Abstract
We have previously shown that high expression levels of the lipid kinase sphingosine kinase-1 (SphK1) correlate with poor survival of glioblastoma (GBM) patients. In this study we examined the regulation of SphK1 expression by epidermal growth factor receptor (EGFR) signaling in GBM cells. As the EGFR gene is often overexpressed and mutated in GBM, and EGFR has been shown to regulate SphK1 in some cell types, we examined the effect of EGF signaling and the constitutively active EGFRvIII mutant on SphK1 in GBM cells. Treatment of glioma cell lines with EGF led to increased expression and activity of SphK1. Expression of EGFRvIII in glioma cells also activated and induced SphK1. In addition, siRNA to SphK1 partially inhibited EGFRvIII-induced growth and survival of glioma cells as well as ERK MAP kinase activation. To further evaluate the connection between EGFR and SphK1 in GBM we examined primary neurosphere cells isolated from fresh human GBM tissue. The GBM-derived neurosphere cell line GBM9, which forms GBM-like tumors intracranially in nude mice, maintained expression of EGFRvIII in culture and had high levels of SphK1 activity. EGFR inhibitors modestly decreased SphK1 activity and proliferation of GBM9 cells. More extensive blockage of SphK1 activity by a SphK inhibitor, potently blocked cell proliferation and induced apoptotic cell death of GBM9 cells. Thus, SphK1 activity is necessary for survival of GBM-derived neurosphere cells, and EGFRvIII partially utilizes SphK1 to further enhance cell proliferation.
Keywords: Sphingosine kinase, Sphingosine-1-phosphate, EGFR, Brain tumor, Glioblastoma
Introduction
Glioblastoma multiforme (GBM) tumors are the most common type of primary brain tumor occurring in adult patients. Despite advances in treatment, GBM patients have a median life span of 12–15 months after diagnosis and a 5-year survival rate of 3% [1, 2]. The effectiveness of treatments including surgery, radiation and chemotherapy is limited due to the high proliferative potential and intrinsic resistance of glioma cells to therapy, as well as the diffusely infiltrating nature of the lesion [3]. Thus, the prognosis for patients remains poor, and a better understanding of the mechanisms regulating the malignant behavior of glioma cells is needed in order to develop effective targeted therapies for these tumors.
We have been investigating the role of the signaling pathway mediated by the lipid sphingosine-1-phosphate (S1P) in regulating malignant behavior of GBM cells. S1P is a bioactive lipid that signals through a set of five G protein-coupled receptors to regulate cell proliferation, survival, motility and differentiation [4]. We have previously shown that high expression levels of the enzyme that generates S1P, sphingosine kinase-1 (SphK1) correlate with an approximately three fold reduced median survival time of GBM patients, suggesting that production of S1P by SphK1 drives malignant behavior of GBM cells [5]. We have also shown that S1P signaling through its receptors can stimulate both proliferation and invasiveness of GBM cells [6]. However, the mechanisms regulating SphK1 expression and activity in GBM cells are largely unknown.
Epidermal growth factor (EGF) plays a critical role in tumorigenesis, and the expression of its receptor (EGFR) correlates with poor patient outlook in several types of cancers [7–9]. In malignant gliomas, EGFR overexpression can be found in 40–50% of patients [10, 11]. Mutations in the EGFR gene are found in many types of tumors and deletions in the extracellular domain are the most frequent. The EGFR mutant EGFRvIII, is the most common truncated receptor mutant in GBM [12, 13]. It has a prevalence of 20–30% in GBM patients and 50–60% of patients whose tumors show amplification of the normal EGFR. This mutant has a deletion of 267 amino acids in the extracellular domain, and is unable to bind any known EGFR ligand [14]. EGFRvIII has a constitutive tyrosine kinase activity which leads to constitutive activation of signaling pathways downstream of the EGFR [12].
Growth factors, including EGF, have been shown to stimulate SphK1 in several cell types [15]. However, little is known regarding the role of SphK1 downstream of EGFR in GBM cells, and no studies have examined the effect of the mutant EGFRvIII on SphK1. In this study we investigate the regulation of SphK1 in GBM cells by EGFR and the role of SphK1 in mediating proliferation and survival of GBM cells, in particular in GBM cells expressing the EGFRvIII mutant. Furthermore, we extend these observations to GBM-derived neurosphere cells [16, 17]. These cells represent a more appropriate model of GBM than traditional human glioma cell lines that have been cultured in serum-containing medium, as they are capable of reforming tumors in mouse models that resemble the original GBM both histologically and genetically, even upon serial transplantation [16–18], while traditional cell lines form tumors that do not resemble GBM histologically and are often not invasive. Our results show that EGFRvIII increases the expression and activity of SphK1, leading to an increase in the proliferation rate of GBM cells. SphK1 or EGFR inhibition results in a decrease of proliferation and cell survival. Thus, SphK1 could be useful as a target for the design of drugs for the treatment of GBM.
Materials and methods
Materials
Antibodies to pERK, ERK2, EGFR, and GAPDH were from Santa Cruz Biotechnology. Antibodies to pAkt, Akt1 and pEGFR were from Cell Signaling Technology. Antibodies to SphK1 were previously described [19]. Cycloheximide and AG1478 were from Sigma. Gefitinib was from LC Laboratories. Doxycycline was from BD Biosciences. EGF was from R&D Systems. Cell culture medium was from Mediatech.
Cell culture
U-251 and U-1242 cell lines used in this study were maintained in Eagle’s minimum essential medium (EMEM) containing 10% fetal bovine serum, non-essential amino acids and sodium pyruvate. The cells were grown at 37°C in 95% air, 5% CO2 and cultures were passaged approximately once per week at a ratio of 1:12. U-251-EGFRvIII-inducible E18 cells [20] were a generous gift of Dr. Amyn Habib of the University of Texas Southwestern Medical Center, and were cultured in EMEM with FBS as described above with the addition of 50 μg/ml zeocin.
All GBM neurosphere cells were cultured from fresh human GBM tissue obtained following surgical resection of tumors at OSU Medical Center. Tissue was cut into small pieces, and then passed several times through an 18 gauge needle and then a 21 gauge needle to achieve a single cell suspension. Cells were cultured in DMEM/F12 containing B27 without vitamin A (Invitrogen), and 20 ng/ml each of EGF and bFGF (R&D Systems). Primary spheres formed in 2–4 weeks. When spheres reached a moderate size they were passaged by trituration with a 21 gauge needle and split at a ratio of 1:4.
Sphingosine kinase assay
Sphingosine kinase activity was measured as described [21] with minor modifications. Briefly, cells were scraped or resuspended into sphingosine kinase buffer (100 mM Tris, pH 7.4, 20% glycerol, 10 mM MgCl2, 1 mM β-mercaptoethanol, 1 mM EDTA, 10 μg/ml leupeptin and aprotinin, 1 mM phenylmethylsulfonylfluoride (PMSF), 15 mM NaF, 1 mM sodium orthovanadate, 40 mM β-glycerophosphate, and 0.5 mM deoxypyridoxine) and lysed by freeze/thawing seven times. Equal amounts of protein were assayed in the presence of 50 μM sphingosine and 1 mM ATP containing 10 μCi [γ-32P]ATP for 1 h at 37°C. Labeled S1P was separated by thin layer chromatography (TLC) on Silica gel G60 plates with CHCl3/Acetone/MeOH/Acetic Acid/water (10/4/3/2/1), and visualized by autoradiography. Radioactive spots were scraped from TLC plates and quantified by scintillation counting.
Western blotting
Cells were washed and lysed in 25 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecylsulfate (SDS), 0.5 mM DTT, 0.5% deoxycholate, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM PMSF, and 10 μg/ml each of aprotinin and leupeptin. Equal amounts of protein were separated by SDS-PAGE before being transferred to nitrocellulose membranes. Membranes were blocked for 1 h at room temperature in PBS + 0.1% Tween-20 (PBS-T) containing 5% (w/v) non-fat dry milk. Membranes were probed overnight with primary antibodies and washed 3 times with PBS-T for 5, 10 and 15 min, followed by incubation for 1 h at room temperature with 1:2,000 horseradish peroxidase-conjugated secondary antibodies (ECM Biosciences) in blocking solution. Membranes were washed as above, followed by incubation in Pierce Super Signal West Pico chemiluminescent substrate.
