Abstract
The B30.2/SPRY domain is present in many proteins, including various members of the tripartite motif (TRIM) protein family such as TRIM5α, which mediates innate intracellular resistance to retroviruses in several primate species. This resistance is dependent on the integrity of the B30.2 domain that evolves rapidly in primates and exhibits species-specific anti-viral activity. TRIM22 is another positively selected TRIM gene. Particularly, the B30.2 domain shows rapid evolution in the primate lineage and recently published data indicate an anti-viral function of TRIM22. We show here that human and rhesus TRIM22 localise to different subcellular compartments and that this difference can be assigned to the positively selected B30.2 domain. Moreover, we could demonstrate that amino acid changes in two variable loops (VL1 and VL3) are responsible for the different subcellular localisations.
Keywords: TRIM22, Retrovirus, B30.2 domain
Introduction
Retroviruses can be potently blocked in target cells shortly after infection as part of an innate immune response (Bieniasz 2004; Towers 2007). Such restrictions are often encountered when species borders are crossed in infections, e.g. the human immunodeficiency virus type 1 (HIV-1) is restrained in cells of catarrhine primates (human, apes, Old World monkeys), or cells of platyrrhine primates (New World monkeys) restrict immunodeficiency viruses of Catarrhini (Himathongkham and Luciw 1996; Hofmann et al. 1999; LaBonte et al. 2002; Shibata et al. 1995). Stremlau et al. (2004) showed that TRIM5α is an essential factor for the inhibition of certain retroviruses and since that, the anti-viral function of TRIM5α has been under intensive investigation. Interestingly, this TRIM5α-mediated restriction is species-specific, meaning that different viruses are blocked in different primate species (Bieniasz 2003; Sharp et al. 2001; Stoye 2002). While the TRIM5α protein of rhesus monkeys (Macaca mulatta, rhTRIM5α) potently inhibits an infection with HIV-1, the human and chimpanzee TRIM5α can only moderately block this virus (Stremlau et al. 2005).
TRIM5α belongs to the family of tripartite motif (TRIM containing) proteins, which consist of three domains: a RING, a B-box and a coiled-coil domain (Reddy et al. 1992). The RING domain is involved in protein–protein interactions and exhibits E3 ubiquitin ligase activity, which is indeed a function of some TRIM proteins (Freemont et al. 1991; Joazeiro and Weissman 2000; Nisole et al. 2005). The B-box domains have so far only been identified in TRIM proteins and might be involved in protein–protein interactions as well (Borden 1998; Nisole et al. 2005). Coiled-coil domains are generally involved in forming homo- and hetero-multimers (Reymond et al. 2001). Approximately 60% of all known human TRIM genes also encode a variable fourth domain at the carboxy terminus, the so-called B30.2 domain (Nisole et al. 2005), which was originally discovered in butyrophilin (Henry et al. 1998).
TRIM proteins form homomultimers through their coiled-coil domains and tend to form higher order protein complexes (Reymond et al. 2001). Such complexes are found in different subcellular compartments, thereby determining the specific functions of the various TRIM proteins. As already pointed out, TRIM proteins are involved in innate immune responses. The transcription of various TRIM genes, e.g. TRIM22, can be induced with type I interferons (Barr et al. 2008; Gongora et al. 2000; James et al. 2007; Orimo et al. 2000; Rajsbaum et al. 2008; Rhodes et al. 2002; Toniato et al. 2002). TRIM22 plays a role in limiting HIV-1 infections by binding to the viral gag protein and interfering with virus budding (Barr et al. 2008). Additionally, TRIM22 is able to repress viral gene transcription in HIV-1 (Tissot and Mechti 1995). TRIM34 can moderately inhibit SIVmac infection (Zhang et al. 2006) and TRIM32 interacts with tat proteins of HIV-1 and HIV-2 (Fridell et al. 1995). TRIM1 moderately restricts infections with N-MLV and overexpression of TRIM19 mediates resistance against the human cytomegalovirus, the vesicular stomatitis virus, herpes simplex virus and influenza virus A (Chelbi-Alix et al. 1998; Everett et al. 2006; Tavalai et al. 2006).
Most TRIM proteins, which were reported in an anti-viral context (e.g. TRIM1, 5, 22, 26 and 34), possess a B30.2 domain at the C-terminus. Stremlau et al. (2005) reported that the B30.2 domain of TRIM5α is responsible for HIV-1 restriction. Song et al. (2005) investigated TRIM5α and other related TRIM genes in several primate species and rodents and identified four regions in the B30.2 domain, which showed species-specific sequence and length variation, referred to as variable regions v1, v2, v3 and v4. The TRIM5α protein of primates is especially polymorphic in v1 and v3, with v1 and v3 being elongated in Catarrhini and Platyrrhini, respectively. Moreover, the v1 region contains exactly the amino acid sequence that is essential for the HIV-1 restriction in rhesus macaques (Stremlau et al. 2005).
