s10495-005-1891-2. [DOI] [PubMed] [Google Scholar]
- 16.Manni A, Mauger D, Gimotty P, Badger B. Prognostic influence on survival of increased ornithine decarboxylase activity in human breast cancer. Clin Cancer Res. 1996;2:1901–1906. [PubMed] [Google Scholar]
- 17.Meyskens FL, Jr, Gerner EW. Development of difluoromethylornithine (DFMO) as a chemoprevention agent. Clin Cancer Res. 1999;5:945–951. [PubMed] [Google Scholar]
- 18.Mohan RR, Challa A, Gupta S, Bostwick DG, Ahmad N, Agarwal R, Marengo SR, Amini SB, Paras F, MacLennan GT, Resnick MI, Mukhtar H. Overexpression of ornithine decarboxylase in prostate cancer and prostatic fluid in humans. Clin Cancer Res. 1999;5:143–147. [PubMed] [Google Scholar]
- 19.O'Brien TG, Megosh LC, Gilliard G, Soler AP. Ornithine decarboxylase overexpression is a sufficient condition for tumor promotion in mouse skin. Cancer Res. 1997;57:2630–2637. [PubMed] [Google Scholar]
- 20.Pan MH, Huang MC, Wang YJ, Lin JK, Lin CH. Induction of apoptosis by hydroxydibenzoylmethane through coordinative modulation of cyclin D3, Bcl-X(L), and Bax, release of cytochrome c, and sequential activation of caspases in human colorectal carcinoma cells. J Agric Food Chem. 2003;51:3977–3984. doi: 10.1021/jf034094i. [DOI] [PubMed] [Google Scholar]
- 21.Pan MH, Sin YH, Lai CS, Wang YJ, Lin JK, Wang M, Ho CT. Induction of apoptosis by 1-(2-hydroxy-5-methylphenyl)-3-phenyl-1,3-propanedione through reactive oxygen species production, GADD153 expression, and caspases activation in human epidermoid carcinoma cells. J Agric Food Chem. 2005;53:9039–9049. doi: 10.1021/jf051476a. [DOI] [PubMed] [Google Scholar]
- 22.Park JK, Chung YM, Kang S, Kim JU, Kim YT, Kim HJ, Kim YH, Kim JS, Yoo YD. c-Myc exerts a protective function through ornithine decarboxylase against cellular insults. Mol Pharmacol. 2002;62:1400–1408. doi: 10.1124/mol.62.6.1400. [DOI] [PubMed] [Google Scholar]
- 23.Ploszaj T, Motyl T, Zimowska W, Skierski J, Zwierzchowski L. Inhibition of ornithine decarboxylase by alpha-difluoromethylornithine induces apoptosis of HC11 mouse mammary epithelial cells. Amino Acids. 2000;19:483–496. doi: 10.1007/s007260070026. [DOI] [PubMed] [Google Scholar]
- 24.Rigas B, Sun Y. Induction of oxidative stress as a mechanism of action of chemopreventive agents against cancer. Br J Cancer. 2008;98:1157–1160. doi: 10.1038/sj.bjc.6604225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith MK, Goral MA, Wright JH, Matrisian LM, Morris RJ, Klein-Szanto AJ, Gilmour SK. Ornithine decarboxylase overexpression leads to increased epithelial tumor invasiveness. Cancer Res. 1997;57:2104–2108. [PubMed] [Google Scholar]
- 26.Sun SY, Hail N, Jr, Lotan R. Apoptosis as a novel target for cancer chemoprevention. J Natl Cancer Inst. 2004;96:662–672. doi: 10.1093/jnci/djh123. [DOI] [PubMed] [Google Scholar]