For densitometry, scanned images were analyzed using LabWorks (UVP BioImaging Systems). Briefly, a monochrome image of the scanned film was created and lanes were outlined. The software locates the bands in the image and the absolute integrated optical density (IOD), i.e. the volume of the band in the lane profile, is obtained. For all western blots, two images were analyzed: loading control (GAPDH or total ERK or Akt for phospho-ERK or phospho-Akt respectively) and the protein of interest. The ratio, protein of interest/loading control of each lane was calculated.
Cell proliferation
U251-E18 cells were plated in 24 well plates and the following day transfected with 3 pmol per well control siRNA (Ambion, negative control siRNA #2) or SphK1-specific siRNA using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. SphK1-specific siRNA sequences were as follows: siSK1-A GGCTGAAATCTCC TTCACG; siSK1-B AAGGATGGGAAAGGTGTGTTT. Four days later cells were trypsinized and counted using a Coulter Z2 particle analyzer. Neurosphere cells were plated in 24 well plates, treated and at the indicated times collected. Cells were passed several times through a 21 gauge needle and counted using a Coulter Z2 particle analyzer or a hemocytometer.
MTT cell proliferation assay
MTT reduction assay was performed using the MTT cell proliferation assay kit from Cayman Chemical. Cells were plated in a 96-well plate at a density of 4 × 103 cells/well in 100 μl of culture medium with or without 1 μg/ml of SphK1 inhibitor. Three hours before the indicated time, 10 μl of MTT reagent was added to each well and incubated at 37°C in a CO2 environment. At the indicated times, the media was aspirated and 100 μl of crystal dissolving solution was added to each well. The absorbance was measured at 570 nm using a BMG LabTech Omega plate reader.
RT-PCR
Total RNA was extracted using Trizol (Invitrogen) according to manufacturer’s instructions, followed by treatment with DNase1 (Ambion) for 20 min at 37°C. The RNA was quantitated and 2.5 μg was used with M-MLV reverse transcriptase (Invitrogen) to synthesize cDNA according to manufacturer’s instructions. PCR amplification was performed using Taq DNA polymerase (New England Biolabs) according to manufacturer’s instructions. Primers used were as follows: EGFRvIII––5′-GAGCTC TTCGGGGAGCAG-3′, and 5′-GTGATCTGTCACCACA TAATTACCTTTCT-3′ [22]; S1P1––5′-CCTCTTCCTGC TAATCAGCG-3′, and 5′-ACAGGTCTTCACCTTGCAGC-3′; S1P2––5′-CTGCCTCTCTACGCCAAGCATTAT-3′, and 5′-ACAGGCATAGTCCAGAAGGAGGAT-3′; S1P3 5′-CATCCTCTTCAAGGCTCAGTGGTT-3′, and 5′-CCATTCTGAAGTGCTGCGTTCTTG-3′; SphK1––5′-AGAGAGTGAGAAGTATCGGCGTCTG-3′, and 5′-GACCACGGGCACATATACCAAGTAG-3′; SphK2––5′-CCTGTTGCTCAACTGCTCACTGTT-3′, and 5′-GATGGCCAACATGAGCACAAAGTC-3′. GAPDH––5′-GACCCCTTCAT TGACCTCAAC-3′ and 5′-AGTGAGCTTCCCGTTCAGC TC-3′. Real time PCR analysis for SphK1 was performed as previously described [5].
Cell viability
Cell viability was measured by trypan blue exclusion. Briefly, cells were plated and at the indicated times, collected. Cells were passed several times through a 21 gauge needle and 20 μl of the cell suspension were diluted with an equal volume of 0.4% Trypan blue. Cells were counted using a hemocytometer. The percentage of viable cells was calculated as follows: the number of viable cells divided by the total number of cells (viable plus dead), multiplied by 100.
SphK1 promoter reporter assay
The SphK1 promoter luciferase reporter construct in pGL3 [23] was a generous gift of Dr. Lina Obeid of the Medical University of South Carolina. Cells were transfected in 100 mm dishes using Lipofectamine 2000. After 6 h incubation cells were trypsinized and replated in 12 well plates. The following day cells were serum starved for 24 h and then stimulated with or without 20 ng/ml EGF for the indicated time. Cells were lysed and luciferase activity was measured using the Promega Luciferase Assay System according to manufacturer’s instructions.
Tumor formation
Neurospheres were disrupted by trituration. Cells were washed three times in PBS and resuspended at a concentration of 66,000 cells/μl. Female athymic mice (nu/nu genotype, 5–6 weeks, Harlan Laboratories) were anesthetized by an intraperitoneal injection of a mixture consisting of Ketamine (10 mg/ml) and Xylazine (1 mg/ml) in PBS and fitted into a stereotactic rodent frame (David Kopf Instruments, Tujunga, CA). A small incision was made just lateral to the midline and bregma exposed. A small (1.0 mm) burr hole was drilled with a #7 drill bit at AP = +1, ML = −2.5 from bregma. A 10 μl Hamilton syringe was filled with 3 μl of the cell suspension. The syringe was lined up with the drilled hole and cells were slowly deposited at a rate of 1 μl/min in the right caudate nucleus at a depth of −3 mm from dura, for a total of 2 × 105 cells/mouse. After injecting the cells the needle remained in place for an additional 3 min before being slowly withdrawn. The incision was sutured closed using 4–0 vicryl with an rb-1 needle.
Annexin V-APC assay
Annexin V staining was performed using Annexin V-APC and the apoptosis detection kit I (BD Biosciences) according to manufacturer’s instructions. Briefly, 2 × 105 cells were plated and treated for the indicated times with 1 μg/ml of SphK inhibitor. Cells were collected and wash twice with cold PBS, then resuspended in 100 μl of 1× binding buffer. Annexin V (5 μl) was added and incubated for 15 min at room temperature in the dark. Cells were plated on slides and analyzed by microscopy. Total number of cells and cells positive for Annexin V staining were counted and percentage was calculated.
Results
Regulation of SphK1 by EGF receptor signaling in glioma cells
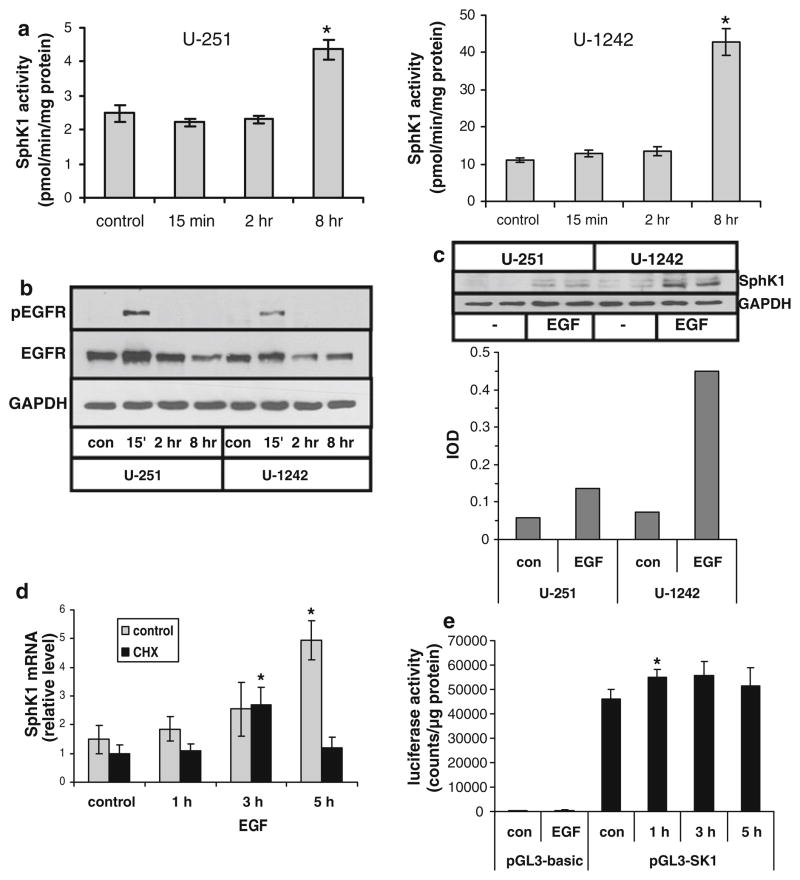
It is well known that EGF plays an important role in the progression of several human cancers, including gliomas [24]. Therefore, we analyzed whether EGF treatment induced SphK1 activation in two human glioma cell lines. Eight hours of EGF (20 ng/ml) treatment increased SphK1 activity by approximately 2-fold in U-251 cells (p < 0.001) and 4-fold in U-1242 cells (p < 0.005) relative to control (Fig. 1a). Both U-1242 and U-251 cells expressed EGFR protein which showed activation, as detected by phospho-EGFR western blots, at 15 min EGF stimulation similarly in both cell lines (Fig. 1b).