Here, we report that human and rhesus macaque TRIM22 differ in their subcellular localisation and this difference could be assigned to the polymorphic B30.2 domain. Furthermore, we could demonstrate that polymorphic amino acid residues of two variable loops in the B30.2 domains, which are expected to mediate contacts to the binding partner, are responsible for the different subcellular localisations.
Material and methods
cDNA constructs and transfection
Full-length human and rhesus macaque TRIM22 cDNA sequences (hTRIM22, rhTRIM22) were obtained with primers 5′aag ctt gca atg gat ttc tca gta aag 3′ (forward human and rhesus macaque), 5′aag ctt cca gga gct cgg tgg gca c 3′ (reverse human) and 5′aag ctt gga gct cgg ttg gca cag 3′ (reverse rhesus macaque), which contain HindIII restriction sites (underlined). PCR products were cloned into the pGEM-T Easy PCR cloning vector (Promega). Clones were sequenced and subcloned in the pAcGFP1-N1 expression vector (Clontech) to allow for expression of a TRIM22-AcGFP fusion protein. For the expression of a V5-His-tagged TRIM22 protein, PCR products were directly cloned in pcDNA3.1/V5-His-TOPO® (Invitrogen). The constructs were transiently transfected in COS-7 or HeLa cells by using Metafectene (Biontex) according to the supplier's manual.
Establishment of chimeric constructs
The chimeric constructs hTRIM-rhB30.2-22 and rhTRIM-hB30.2-22 were established by exchanging the exons coding for the TRIM22 B30.2 domains of human and rhesus macaque. The exons were excised from the expression constructs using ScaI and SacII restriction enzymes. In the rhesus macaque construct, an internal ScaI restriction site had to be generated using site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Stratagene), which did not change the amino acid sequence. The excised exons were ligated to the opposed remaining TRIM22 constructs.
As TRIM22 possesses an internal BamHI restriction site, the chimeric constructs hTRIM-rhB30.2(368-498)-22 and rhTRIM-hB30.2(368-498)-22 could be established by excising the B30.2 segments and ligating the excised part to the remaining opposite TRIM22 construct. All other chimeric constructs were established by using site-directed mutagenesis according to the supplier's manual (QuikChange Site Directed Mutagenesis Kit, Stratagene).
Immunoblotting and immunoprecipitation
For co-immunoprecipitation experiments HEK293T cells were transiently co-transfected with hTRIM22-V5-His and rhTRIM22-AcGFP expression constructs. Twenty-four hours post-transfection, 0.5 × 107 cells were lysed with 500 µl NP-40 lysis buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1% NP-40) containing protease inhibitor cocktail (Roche). After incubation overnight at 4°C with 1 µg monoclonal anti-GFP antibody (Jl-8, BD), 50 µl of precleared protein G sepharose beads was added to the cell lysate and the suspension was incubated for an additional 2 h at 4°C on a rotator. Protein G sepharose beads were washed five times with ice-cold NP-40 lysis buffer to remove unbound protein. Proteins were eluted by adding sample buffer (1% SDS, 100 mM DTT, 50 mM TRIS pH 7.5) to the beads, and after incubation at 95°C for 10 min, proteins were resolved by SDS-PAGE and immunoblotted with horseradish peroxidase-conjugated anti-V5 antibody (Invitrogen).
Immunocytochemistry and microscopy
Transiently transfected cells (HeLa or COS-7) were grown on tissue culture coverslips (Sarstedt) for 24–48 h and fixed with 3% paraformaldehyde for 10 min. Human and rhesus macaque peripheral blood mononuclear cells (PBMC) were isolated from whole blood using Leucosep tubes (Greiner) and fixed with 1.5% paraformaldehyd for 10 min.
After three washes with phosphate-buffered saline (PBS), cells were permeabilised with 0.5% Triton X-100 in PBS for 5 min at room temperature. Non-specific binding was blocked by incubation with 5% bovine serum albumin (BSA) in PBS for 30 min at room temperature. Antibodies were diluted in 0.5% BSA in PBS; primary antibody (anti-α-tubulin, 1:5,000; polyclonal anti-TRIM22, 1:500) was applied overnight at 4°C and secondary antibody (polyclonal rabbit–anti-mouse–TRITC, DAKO, 1:40) for 1 h at room temperature. Nuclei were stained with 4′,6-diamidino-2-phenylindoledihydrochloride (DAPI; Sigma). Cells were rinsed with PBS and mounted with fluorescent mounting medium (DAKO). Fluorescent cells were analysed with a Zeiss Axio Observer confocal microscope through ×20 or ×40 objectives or with a confocal laser scanning microscope LSM 5 PASCAL (Zeiss) through ×40 or ×63 objectives.