Fig. 1.
Regulation of SphK1 activity and expression by EGF in glioma cell lines. U-251 or U-1242 human glioma cells were stimulated with 20 ng/ml EGF for the indicated time. a SphK1 activity was measured as described in Materials and Methods. b Cell lysates were immunoblotted for EGFR and tyrosine phosphorylated EGFR. c Cells were treated for 8 h with or without 20 ng/ml EGF. Duplicate samples were immunoblotted for SphK1. Bands were quantitated by scanning densitometry, and results are ratios of IOD for SphK1 relative to GAPDH. d U-251 cells were pretreated for 30 min with 10 μg/ml cycloheximide or with ethanol as a vehicle control and then stimulated with EGF for the indicated time. RNA was isolated and SphK1 mRNA level was determined by real time PCR analysis. e U-251 cells were transfected with pGL3-basic or pGL3-SK1 and then replated in multiwell plates. Cells were stimulated with 20 ng/ml EGF for 3 h (pGL3-basic) or the indicated time (pGL3-SK1) and luciferase activity was measured. Results for panels a, d and e are means ± s.d. of triplicate samples. The * indicates statistically significant difference by Student’s T test, p < 0.05. Two independent experiments provided similar results for all panels
We next determined whether the increased SphK activity was due to elevated SphK1 expression. SphK1 protein as detected by Western analysis was increased after 8 h stimulation with EGF in both cell lines (Fig. 1c). Densitometric scanning showed that the extent of the increase in SphK1 protein expression was similar to the increase in SphK activity at 8 h EGF treatment, approximately 2.5 fold in U-251 cells and 5 fold in U-1242. Real time PCR analysis revealed that SphK1 mRNA levels were slightly increased 3 h after EGF stimulation and had increased ~4 fold by 5 h (p < 0.005) (Fig. 1d). Interestingly cycloheximide (CHX) pretreatment to block protein synthesis did not inhibit the increase in SphK1 mRNA at 3 h (p < 0.02), but did block the stronger induction seen at 5 h.
To further examine the mechanism of induction of SphK1 expression by EGF in glioma cells we performed reporter assays using a construct containing the human SphK1 promoter linked to a luciferase gene. U-251 cells transfected with a vector encoding luciferase but lacking the SphK1 promoter (pGL3-basic) displayed only very modest luciferase activity which was not affected by EGF treatment (Fig. 1e). Transfection with the SphK1-promoter luciferase vector (pGL3-SK1) led to a strong basal luciferase activity. EGF treatment for 1 h only modestly enhanced promoter activity (p < 0.05) with no additional increase seen up to 5 h. Additional experiments examining promoter activity up to 8 h after EGF stimulation did not show any further increase (data not shown).
Taken together the above data suggest that there is a modest immediate early response of SphK1 induction in response to EGF which is mediated through activation of the SphK1 promoter, and a later, more pronounced, delayed response induction which is mediated by an indirect mechanism.
Regulation of SphK1 by EGFRvIII in glioma cells
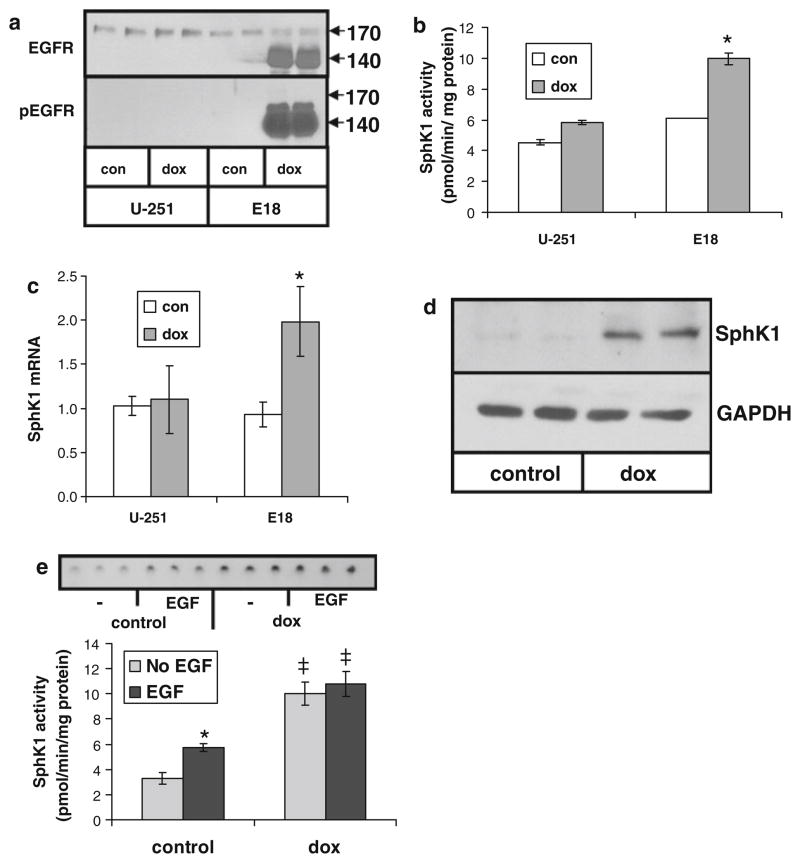
EGFRvIII is a common mutation occurring in GBM [12, 25]. To determine the effect of this mutation on SphK1 activity we used U-251-E18 cells, which are stably transfected with an EGFRvIII expression construct in a TET-on inducible expression system [20]. EGFRvIII is overexpressed in these cells in doxycycline (dox)-containing medium but not in medium without dox [20]. Treatment with 10 ng/ml dox potently induced expression of a protein reacting with EGFR antibodies and migrating at the expected molecular weight of EGFRvIII in U-251-E18 but not in parental U-251 cells (Fig. 2a). Furthermore, the EGFRvIII expressed in dox-treated U-251-E18 cells was active as indicated by Western analysis with an antibody specific for tyrosine-phosphorylated EGFR.
Fig. 2.
Regulation of SphK1 activity and expression by EGFRvIII in glioma cells. Parental U-251 cells or U-251 cells containing a doxycycline (dox)-inducible EGFRvIII construct (E18) were grown for 3 days in the presence or absence of 10 ng/ml dox. a Cell lysates were immunoblotted for EGFR and tyrosine phosphorylated EGFR, b SphK1 activity was measured, c SphK1 mRNA level was determined by real time PCR, or d SphK1 protein expression was measured by Western blot. e U-251-E18 cells grown for 3 days in the presence or absence of 10 ng/ml dox were treated for 8 h with or without 20 ng/ml EGF and SphK1 activity was measured. The inset shows a thin layer chromatogram. Data are means ± s.d. of triplicate determinations. The * indicates statistically significant difference by Student’s T test, p < 0.05. The ‡ indicates statistically significant difference of dox-treated cells compared to control cells by Student’s T test, p < 0.05. At least two independent experiments provided similar results for all panels
While dox treatment had a slight effect on SphK activity in parental U-251 cells it much more potently induced increased SphK activity in U-251-E18 cells (p < 0.005) (Fig. 2b). SphK1 expression was increased by dox treatment on both the mRNA (p < 0.02) (Fig. 2c) and protein level (Fig. 2d) in U-251-E18 cells but not in parental U-251 cells. Figure 2e shows that, in the absence of dox, EGF treatment of U-251-E18 cells increases SphK1 activity (p < 0.05), consistent with the expression of endogenous wt-EGFR expression in these cells (Fig. 2a) and with the results of Fig. 1a. However, when U-251-E18 cells are treated with dox to induce EGFRvIII expression, SphK1 activity increases more dramatically, independently of EGF (p < 0.05), in agreement with the robust induction and high level of tyrosine phosphorylation of the EGFRvIII (Fig. 2a). Together these results indicate that overexpression of EGFRvIII induces SphK1 expression and thus an increase in activity.