Establishment of a mouse anti-human TRIM22 antiserum
A bacterial expression construct of human TRIM22 that lacks the coiled-coil domain was established using the prokaryotic expression vector pGEX-4T3, which allows the expression of a GST-tagged protein. After overexpression of TRIM22-GST, inclusion bodies were isolated and 100 µg was used to immunise C3H mice. Serum was obtained after three immunisations with 2-week intervals. The immunisations had been approved by the local government and were in accordance with institutional guidelines for the welfare of animals. Specificity of the antiserum was analysed by immunoblotting and immunocytochemistry. The antiserum also cross-reacts with rhesus macaque TRIM22.
Results
Subcellular localisation of TRIM22-AcGFP fusion constructs
Recent data indicated that the B30.2 domain of TRIM22 is subject to positive selection in primates (Sawyer et al. 2007). We therefore hypothesised that TRIM22 might show species-specific differences in function, similar to what was published for TRIM5α. In a first approach, we compared the subcellular localisation of human and rhesus macaque TRIM22, as different subcellular localisations are frequently associated with functional differences.
COS-7 cells were transiently transfected with pAcAcGFP-hTRIM22 or pAcGFP-rhTRIM22 constructs to allow for expression of AcGFP-tagged proteins, respectively. Protein expression was visualised 48 h post-transfection by confocal microscopy. Interestingly, human and rhesus macaque TRIM22 showed indeed a different subcellular localisation (Fig. 1a). Human TRIM22 was localised in diffuse accumulation around the nucleus, while rhesus macaque TRIM22 formed cytoplasmic bodies (speckles). These speckles of rhesus macaque TRIM22 are hollow structures with diameters up to 2 µm (Fig. 1b). Similar structures were also described for human TRIM5α (Campbell et al. 2007). The possibility that the subcellular localisation is influenced by the AcGFP tag was excluded by using respective V5/His-tagged TRIM22 expression constructs (Fig. 1c). We also transiently transfected human cell lines HeLa and HEK293T with the human and rhesus macaque TRIM22 expression constructs and found the same intracellular distribution as for COS-7 cells (data not shown).
Fig. 1.
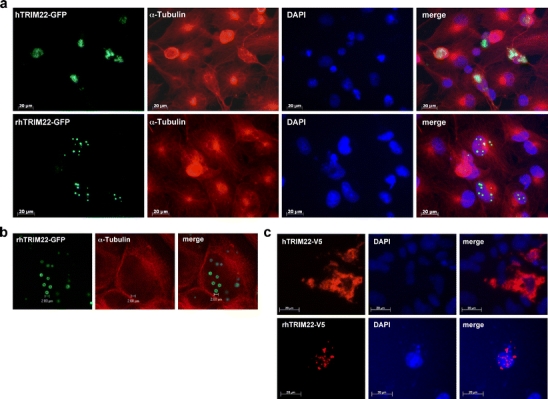
Subcellular localisation of TRIM22. a COS-7 cells were transiently transfected with human (hTRIM22-AcGFP) or rhesus macaque (rhTRIM22-AcGFP) TRIM22 expression constructs (green). After 24 h, cells were fixed and α-tubulin was stained with anti-α-tubulin antibody (red) and nuclei were stained with DAPI (blue). Fluorescent microscopy (×40 magnification) reveals that human TRIM22 is localised diffuse in the cytoplasm, whereas rhesus macaque TRIM22 forms speckles. b COS-7 cells were transiently transfected with rhTRIM22-AcGFP fusion construct. Twenty-four hours post-transfection, cells were fixed and α-tubulin was stained with anti-α-tubulin antibody (red). Fluorescent cells were analysed with a confocal laser-scanning microscope with ×100 magnification. The rhTRIM22 speckles (green) are hollow structures with diameter up to 2 µm. c HeLa cells were transiently transfected with hTRIM22-V5 and rhTRIM22-V5. Forty-eight hours post-transfection, cells were fixed and TRIM22 was detected by anti-V5 antibody (red). Nuclei were stained with DAPI (blue). TRIM22-V5 molecules show comparable subcellular localisation as TRIM22-AcGFP: hTRIM22 localises diffuse in the cytoplasm; rhTRIM22 forms speckles
As the localisation results were obtained from transient transfection experiments and overexpression of TRIM22-AcGFP expression proteins, we investigated the endogenous TRIM22 expression with an anti-TRIM22 antiserum, which we raised in mice. As shown in Fig. 2a, the endogenous hTRIM22 in HeLa cells exhibited the same diffuse cytoplasmic distribution as hTRIM22-AcGFP-transfected HeLa cells. Moreover, we compared human and rhesus macaque endogenous TRIM22 expression in PBMC by indirect immunofluorescence with the specific antiserum. The endogenous rhesus macaque TRIM22 was localised in comparable cytoplasmic speckles as rhTRIM22-AcGFP-transfected HeLa cells while the endogenous human TRIM22 showed diffuse cytoplasmic localisation (Fig. 2b).
Fig. 2.