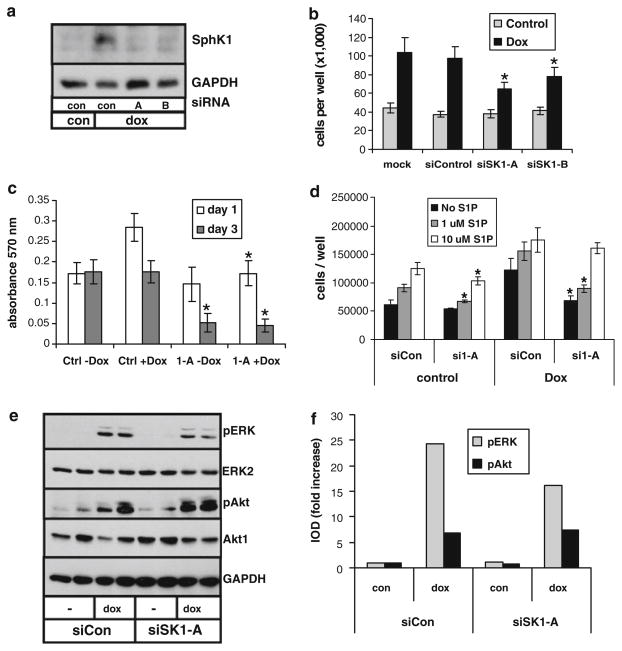
We next wished to determine the role of SphK1 induction in the biological responses of glioma cells to EGFRvIII. We thus examined the effect of EGFRvIII on cell proliferation in the absence and presence of SphK1 knockdown by RNA interference. U251-E18 cells were transfected with control siRNA or two different SphK1-specific siRNAs that knocked down dox-induced SphK1 expression to approximately control levels (Fig. 3a).
Fig. 3.
Role of SphK1 in EGFRvIII-mediated glioma cell proliferation. U-251-E18 cells were transfected with control random sequence siRNA (con) or two different siRNAs specific to SphK1 (A and B) and grown for 2 days in the absence or presence of 10 ng/ml dox. a Cell lysates were immunoblotted for SphK1. b Cell number was determined after 3 days in culture. c Cell proliferation was examined by MTT assay. d Cells were transfected with control or SphK1-specific siRNA and grown for 3 days in the absence or presence of dox and the indicated concentration of S1P. The * indicate statistically significant difference of SphK1-specific siRNA-transfected cells from control siRNA transfected cells, p < 0.05. e U-251-E18 cells were transfected with control siRNA or SphK1-specific siRNA and grown for 1 day in the absence or presence of dox. Duplicate cell lysates were immunoblotted for phosphorylated ERK or Akt, after which blots were stripped and reblotted for total ERK or Akt. f Western blots shown in panel d were quantitated by densitometry. Results are expressed as doxycycline-induced fold increase in the ratio of the integrated optical density (IOD) of phosphorylated to total ERK or Akt. At least two independent experiments provided similar results for all panels
Cell proliferation was examined in medium containing charcoal-stripped, delipidated serum (CS-FBS) to avoid effects of S1P normally present in FBS. In these conditions, mock and control siRNA-transfected U-251-E18 cells demonstrated an approximately two fold increase in cell number after 4 days of dox treatment (Fig. 3b). Dox treatment of parental U-251 cells did not stimulate proliferation (data not shown). Dox-induced accumulation of U-251-E18 was partially inhibited by knockdown of SphK1 with either of the two siRNAs (p < 0.05) (Fig. 3b). To further examine U-251-E18 cell proliferation MTT assays were performed. Figure 3c shows that dox stimulated proliferation after 1 day treatment. After 3 days dox treatment U-251-E18 cell proliferation had returned to basal levels. This slowing of proliferation at later times is commonly seen in these experiments and is likely due to cells becoming over confluent quickly due to enhanced proliferation early in the assay, as it often occurs faster in dox-treated wells. Transfection with SphK1-specific siRNA blocked dox-induced proliferation of U-251-E18 cells at 1 day (p < 0.05) while proliferation of cells in the absence of dox was unaffected. Three days following siRNA transfection the number of metabolically active cells as determined by MTT assay is dramatically decreased by SphK1-specific siRNA in both the absence (p < 0.005) and presence (p < 0.002) of dox, suggesting that SphK1 knock down causes an initial decrease in EGFRvIII-induced proliferation, followed by cell death at later times.
To assess the role of S1P downstream of SphK1 in EGFRvIII-induced glioma cell proliferation, we next examined the effect of SphK1 knockdown in the presence or absence of added exogenous S1P. As above, in the absence of exogenous S1P dox enhanced proliferation of U-251-E18 cells and SphK1 knockdown decreased the dox-induced effect. While addition of 1 μM exogenous S1P was without effect, 10 μM exogenous S1P eliminated the decrease in doxycycline-induced proliferation caused by SphK1 siRNA (Fig. 3d).
To examine the mechanism by which SphK1 contributes to EGFRvIII-induced effects on glioma cells, we examined changes in activation of key signaling molecules. Figure 3e shows that SphK1 knock down by siRNA partially inhibited dox-induced activation of ERK in U-251-E18 cells, but did not affect Akt activation. Densitometric quantitation showed a 34% decrease of dox-induced phospho-ERK in SphK1 siRNA-transfected cells compared to control siRNA-transfected cells (Fig. 3f). Thus, SphK1 partially contributes to EGFRvIII regulation of glioma cell growth by enhancing activation of ERK signaling.
EGFRvIII and SphK1 in primary brain tumor neurosphere cells
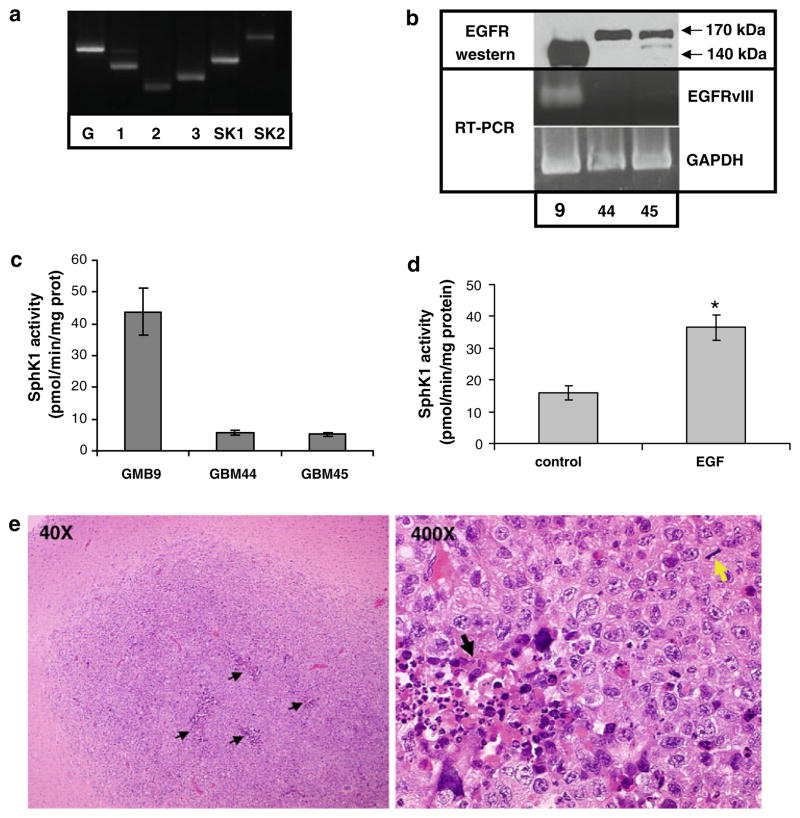
As discussed above, the use of traditional glioma cell lines grown in serum-containing medium, such as U-251, has drawbacks in that the tumors formed by such cells do not histologically resemble GBM. In addition, such cell lines commonly lose EGFR overexpression over time in culture. Thus to examine the role of SphK1 downstream of EGFR signaling in a more appropriate model we utilized primary neurosphere cells grown from fresh human GBM tissue. Cells were isolated from GBM tissue obtained following surgical resection and cultured in defined medium containing EGF and bFGF. Under these conditions GBM cells grow in suspension as neurospheres [17]. To evaluate whether our neurosphere cells express the elements of the SphK pathway we isolated total RNA and RT-PCR was performed using primers specific for SphK1, SphK2 and S1P receptors 1, 2, and 3, which are commonly expressed in human GBM tissue [5]. Figure 4a shows a representative result from one neurosphere cell line, GBM9. Both SphK isoforms and all three S1P receptors examined are expressed in GBM9 cells. Similar results were seen for two additional neurosphere lines examined (data not shown). We then evaluated the expression of EGFR and EGFRvIII by Western blot and RT-PCR analysis using primers specific for the EGFRvIII mutant. Figure 4b shows that one neurosphere line, GBM9, expresses a large amount of a protein that reacts with an EGFR-specific antibody and migrates at approximately 140 kDa, the expected size of the EGFRvIII mutant. The other two neurosphere lines examined, GBM44 and GBM45, expressed a moderate amount of protein migrating at 170 kDa, consistent with the wt-EGFR. RT-PCR analysis using primers specific for the mutational junction found in the EGFRvIII deletion confirmed expression of the EGFRvIII in GBM9 cells. We have examined EGFRvIII in GBM9 cells after 15 passages and have not detected any loss of expression. Thus, our neurosphere lines are capable of maintaining EGFRvIII expression for at least 15 passages. Interestingly the same line, GBM9, also expressed the highest level of SphK1 activity (Fig. 4c). In addition, EGF treatment of GBM44 cells, which express wt-EGFR but not EGFRvIII, enhanced SphK1 activity (p < 0.05) (Fig. 4d). Thus, EGFR signaling also activates SphK1 in GBM neurosphere cells.