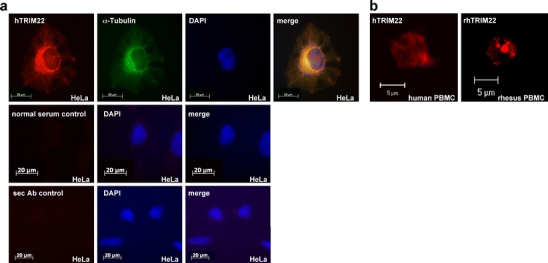
Subcellular localisation of endogenous TRIM22. a HeLa cells were fixed and stained with the mouse anti-human TRIM22 antiserum and with secondary rabbit anti-mouse TRITC-coupled antibody. α-Tubulin was stained with FITC-coupled anti-α-tubulin antibody, and the nucleus was visualised with DAPI. The endogenous hTRIM22 is localised diffuse in the cytoplasm (×40 magnification). No staining was observed in control experiments, where cells were incubated with normal mouse serum obtained before immunisation (normal serum control) or with the secondary antibody (sec Ab control) alone. b After staining of human and rhesus macaque PBMC with the mouse anti-human TRIM22 antiserum and with secondary rabbit anti-mouse TRITC-coupled antibody, the endogenous hTRIM22 can be found diffuse in the cytoplasm while the endogenous rhTRIM22 forms speckles (×63 magnification, LSM). Note that small lymphocytes were used for staining in comparison to large HeLa cells
Interaction of hTRIM22 and rhTRIM22
Upon co-transfection of human and rhesus macaque TRIM22 constructs in HeLa cells, co-localisation of both proteins was found in most cells (Fig. 3a). TRIM22-expressing cells either showed a diffuse cytoplasmic localisation or speckles, which is most likely depending on the individual concentration of both constructs in a single co-transfected cell. These data suggest interaction of human and rhesus macaque TRIM22. As it is known that TRIM proteins can form multimers via their coiled-coil domain (Reymond et al. 2001), we performed co-immunoprecipitations to investigate whether this observed co-localisation upon co-transfection of both constructs is due to molecular interaction. HEK293T cells were co-transfected with expression constructs coding for a V5/His-tagged human TRIM22 and an AcGFP-tagged rhesus macaque TRIM22. Immunoprecipitation with the anti-AcGFP antibody and subsequent immunoblotting with the anti-V5 antibody detected human V5-tagged TRIM22 (Fig. 3b). These results showed that hTRIM22 and rhTRIM22 indeed interact, most likely via their coiled-coil domains, and therefore co-localise upon co-expression.
Fig. 3.
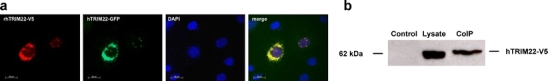
Co-expression of human and rhesus macaque TRIM22. a HeLa cells were co-transfected with both hTRIM22-AcGFP and rhTRIM22-V5/His expression constructs. Twenty-four hours post-transfection, cells were fixed and rhTRIM22-V5-His was stained with anti-V5 antibody (red). Fluorescent microscopy (×40 magnification) reveals that hTRIM22 and rhTRIM22 co-localise upon co-expression. Both diffuse localisation and formation of speckles could be observed. b Interaction of human and rhesus macaque TRIM22. HEK293T cells were transfected with both hTRIM22-V5-His and rhTRIM22-AcGFP expression constructs. Twenty-four hours later, cell lysates were immunoprecipitated with anti-GFP antibody. Immunoblotting with anti-V5 antibody detected co-immunoprecipitated hTRIM22. Untransfected HEK293T cells were used as a control
Differences in subcellular localisation are due to polymorphisms in the variable loops of the B30.2 domain
We postulated that the positively selected B30.2 domain might be responsible for the diverse subcellular distribution of human and rhesus macaque TRIM22. To test this hypothesis, the human B30.2 domain was fused to the tripartite motif of the rhesus macaque TRIM22 and vice versa (Fig. 4a). After transient transfection in COS-7 cells, the chimeric human TRIM22 with the rhesus macaque B30.2 domain is localised in speckles (Fig. 4b) like the parental rhesus macaque TRIM22 (see Fig. 1). In contrast, the chimeric rhesus macaque TRIM22 with the human B30.2 domain shows diffuse cytoplasmic distribution (Fig. 4b) like the parental human TRIM22 (see Fig. 1). Thus, the rapidly evolving B30.2 domain is responsible for the diverse subcellular localisations.
Fig. 4.
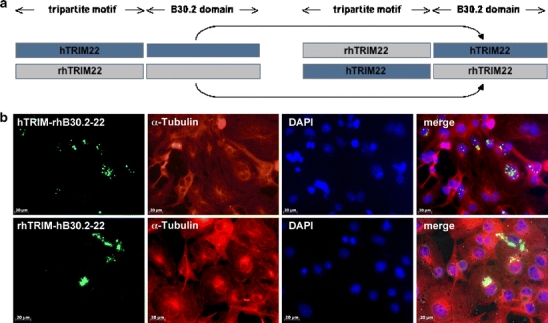
Establishment of chimeric TRIM22 constructs and transient transfection in COS-7 cells. a The human B30.2 domain of TRIM22 was fused to the tripartite motif of rhesus macaque TRIM22 (rhTRIM-hB30.2-22) and vice versa (hTRIM-rhB30.2-22). b COS-7 cells were fixed 24 h after transient transfection of the chimeric TRIM22 constructs, α-tubulin was visualised with anti-α-tubulin antibody (red) and the nuclei were stained with DAPI (blue). rhTRIM-hB30.2-22 is localised diffuse around the nucleus like the parental hTRIM22, whereas hTRIM-rhB30.2-22 forms comparable cytoplasmic speckles like the parental rhTRIM22, indicating that the B30.2 domain influences the subcellular localisation
The next question was then to identify the amino acid residues in the B30.2 domain that mediate the different subcellular localisations. Several chimeric constructs were established (Fig. 5) and transiently transfected in HeLa and COS-7 cells. Construct hTRIM-rhB30.2(368-498)-22 showed similar subcellular localisation as rhTRIM22, while construct rhTRIM-hB30.2(368-498)-22 exhibited diffuse localisation as hTRIM22 (Fig. 5). These data indicate that differences in the amino acid sequence of the region 368–498 of human and rhesus macaque TRIM22 are likely to be responsible for the observed differential localisation. We further narrowed down the responsible amino acid residues by site-directed mutagenesis and established three further constructs of the complete human TRIM22 in which either codon 476, codons 493 and 495 or codons 395, 396 and 400 were replaced by the respective rhesus macaque codons. However, all three mutated human TRIM22 constructs showed the same localisation as the parental hTRIM22 (Fig. 5). Therefore, none of the tested amino acids alone is responsible for the subcellular localisation. We then tested combinations of mutated amino acids. Indeed, the chimeric construct hTRIM-rhB30.2(368-498)h395,396,400-22 is localised diffuse in the cytoplasm as the parental hTRIM22, while the chimeric construct hTRIM-rhB30.2(368-498)-22 (see also Fig. 4) shows the same cytoplasmic speckles as rhTRIM22 (Figs. 5 and 6). Thus, the amino acids located in the region 301–368 in combination with the amino acids 395, 396 and 400 are responsible for the subcellular localisation of TRIM22.
Fig. 5.
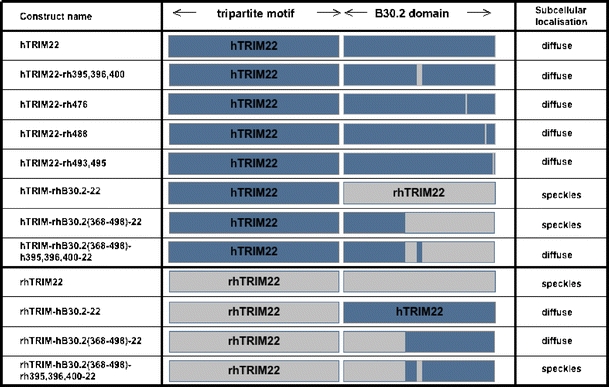
Schematic illustration of TRIM22 chimeric constructs. The established chimeric TRIM22 constructs are named according to their mutated amino acids. Human TRIM22 is marked in blue; rhesus macaque TRIM22 is marked in grey. The observed subcellular localisation upon transfection of the respective construct in HeLa and COS-7 cells is listed
Fig. 6.
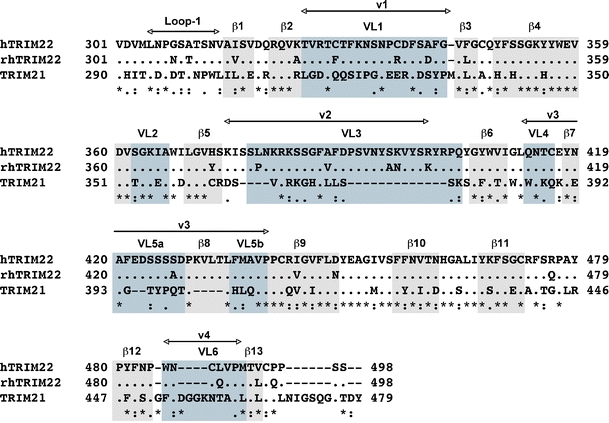
Alignment of the B30.2 domain amino acid sequence of hTRIM22 and rhTRIM22 with TRIM21. The β-sheets (grey) and the variable loops (blue) are outlined as indicated by James et al. (2007). The variable regions v1–v4 described by Song et al. (2005) are marked with arrows. Dots represent identical amino acid residues, dashes indicate gaps introduced to maximise homology
Localisation of TRIM22 polymorphisms in a 3D model
As 3D data of TRIM22 are currently not available, we aligned its B30.2 domain with that of TRIM21 for which the crystal structure is already known (James et al. 2007; Keeble et al. 2008). The B30.2 domain of TRIM21 forms β-sheets that alternate with variable loops, consistent with the variable regions of TRIM5α identified by Song et al. (2005). Interestingly, most polymorphic residues between the hTRIM22 and rhTRIM22 B30.2 domains are located in the variable loops VL1 and VL3 (Fig. 7). Moreover, our experiments with the chimeric and mutated TRIM22 constructs show that the polymorphic amino acids, which are responsible for the different subcellular localisations, are located in these two variable loops.