Fig. 4.
SphK and EGFR in brain tumor stem cells. a RNA was extracted from GBM9 neurosphere cells and RT-PCR analysis was performed to detect expression of SphK isoforms 1 and 2 (SK1 and SK2) and S1P receptors 1, 2, and 3. b Cell lysates from three neurosphere lines were immunoblotted with an antibody that recognizes wt-EGFR and EGFRvIII (top panel). RNA was extracted from the same lines and RT-PCR analysis for expression of the EGFRvIII mutant was performed. c SphK1 activity was measured in lysates from neurosphere lines. d GBM44 cells were treated with or without 20 ng/ml EGF for 8 h and SphK1 activity was measured. Results for panels c and d are means ± s.d. of triplicate samples. Two independent experiments provided similar results. The * indicates statistically significant difference by Student’s T test, p < 0.05. e GBM9 cells were injected into the brains of nude mice and tumors were allowed to develop. Images are micrographs of hematoxylin and eosin stained tissue from a representative mouse brain showing GBM tumors. Black arrows = areas of focal necrosis. Yellow arrow = mitotic figure
To further validate these cells as an appropriate model of GBM, we injected GBM9 cells into the brains of nude mice. Mice were sacrificed when they began to show signs of neurological deficit and brains were dissected. All mice injected intracranially developed tumors (Fig. 4e). Tumors were examined by a board certified neuropathologist (ARC). All tumors were highly cellular neoplasms displaying cellular pleomorphism, diffuse invasion into surrounding brain tissue, focal areas of necrosis (black arrows), and numerous mitotic figures (yellow arrow), and were thus diagnosed as GBM. Taken together, GBM9 cells maintain expression of EGFRvIII in culture and create GBM-like tumors in vivo, and are thus an excellent model in which to study the role of EGFRvIII in GBM.
Role of EGFRvIII and SphK1 in GBM neurosphere cells
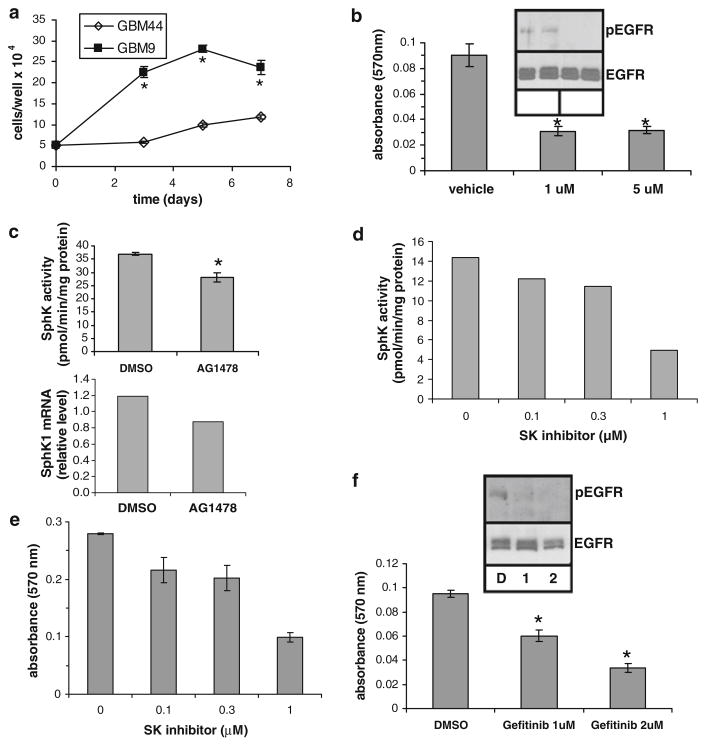
We first evaluated the growth of two neurosphere lines: one that expressed the wild type EGFR (GBM44), and one that expresses the EGFRvIII mutant (GBM9). Neurospheres were disrupted and cells were plated at a low concentration and allowed to grow for 1 week. Figure 5a shows that GBM9 cells have a much higher proliferation rate than GBM44 cells, which do not express EGFRvIII, and have much lower SphK1 activity (p < 0.05).
Fig. 5.
The role of EGFRvIII in growth and SphK1 regulation in GBM-derived neurosphere cells. a GBM9 and GBM44 were plated at 50,000 cells per well and cultured for the indicated time. Cells per well were counted. b GBM9 cells were treated with DMSO as a vehicle control or the indicated concentration of AG1478 for 2 days and cell proliferation was measured by MTT assay. The insets in panels b and f show the effects of AG1478 and gefitinib on EGFRvIII tyrosine phosphorylation as determined by phospho-EGFR-specific Western blot analysis. c GBM9 cells were treated with 1 μM AG1478 for 2 days and SphK1 activity was measured, or real time PCR was performed to measure SphK1 mRNA level. d and e GBM9 cells were grown for 2 days in the presence of vehicle or the indicated concentration of SphK inhibitor and SphK activity was assayed (d) or cell proliferation was measured by MTT assay (e) in parallel wells. f GBM9 cells were grown for 3 days in the presence of the indicated concentration of gefitinib or DMSO as a vehicle control and cell proliferation was measured by MTT assay. Results for all panels are means ± s.d. of triplicate samples. The * indicate statistically significant difference by Student’s T test, p < 0.05
To examine the role of EGFRvIII in the rapid growth of GBM9 cells we utilized the selective EGFR tyrosine kinase inhibitor AG1478. AG1478 treatment of GBM9 cells effectively blocked tyrosine phosphorylation of EGFRvIII (Fig. 5b). AG14787 treatment also significantly decreased growth of GBM9 cells as determined by MTT assay (p < 0.001) (Fig. 5b). To examine the possible involvement of SphK1 we also examined the effect of AG1478 on SphK1 activity in these cells. AG1478 induced a small but significant reduction of SphK1 activity (p < 0.001) and a similar decrease in SphK1 mRNA level (Fig. 5c). To determine whether such a modest decrease in SphK1 activity could affect cell proliferation, we treated GBM9 cells with a SphK inhibitor [26] in a dose response fashion and measured both SphK activity and cell proliferation in parallel wells (Fig. 5d, e). We observed an excellent correlation of SphK1 activity with cell proliferation, even at very low concentrations of SphK inhibitor, which decreased SphK activity modestly to similar levels as did AG1478. Thus, a modest inhibition of SphK activity is sufficient to decrease GBM neurosphere cell proliferation.
We confirmed the role of EGFRvIII in proliferation of GBM9 cells using the EGFR inhibitor gefitinib. Gefitinib blocked EGFR tyrosine phosphorylation and decreased proliferation of GBM9 cells (p < 0.01 at 1 μM and p < 0.002 at 2 μM) (Fig. 5f). Taken together these results suggest that EGFRvIII drives enhanced proliferation of GBM9 cells and that SphK1 may partially mediate this response.