Fig. 7.
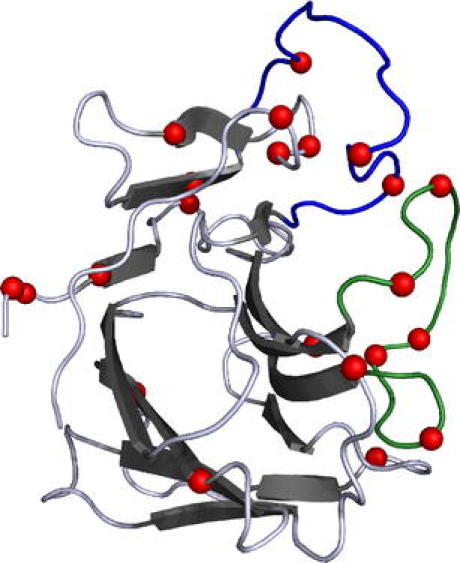
3D model of the B30.2 domain of TRIM21 according to James et al. (2007). The B30.2 domain exhibits variable loops between conserved β-sheets. The amino acid sequence of TRIM22 was aligned to TRIM21. The differences between human and rhesus macaque TRIM22 were marked in the 3D model according to the corresponding amino acids in TRIM21 (red). As TRIM22 possesses sequence elongations in VL3, the marked amino acid residues in this region do not represent the corresponding amino acids of TRIM21. Most amino acid substitutions between human and rhesus macaque TRIM22 are found in the variable regions VL1 (blue) and VL3 (green)
Discussion
It was previously shown that the anti-viral TRIM5α and TRIM22 genes are subject to positive selection (Sawyer et al. 2005, 2007). In particular the B30.2 domains show signs of positive selection in the primate lineage, including amino acid changes and sequence elongations. These changes lead to species-specific anti-viral activity of TRIM5α (Sawyer et al. 2005; Song et al. 2005). Other TRIM proteins were also shown to exhibit anti-viral activity, although these activities are less specific against particular retroviruses as was reported for TRIM5α (Bonilla et al. 2002; Turelli et al. 2001; Zhang et al. 2006).
We focused our analysis on the subcellular localisation of TRIM22, because variations in localisation are likely associated with functional differences. Interestingly, we found that human and rhesus macaque TRIM22 differ in their subcellular localisation. These differences could be assigned to the positively selected B30.2 domain. Human TRIM22 localises diffuse in the cytoplasm, while rhesus macaque TRIM22 forms cytoplasmic bodies (speckles). TRIM proteins form high molecular weight complexes via the coiled-coil domain and this multimerising is necessary for the correct subcellular localisation. The RING and B-box domains are also involved in the subcellular targeting of TRIM proteins (Reymond et al. 2001). Our data clearly indicate that in addition the B30.2 domain is important for the cellular distribution, at least in TRIM22.
Human TRIM5α tend to form speckles as rhesus macaque TRIM22 does. These speckles of TRIM5α are highly dynamic structures and move along microtubules (Campbell et al. 2007). The structure of rhesus macaque TRIM22 speckles resembles the nuclear bodies formed by TRIM19 (PML), the best characterised TRIM family member. For PML, it was hypothesised that nuclear body formation is necessary for PML function (Dong et al. 2004; Zhu et al. 1997). Mutations in the B30.2 domain of TRIM9 and TRIM18 (MID1) also lead to changes in their subcellular localisation: while both proteins are normally located in microtubuli-associated structures, mutations lead to localisation into cytoplasmic speckles (Short and Cox 2006). The described mutations in the B30.2 domain of TRIM18 are associated with the Opitz syndrome (Cox et al. 2000). Reymond et al. (2001) hypothesised that TRIM domains provide the protein–protein interfaces for the recruitment of other proteins, thereby specifying cellular compartments. The rapid evolution of TRIM22, and in particular its B30.2 domain, might have led to differences in protein–protein interaction interfaces and to species-specific functional differences of TRIM22 as a means to adapt to pathogenic threats.
Song et al. (2005) identified four variable regions (v1–v4) in the B30.2 domain of TRIM proteins. For TRIM5α, several amino acid substitutions and sequence elongations were found in the v1 and v3 regions. Moreover, it was shown that those amino acids responsible for the restriction of HIV are located in the v1 region. James et al. (2007) analysed the crystal structure of the B30.2 domain of TRIM21 and found that the B30.2 domain comprises several conserved β-sheets alternating with variable loops (VL), which correspond to the v1–v4 regions identified by Song et al. (2005). These variable regions are thought to be involved in protein–protein interactions. Interestingly, most amino acid differences between human and rhesus macaque TRIM22 map to VL1 and VL3. We could narrow down the amino acids, which are responsible for the subcellular localisation, to the VL1 and VL3 regions. Those amino acid substitutions might lead to conformational changes that alter the functional properties of TRIM22. Alignment of the TRIM22 VL1 and VL3 amino acid residues with the known 3D structure of the TRIM21 B30.2 domain (James et al. 2007) suggests that these variable loops are in close contact (Fig. 7).
It has been shown by several groups that human TRIM22 is inducible by type I interferons and is able to block HIV-1 replication (Barr et al. 2008; Bouazzaoui et al. 2006; Tissot and Mechti 1995). Moreover, Barr et al. (2008) could demonstrate binding of TRIM22 to HIV-Gag, resulting in alteration of Gag's intracellular trafficking from punctuate structures near the cell membrane to diffuse cytoplasmic localisation. Interestingly, differences in the subcellular localisations were also reported for human and rhesus macaque TRIM5α (Campbell et al. 2007; Stremlau et al. 2004). In addition, the positive selection of the closely linked TRIM5α and TRIM22 genes was found to be mutually exclusive in primates and mutually exclusive presence/absence of these genes is evident in certain non-primate mammals (Sawyer et al. 2007). These data indicate that rhesus macaque TRIM5α and human TRIM22 have assumed similar anti-retroviral functions, but that either TRIM5 or TRIM22 predominate in the anti-retroviral activity of primates.
In summary, our findings showed a difference in the subcellular localisation of human and rhesus macaque TRIM22. This difference could be assigned to the B30.2 domain and was further narrowed down to few polymorphic amino acid residues in the variable loops VL1 and VL3. We hypothesise that these differences in the subcellular localisation are associated with functional variations of TRIM22 and reflect evolutionary adaptations to species-specific anti-retroviral functions.
Acknowledgements
The authors are grateful to Nicole Otto, Christiane Schwarz and Leslie Elsner for expert technical assistance. This study was supported by the Volkswagenstiftung (‘Evolutionsbiologie’) by a Ph.D. fellowship to A.H.
References
- Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD. Restriction factors: a defense against retroviral infection. Trends Microbiol. 2003;11:286–291. doi: 10.1016/S0966-842X(03)00123-9. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- Bonilla WV, Pinschewer DD, Klenerman P, Rousson V, Gaboli M, Pandolfi PP, Zinkernagel RM, Salvato MS, Hengartner H. Effects of promyelocytic leukemia protein on virus-host balance. J Virol. 2002;76:3810–3818. doi: 10.1128/JVI.76.8.3810-3818.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL. RING fingers and B-boxes: zinc-binding protein–protein interaction domains. Biochem Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- Bouazzaoui A, Kreutz M, Eisert V, Dinauer N, Heinzelmann A, Hallenberger S, Strayle J, Walker R, Rubsamen-Waigmann H, Andreesen R, Briesen H. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology. 2006;356:79–94. doi: 10.1016/j.virol.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Campbell EM, Dodding MP, Yap MW, Wu X, Gallois-Montbrun S, Malim MH, Stoye JP, Hope TJ. TRIM5 alpha cytoplasmic bodies are highly dynamic structures. Mol Biol Cell. 2007;18:2102–2111. doi: 10.1091/mbc.E06-12-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix MK, Quignon F, Pelicano L, Koken MH, The H. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol. 1998;72:1043–1051. doi: 10.1128/jvi.72.2.1043-1051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TC, Allen LR, Cox LL, Hopwood B, Goodwin B, Haan E, Suthers GK. New mutations in MID1 provide support for loss of function as the cause of X-linked Opitz syndrome. Hum Mol Genet. 2000;9:2553–2562. doi: 10.1093/hmg/9.17.2553. [DOI] [PubMed] [Google Scholar]
- Dong S, Stenoien DL, Qiu J, Mancini MA, Tweardy DJ. Reduced intranuclear mobility of APL fusion proteins accompanies their mislocalization and results in sequestration and decreased mobility of retinoid X receptor alpha. Mol Cell Biol. 2004;24:4465–4475. doi: 10.1128/MCB.24.10.4465-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont PS, Hanson IM, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64:483–484. doi: 10.1016/0092-8674(91)90229-R. [DOI] [PubMed] [Google Scholar]
- Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- Gongora C, Tissot C, Cerdan C, Mechti N. The interferon-inducible Staf50 gene is downregulated during T cell costimulation by CD2 and CD28. J Interferon Cytokine Res. 2000;20:955–961. doi: 10.1089/10799900050198390. [DOI] [PubMed] [Google Scholar]