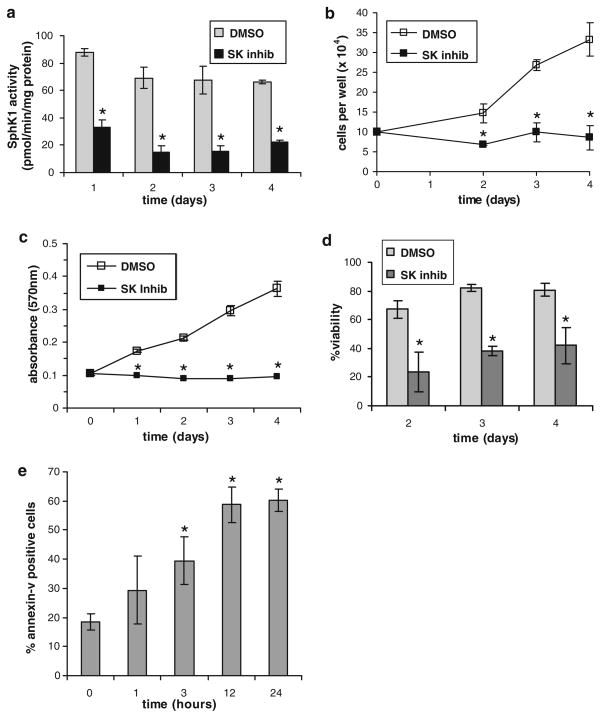
We further explored the role of SphK1 in GBM neurosphere cell proliferation and survival using the SphK inhibitor. As shown in Fig. 6a, SphK inhibition potently decreased SphK1 activity in GBM9 cells in comparison to DMSO vehicle-treated cells (p < 0.002, at all time points), and the inhibition lasted at least 4 days. SphK inhibition also eliminated accumulation of GBM9 cells over a 4 day period as determined by cell counts (p < 0.01, at all time points) (Fig. 6b), and MTT analysis showed a complete inhibition of GBM9 cell proliferation by SphK inhibition (p < 0.001, at all time points) (Fig. 6c). Furthermore, cell viability was decreased approximately 50% by SphK inhibition throughout the course of the experiment (p < 0.01, at all time points) (Fig. 6d). Thus, SphK is crucial for GBM9 cell proliferation and may also be important for survival. To examine the effect of SphK inhibition on GBM9 cell survival we stained vehicle- and SphK inhibitor-treated cells with annexin-V. The percentage of annexin-V positive cells was significantly increased after 3 h treatment with SphK inhibitor (p < 0.05) and peaked at approximately 60% annexin-V positivity after 12 h (p < 0.001) (Fig. 6e). Together, these results indicate that SphK1 is crucial to maintain GBM neurosphere cell survival and proliferation.
Fig. 6.
The role of SphK1 in GBM neurosphere cell proliferation. GBM9 cells were treated for the indicated time with 3 μM SphK inhibitor (SK inhib) or an equal volume of DMSO as a vehicle control and a SphK1 activity was measured, b cells per well were counted, c cell proliferation was examined by MTT assay, d cell viability was measured, or e apoptosis was measured by annexin-V staining. Results are means ± s.d. of triplicate samples. The * indicate statistically significant difference by Student’s T test, p < 0.05. Duplicate experiments provided similar results
Discussion
Recent studies have linked SphK1 to cancer development and progression [27–29]. Up-regulation of SphK1 expression has been shown in several types of tumors [30–32]. In previous studies we established that high levels of SphK1 expression correlate with decreased survival of GBM patients [5]. A recent study confirmed this observation and additionally showed that SphK1 is upregulated in astrocytomas compared to adjacent non-malignant tissue and that SphK1 upregulation correlates with histological grade of astrocytomas [33]. However, the mechanisms by which glioma cells upregulate the expression of this enzyme are poorly understood.
In this paper we demonstrate that EGF signaling or expression of the constitutively active mutant receptor EGFRvIII increases SphK1 activity in glioma cell lines and in primary neurosphere cells isolated from human GBM tissue. Furthermore, both wt-EGFR and EGFRvIII signaling lead to enhanced SphK1 expression. A modest increase in SphK1 promoter activity occurred at early times after EGF stimulation and modestly increased mRNA levels also were seen at a similar time point by a mechanism that did not require protein synthesis. However, no significant increase in SphK1 activity was seen until later times. Furthermore, the later, more pronounced increase in SphK1 mRNA did require protein synthesis, indicating that this is due to an indirect mechanism. This late increase in SphK1 mRNA did not correlate with a further increase in SphK1 promoter activity. Note that interleukin-1 has been shown to induce SphK1 transcription in glioma cells, also at late times following stimulation, by activating an AP-1 element within the first intron [34]. Thus, EGFR signaling may work by a similar mechanism to cause the late, indirect induction of SphK1 expression.
While EGF signaling has been previously reported to induce SphK1 expression [27, 35, 36], this is the first demonstration of a constitutively active mutant EGFR leading to a long term increase in SphK1 expression. EGFRvIII, the most common EGFR mutant, is expressed in 31–50% of gliomas [12, 25, 37]. Experiments were performed to examine SphK1 levels after several days of EGFRvIII induction. This is relevant to the in vivo situation for GBMs expressing the EGFRvIII mutant. As this mutant is constitutively active, the biological responses mediated by it should consist of long term changes rather than acute activation of signaling pathways. Thus, the fact that increased SphK1 expression in response to EGFRvIII induction is maintained over at least several days in culture suggests that SphK1 may be important for prolonged biological effects of EGFRvIII seen in GBM.
A number of recent studies have shown that SphK1 and its product S1P interact in several ways with EGF signaling. For example, Shida and collaborators reported that in gastric cancer cells S1P transactivates EGFR and c-Met [38], and that the lysophosphatidic acid receptor LPA1 transactivates EGFR to induce SphK1 upregulation [39]. Furthermore, S1P signaling has been shown to induce EGFR expression in vascular smooth muscle cells, [40], and activation of SphK1 by estrogen signaling leads to transactivation of EGFR via S1P3 in breast cancer cells [41]. Thus, the interactions of these pathways are clearly complex and multidirectional, and the detailed mechanisms at work are likely specific to given cell types.
Examination of the levels of SphK1 expression in comparison to EGFR mutations in data from GBM cases available from The Cancer Genome Atlas (TCGA) failed to show a correlation (data not shown). Note that SphK1 expression can be stimulated downstream of a number of growth factors including EGF, PDGF, bFGF, GDNF and NGF, and by signaling through a number of common growth factor-activated signaling pathways [27, 34, 42–45]. Thus it is likely that EGFRvIII signaling is only one mechanism by which SphK1 can be upregulated in GBM cells. An important lesson learned from large sequencing studies such as TCGA is that most GBM tumors have genetic alterations in some elements of several key signaling pathways including growth factor receptor signaling pathways [46, 47]. Therefore, it is possible that in tumors lacking EGFR mutations other genetic alterations that affect common growth factor signaling pathways induce SphK1 expression. Thus the fact that a correlation of SphK1 level with EGFR mutation may not be demonstrable in GBM cases, does not necessarily mean that an important connection does not exist.
EGFRvIII is known to provide a growth advantage to glioblastoma cells both in vitro and in vivo by cooperating with other oncogenic signaling pathways [48, 49]. In agreement, EGFRvIII induction in U-251-E18 cells enhanced cell proliferation as determined by both cell number increase and MTT assays, while inhibition of EGFRvIII in GBM neurosphere cells, using either AG1478 or gefitinib, decreased proliferation. The increase in proliferation afforded by EGFRvIII is partially dependent upon SphK1, as evidenced by the following: (1) SphK1 knockdown by siRNA decreases cell accumulation caused by EGFRvIII induction in U-251-E18. (2) Addition of 10 μM S1P overrides the effect of SphK1 knockdown on EGFRvIII-induced proliferation. (3) SphK1 knockdown also blocks EGFRvIII-induced proliferation as determined by MTT assay. (4) AG1478 treatment of GBM neurosphere cells decreased SphK1 activity in parallel with decreased cell growth. (5) Pharmacological inhibition of SphK1 effectively blocked GBM neurosphere cell growth.
It should be noted that although AG1478 decreased SphK1 activity to a similar extent as did 0.1–0.3 μM SphK inhibitor, it much more effectively blocked GBM9 neurosphere cell proliferation, than did these low concentrations of SphK inhibitor. This is consistent with the idea that SphK1 is only partially responsible for mediating EGFRvIII-induced proliferation, as AG1478 would be expected to block multiple EGFRvIII-activated pathways in addition to SphK1 induction.
Interestingly, the decrease in dox-induced U-251-E18 cell proliferation caused by SphK1 knockdown was overridden by a high concentration of exogenous S1P, 10 μM, but not by 1 μM S1P. In this regard, a recent study showed that 10 μM S1P is able to activate the intracellular S1P target TRAF2, while lower concentrations of S1P, which are known to activate S1P receptors, are not [50]. Thus it is possible that the contribution of S1P to GBM neurosphere cell proliferation downstream of EGFRvIII-induced SphK activation is mediated by an intracellular S1P target.