- Henry J, Mather IH, McDermott MF, Pontarotti P. B30.2-like domain proteins: update and new insights into a rapidly expanding family of proteins. Mol Biol Evol. 1998;15:1696–1705. doi: 10.1093/oxfordjournals.molbev.a025896. [DOI] [PubMed] [Google Scholar]
- Himathongkham S, Luciw PA. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology. 1996;219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- Hofmann W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferrigno P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James LC, Keeble AH, Khan Z, Rhodes DA, Trowsdale J. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc Natl Acad Sci U S A. 2007;104:6200–6205. doi: 10.1073/pnas.0609174104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/S0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Keeble AH, Khan Z, Forster A, James LC. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proc Natl Acad Sci U S A. 2008;105:6045–6050. doi: 10.1073/pnas.0800159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonte JA, Babcock GJ, Patel T, Sodroski J. Blockade of HIV-1 infection of New World monkey cells occurs primarily at the stage of virus entry. J Exp Med. 2002;196:431–445. doi: 10.1084/jem.20020468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- Orimo A, Tominaga N, Yoshimura K, Yamauchi Y, Nomura M, Sato M, Nogi Y, Suzuki M, Suzuki H, Ikeda K, Inoue S, Muramatsu M. Molecular cloning of ring finger protein 21 (RNF21)/interferon-responsive finger protein (ifp1), which possesses two RING-B box-coiled coil domains in tandem. Genomics. 2000;69:143–149. doi: 10.1006/geno.2000.6318. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R, Stoye JP, O'Garra A. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur J Immunol. 2008;38:619–630. doi: 10.1002/eji.200737916. [DOI] [PubMed] [Google Scholar]
- Reddy BA, Etkin LD, Freemont PS. A novel zinc finger coiled-coil domain in a family of nuclear proteins. Trends Biochem Sci. 1992;17:344–345. doi: 10.1016/0968-0004(92)90308-V. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DA, Ihrke G, Reinicke AT, Malcherek G, Towey M, Isenberg DA, Trowsdale J. The 52 000MW Ro/SS-A autoantigen in Sjogren's syndrome/systemic lupus erythematosus (Ro52) is an interferon-gamma inducible tripartite motif protein associated with membrane proximal structures. Immunology. 2002;106:246–256. doi: 10.1046/j.1365-2567.2002.01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. Discordant Evolution of the Adjacent Antiretroviral Genes TRIM22 and TRIM5 in Mammals. PLoS Pathog. 2007;3:e197. doi: 10.1371/journal.ppat.0030197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Bailes E, Chaudhuri RR, Rodenburg CM, Santiago MO, Hahn BH. The origins of acquired immune deficiency syndrome viruses: where and when? Philos Trans R Soc Lond B Biol Sci. 2001;356:867–876. doi: 10.1098/rstb.2001.0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Sakai H, Kawamura M, Tokunaga K, Adachi A. Early replication block of human immunodeficiency virus type 1 in monkey cells. J Gen Virol. 1995;76(Pt 11):2723–2730. doi: 10.1099/0022-1317-76-11-2723. [DOI] [PubMed] [Google Scholar]
- Short KM, Cox TC. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J Biol Chem. 2006;281:8970–8980. doi: 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- Song B, Gold B, O'Huigin C, Javanbakht H, Li X, Stremlau M, Winkler C, Dean M, Sodroski J. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5alpha exhibits lineage-specific length and sequence variation in primates. J Virol. 2005;79:6111–6121. doi: 10.1128/JVI.79.10.6111-6121.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye JP. An intracellular block to primate lentivirus replication. Proc Natl Acad Sci U S A. 2002;99:11549–11551. doi: 10.1073/pnas.192449399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Welikala S, Sodroski J. Species-specific variation in the B30.2(SPRY) domain of TRIM5alpha determines the potency of human immunodeficiency virus restriction. J Virol. 2005;79:3139–3145. doi: 10.1128/JVI.79.5.3139-3145.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol. 2006;80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot C, Mechti N. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J Biol Chem. 1995;270:14891–14898. doi: 10.1074/jbc.270.25.14891. [DOI] [PubMed] [Google Scholar]
- Toniato E, Chen XP, Losman J, Flati V, Donahue L, Rothman P. TRIM8/GERP RING finger protein interacts with SOCS-1. J Biol Chem. 2002;277:37315–37322. doi: 10.1074/jbc.M205900200. [DOI] [PubMed] [Google Scholar]
- Towers GJ. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 2007;4:40. doi: 10.1186/1742-4690-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Doucas V, Craig E, Mangeat B, Klages N, Evans R, Kalpana G, Trono D. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol Cell. 2001;7:1245–1254. doi: 10.1016/S1097-2765(01)00255-6. [DOI] [PubMed] [Google Scholar]
- Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology. 2006;353:396–409. doi: 10.1016/j.virol.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Zhu J, Koken MH, Quignon F, Chelbi-Alix MK, Degos L, Wang ZY, Chen Z, The H. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc Natl Acad Sci U S A. 1997;94:3978–3983. doi: 10.1073/pnas.94.8.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]