AG1478 only modestly decreased SphK1 activity, while the SphK inhibitor much more effectively blocked activity. This suggests that a significant level of basal SphK1 activity is present in GBM cells, in agreement with our data showing that the SphK1 promoter reporter complex is highly active in U-251 cells under serum starvation conditions. Thus SphK1 upregulation in response to EGFRvIII signaling adds a moderate additional amount of SphK1 activity which plays a role in EGFRvIII-enhancement of cell proliferation. On the other hand inhibition of SphK1 levels below that achieved by AG1478 with the SphK inhibitor led to decreased viability and increased annexin-V positivity indicating the induction of apoptosis. Together these data suggest that the basal SphK1 activity in these cells is necessary to maintain survival, while EGFRvIII-enhanced SphK1 activity contributes to increased growth. Thus it should be stressed that the contribution of SphK1 to EGFRvIII-driven growth of glioma cells is likely moderate. Nevertheless, as glioma is not a disease caused by a single genetic abnormality, it is likely that effective therapeutic targeting of gliomas will require drugs that target numerous pathways used simultaneously. Therefore it is useful to understand partial contributions from several pathways. Current and future work in our laboratory is aimed at determining which glioma cases will be sensitive to drugs targeting the SphK1 pathway and which drugs might provide synergistic effects in combination with SphK1-targeting drugs.
The contribution of basal SphK1 activity to glioma cell survival may be of more import particularly in regard to resistance of glioma cells to chemotherapeutic drug-induced apoptosis. Several reports have suggested that GBM-derived neurosphere cells have properties of enhanced resistance to standard therapies [51–53]. Interestingly, a number of chemotherapeutic drugs induce cancer cell apoptosis through production of the pro-apoptotic sphingolipid ceramide [54, 55]. It is well known that SphK alters the balance of the “sphingolipid rheostat” by increasing S1P levels while reducing ceramide levels [56], and thus targeting SphK to enhance sensitivity of cancer cells to therapy has been proposed [57, 58]. Therefore, the fact that SphK inhibition potently decreases neurosphere cell viability and enhances apoptosis suggests that SphK1 may be an important contributor to the resistance of GBM to chemotherapy, and a useful molecule to target in these cells.
In summary, we show that SphK1 is induced by EGFR and EGFRvIII signaling in glioblastoma cells and contributes to enhanced cell proliferation. Furthermore, we show evidence that SphK1 is necessary to maintain survival of glioblastoma cells. Thus this enzyme could be useful as a potential therapeutic target to decrease excess proliferation of glioma cells and possibly to enhance the pro-apoptotic effect of other drugs.
Acknowledgments
The authors are grateful to Dr. Amyn Habib of the University of Texas Southwestern Medical Center for the gift of U251-E18 cells, and to Dr. Lina Obeid of the Medical University of South Carolina for the gift of the SphK1 promoter luciferase construct. This work was supported by Grant # R21CA124685 from the National Cancer Institute (NCI) to JRVB and by the Department of Pathology, The Ohio State University.
Contributor Information
Adriana Estrada-Bernal, Department of Pathology, The Ohio State University, 4164 Graves Hall, 333 W. 10th Ave., Columbus, OH 43210, USA.
Sean E. Lawler, Department of Neurosurgery, The Ohio State University, Columbus, OH, USA
Michal O. Nowicki, Department of Neurosurgery, The Ohio State University, Columbus, OH, USA
Abhik Ray Chaudhury, Department of Pathology, The Ohio State University, 4164 Graves Hall, 333 W. 10th Ave., Columbus, OH 43210, USA.
James R. Van Brocklyn, Email: James.vanbrocklyn@osumc.edu, Department of Pathology, The Ohio State University, 4164 Graves Hall, 333 W. 10th Ave., Columbus, OH 43210, USA
References
- 1.Bansal K, Liang ML, Rutka JT. Molecular biology of human gliomas. Technol Cancer Res Treat. 2006;5:185–194. doi: 10.1177/153303460600500302. [DOI] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 3.Nakada M, Nakada S, Demuth T, Tran NL, Hoelzinger DB, Berens ME. Molecular targets of glioma invasion. Cell Mol Life Sci. 2007;64:458–478. doi: 10.1007/s00018-007-6342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young N, Van Brocklyn JR. Signal transduction of sphingosine-1-phosphate G protein-coupled receptors. Scientific World Journal. 2006;6:946–966. doi: 10.1100/tsw.2006.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Brocklyn JR, Jackson CA, Pearl DK, Kotur MS, Snyder PJ, Prior TW. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme. Roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. J Neuropathol Exp Neurol. 2005;64:695–705. doi: 10.1097/01.jnen.0000175329.59092.2c. [DOI] [PubMed] [Google Scholar]
- 6.Young N, Van Brocklyn JR. Roles of sphingosine-1-phosphate (S1P) receptors in malignant behavior of glioma cells. Differential effects of S1P2 on cell migration and invasiveness. Exp Cell Res. 2007;313:1615–1627. doi: 10.1016/j.yexcr.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 8.Huang SM, Harari PM. Epidermal growth factor receptor inhibition in cancer therapy: biology, rationale and preliminary clinical results. Invest New Drugs. 1999;17:259–269. doi: 10.1023/a:1006384521198. [DOI] [PubMed] [Google Scholar]
- 9.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(Suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 10.Frederick L, Wang XY, Eley G, James CD. Diversity and frequency of epidermal growth factor receptor mutations in human glioblastomas. Cancer Res. 2000;60:1383–1387. [PubMed] [Google Scholar]
- 11.Voelzke WR, Petty WJ, Lesser GJ. Targeting the epidermal growth factor receptor in high-grade astrocytomas. Curr Treat Options Oncol. 2008;9:23–31. doi: 10.1007/s11864-008-0053-5. [DOI] [PubMed] [Google Scholar]
- 12.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16:748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen MW, Meltorn M, Damstrup L, Poulsen HS. The type III epidermal growth factor receptor mutation. Biological significance and potential target for anti-cancer therapy. Ann Oncol. 2001;12:745–760. doi: 10.1023/a:1011177318162. [DOI] [PubMed] [Google Scholar]
- 14.Voldborg BR, Damstrup L, Spang-Thomsen M, Poulsen HS. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann Oncol. 1997;8:1197–1206. doi: 10.1023/a:1008209720526. [DOI] [PubMed] [Google Scholar]
- 15.Maceyka M, Milstien S, Spiegel S. Sphingosine-1-phosphate: the Swiss army knife of sphingolipid signaling. J Lipid Res. 2009;50(Suppl):S272–S276. doi: 10.1194/jlr.R800065-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, Black KL, Yu JS. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–9400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 17.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 18.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Machesky NJ, Zhang G, Raghavan B, Zimmerman P, Kelly SL, Merrill AH, Jr, Waldman WJ, Van Brocklyn JR, Trgovcich J. Human cytomegalovirus regulates bioactive sphingolipids. J Biol Chem. 2008;283:26148–26160. doi: 10.1074/jbc.M710181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramnarain DB, Park S, Lee DY, Hatanpaa KJ, Scoggin SO, Otu H, Libermann TA, Raisanen JM, Ashfaq R, Wong ET, Wu J, Elliott R, Habib AA. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- 21.Kohama T, Olivera A, Edsall L, Nagiec MM, Dickson R, Spiegel S. Molecular cloning and functional characterization of murine sphingosine kinase. J Biol Chem. 1998;273:23722–23728. doi: 10.1074/jbc.273.37.23722. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto K, Dang J, Zhu S, Nathanson D, Huang T, Dumont R, Seligson DB, Yong WH, Xiong Z, Rao N, Winther H, Chakravarti A, Bigner DD, Mellinghoff IK, Horvath S, Cavenee WK, Cloughesy TF, Mischel PS. Development of a real-time RT-PCR assay for detecting EGFRvIII in glioblastoma samples. Clin Cancer Res. 2008;14:488–493. doi: 10.1158/1078-0432.CCR-07-1966. [DOI] [PubMed] [Google Scholar]
- 23.Anelli VV, Gault CR, Cheng AB, Obeid LM. Sphingosine kinase 1 is up-regulated during hypoxia in U87MG glioma cells: role of hypoxia-inducible factors 1 and 2. J Biol Chem. 2008;283:3365–3375. doi: 10.1074/jbc.M708241200. [DOI] [PubMed] [Google Scholar]
- 24.Omuro AM. Exploring multi-targeting strategies for the treatment of gliomas. Curr Opin Investig Drugs. 2008;9:1287–1295. [PubMed] [Google Scholar]
- 25.Sampson JH, Archer GE, Mitchell DA, Heimberger AB, Bigner DD. Tumor-specific immunotherapy targeting the EGFRvIII mutation in patients with malignant glioma. Semin Immunol. 2008;20:267–275. doi: 10.1016/j.smim.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.French KJ, Schrecengost RS, Lee BD, Zhuang Y, Smith SN, Eberly JL, Yun JK, Smith CD. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 2003;63:2969–5962. [PubMed] [Google Scholar]
- 27.Döll F, Pfeilschifter J, Huwiler A. The epidermal growth factor stimulates sphingosine kinase-1 expression and activity in the human mammary carcinoma cell line MCF7. Biochim Biophys Acta. 2005;1738:72–81. doi: 10.1016/j.bbalip.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Yu CP, Xia JT, Zhang L, Weng GX, Zheng HQ, Kong QL, Hu LJ, Zeng MS, Zeng YX, Li M, Li J, Song LB. Sphingosine kinase 1 is associated with gastric cancer progression and poor survival of patients. Clin Cancer Res. 2009;15:1393–1399. doi: 10.1158/1078-0432.CCR-08-1158. [DOI] [PubMed] [Google Scholar]
- 29.Ruckhäberle E, Rody A, Engels K, Gaetje R, von Minckwitz G, Schiffmann S, Grösch S, Geisslinger G, Holtrich U, Karn T, Kaufmann M. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res Treat. 2007;112:41–52. doi: 10.1007/s10549-007-9836-9. [DOI] [PubMed] [Google Scholar]
- 30.Kawamori T, Osta W, Johnson KR, Pettus BJ, Bielawski J, Tanaka T, Wargovich MJ, Reddy BS, Hannun YA, Obeid LM, Zhou D. Sphingosine kinase 1 is up-regulated in colon carcinogenesis. FASEB J. 2006;20:386–388. doi: 10.1096/fj.05-4331fje. [DOI] [PubMed] [Google Scholar]
- 31.Kohno M, Momoi M, Oo ML, Paik JH, Lee YM, Venkataraman K, Ai Y, Ristimaki AP, Fyrst H, Sano H, Rosenberg D, Saba JD, Proia RL, Hla T. Intracellular role for sphingosine kinase 1 in intestinal adenoma cell proliferation. Mol Cell Biol. 2006;26:7211–7223. doi: 10.1128/MCB.02341-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Scolan E, Pchejetski D, Banno Y, Denis N, Mayeux P, Vainchenker W, Levade T, Moreau-Gachelin F. Overexpression of sphingosine kinase 1 is an oncogenic event in erythroleukemic progression. Blood. 2005;106:1808–1816. doi: 10.1182/blood-2004-12-4832. [DOI] [PubMed] [Google Scholar]
- 33.Li J, Guan HY, Gong LY, Song LB, Zhang N, Wu J, Yuan J, Zheng YJ, Huang ZS, Li M. Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin Cancer Res. 2008;14:6996–7003. doi: 10.1158/1078-0432.CCR-08-0754. [DOI] [PubMed] [Google Scholar]
- 34.Paugh BS, Bryan L, Paugh SW, Wilczynska KM, Alvarez SM, Singh SK, Kapitonov D, Rokita H, Wright S, Griswold-Prenner I, Milstien S, Spiegel S, Kordula T. Interleukin-1 regulates the expression of sphingosine kinase 1 in glioblastoma cells. J Biol Chem. 2009;284:3408–3417. doi: 10.1074/jbc.M807170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paugh BS, Paugh SW, Bryan L, Kapitonov D, Wilczynska KM, Gopalan SM, Rokita H, Milstien S, Spiegel S, Kordula T. EGF regulates plasminogen activator inhibitor-1 (PAI-1) by a pathway involving c-Src, PKCδ, and sphingosine kinase 1 in glioblastoma cells. FASEB J. 2007;22:455–465. doi: 10.1096/fj.07-8276com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar S, Maceyka M, Hait NC, Paugh SW, Sankala H, Milstien S, Spiegel S. Sphingosine kinase 1 is required for migration, proliferation and survival of MCF-7 human breast cancer cells. FEBS Lett. 2005;579:5313–5317. doi: 10.1016/j.febslet.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 37.Heimberger AB, Hlatky R, Suki D, Yang D, Weinberg J, Gilbert M, Sawaya R, Aldape K. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11:1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 38.Shida D, Kitayama J, Yamaguchi H, Yamashita H, Mori K, Watanabe T, Yatomi Y, Nagawa H. Sphingosine 1-phosphate transactivates c-Met as well as epidermal growth factor receptor (EGFR) in human gastric cancer cells. FEBS Lett. 2004;577:333–338. doi: 10.1016/j.febslet.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Shida D, Fang X, Kordula T, Takabe K, Lepine S, Alvarez SE, Milstien S, Spiegel S. Cross-talk between LPA1 and epidermal growth factor receptors mediates up-regulation of sphingosine kinase 1 to promote gastric cancer cell motility and invasion. Cancer Res. 2008;68:6569–6577. doi: 10.1158/0008-5472.CAN-08-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh HL, Sun CC, Wu CB, Wu CY, Tung WH, Wang HH, Yang CM. Sphingosine 1-phosphate induces EGFR expression via Akt/NF-κB and ERK/AP-1 pathways in rat vascular smooth muscle cells. J Cell Biochem. 2007;103:1732–1746. doi: 10.1002/jcb.21563. [DOI] [PubMed] [Google Scholar]
- 41.Sukocheva O, Wadham C, Holmes A, Albanese N, Verrier E, Feng F, Bernal A, Derian CK, Ullrich A, Vadas MA, Xia P. Estrogen transactivates EGFR via the sphingosine 1-phosphate receptor Edg-3: the role of sphingosine kinase-1. J Cell Biol. 2006;173:301–310. doi: 10.1083/jcb.200506033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edsall LC, Pirianov GG, Spiegel S. Involvement of sphingosine 1-phosphate in nerve growth factor-mediated neuronal survival and differentiation. J Neurosci. 1997;17:6952–6960. doi: 10.1523/JNEUROSCI.17-18-06952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rius RA, Edsall LC, Spiegel S. Activation of sphingosine kinase in pheochromocytoma PC12 neuronal cells in response to trophic factors. FEBS Lett. 1997;417:173–176. doi: 10.1016/s0014-5793(97)01277-5. [DOI] [PubMed] [Google Scholar]
- 44.Francy JM, Nag A, Conroy EJ, Hengst JA, Yun JK. Sphingosine kinase 1 expression is regulated by signaling through PI3 K, AKT2, and mTOR in human coronary artery smooth muscle cells. Biochim Biophys Acta. 2007;1769:253–265. doi: 10.1016/j.bbaexp.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 45.Murakami M, Ichihara M, Sobue S, Kikuchi R, Ito H, Kimura A, Iwasaki T, Takagi A, Kojima T, Takahashi M, Suzuki M, Banno Y, Nozawa Y, Murate T. RET signaling-induced SPHK1 gene expression plays a role in both GDNF-induced differentiation and MEN2-type oncogenesis. J Neurochem. 2007;102:1585–1594. doi: 10.1111/j.1471-4159.2007.04673.x. [DOI] [PubMed] [Google Scholar]
- 46.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu H, Acquaviva J, Ramachandran P, Boskovitz A, Woolfenden S, Pfannl R, Bronson RT, Chen JW, Weissleder R, Housman DE, Charest A. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci USA. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, Milstien S, Spiegel S. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 52.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 53.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-1185-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem. 2008;49:413–440. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Brocklyn JR. Lipids in neural tumors. In: Lajtha A, editor. Handbook of neurochemistry and molecular neurobiology, neural lipids. 3. Springer-Verlag; Heidelberg, Germany: 2009. pp. 535–561. [DOI] [Google Scholar]
- 56.Hait NC, Oskeritzian CA, Paugh SW, Milstien S, Spiegel S. Sphingosine kinases, sphingosine 1-phosphate, apoptosis and diseases. Biochim Biophys Acta. 2006;1758:2016–2026. doi: 10.1016/j.bbamem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 57.Cuvillier O. Downregulating sphingosine kinase-1 for cancer therapy. Expert Opin Ther Targets. 2008;12:1009–1020. doi: 10.1517/14728222.12.8.1009. [DOI] [PubMed] [Google Scholar]
- 58.Shida D, Takabe K, Kapitonov D, Milstien S, Spiegel S. Targeting SphK1 as a new strategy against cancer. Curr Drug Targets. 2008;9:662–673. doi: 10.2174/138945008785132402. [DOI] [PMC free article] [PubMed] [Google Scholar]