Abstract
Cell-cell interactions through direct contact are very important for cellular communication and coordination – especially for immune cells. The human immunodeficiency virus type I (HIV-1) induces immune cell interactions between CD4+ cells to shuttle between T cells via a virological synapse. A goal to understand the process of cell-cell transmission through virological synapses is to determine the cellular states that allow a chance encounter between cells to become a stable cell-cell adhesion. Here we demonstrate the use of optical tweezers to manipulate uninfected primary CD4+ T cells near HIV Gag-iGFP transfected Jurkat T cells to probe the determinants that induce stable adhesion. When combined with fast 4D confocal fluorescence microscopy, optical tweezers can be utilized to not only facilitate cell-cell contact, but to also allow one to simultaneously track the formation of a virological synapse, and ultimately to enable us to precisely determine all events preceding virus transfer.
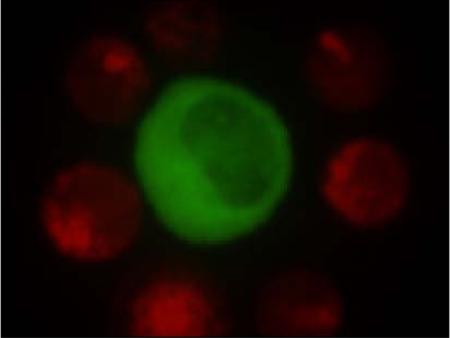
HIV-1 infected T cell (green) decorated with uninfected primary T cells (red) by manipulating the primary cells with an optical tweezers system
Keywords: Optical tweezers, micromanipulation, HIV-1, cell-cell transmission, fluorescence microscopy
1. Introduction
Direct cell-cell contact is not only important for scaffolding cellular structures (i.e. tissue), but also for inter-cellular communication and coordination [1–4]. Neuronal cells, for example, communicate by releasing neuro-transmitters at synaptic junctions between neurons to stimulate each other [5, 6]. Synaptic function and plasticity is also directly aided by an underlying complex neuron-astrocyte interaction [7–9]. The neuronal synapse formation and function is readily tracked over time because neurons are adherent cells, their synapses have a suitably long lifetime, and their function is susceptible to external stimuli [10–12]. A very different, much larger type of cell-cell synapse is of great importance for the adaptive immune system and facilitates antigen presentation. Here, an antigen-presenting cell, such as a dendritic cell or B cell, participates in an immunological synapse with a T lymphocyte to present a foreign antigen to it. Depending on the outcome of this interaction, the lymphocyte may subsequently trigger a specific immune reaction [13–16]. A particular difficulty in studying this complex cell-cell interaction is its seemingly random and short-lived nature, since immune cells, like T lymphocytes are non-adherent cells that freely circulate in the blood [17]. There arises the need for novel tools that enable the gentle micromanipulation of these cells. The manipulation process should ideally not harm the cells, but still enable the triggering of cell-cell contact, thus permitting the controlled and efficient observation of how the interaction process unfolds. Finally, a clever marker or indicator of the interaction process would be useful for visual tracking.
The discovery and utilization of the jellyfish green fluorescent protein (GFP) as a probe to track recombinant proteins has truly revolutionized cell biology [18]. By engineering genetic fusion proteins that are fluorescently tagged upon synthesis we can locate and track the expression and movement of these proteins in living cells – a task that would be nearly impossible using conventional antibody-based labels and synthetic fluorophores [19–22]. The recent successful creation of a replication-competent clone of the human immunodeficiency virus (HIV-1) containing GFP (HIV Gag-iGFP) has enabled, for the first time, the direct visualization of viral spread between live cells [23, 24]. When a HIV-1 infected T cell and an uninfected CD4+ T cell engage each other through interactions of HIV Env and CD4 they can form a virological synapse - a synaptic junction that draws comparisons to the immunological synapse [25–30]. HIV-1 viral proteins are then observed to rapidly and focally assemble virus particles at the point of cell-cell contact. Virus is subsequently transferred to the target cell, leading to productive infection. This process is highly efficient and very rapid. However, while we can directly visualize the transfer process in cells that have already established a virological synapse, the specific kinetics of synapse formation and viral transfer remain unclear as these early events are not easily captured because the initiation of new adhesions between cells is a relatively rare event to capture. Using current laboratory methods, this process is initiated in bulk, by mixing infected and uninfected cells and allowing contacts to be made during co-incubation. As a consequence, a significant amount of time and effort is spent searching through the volume of the cell chamber for a suitable cell pair to image using high-resolution video microscopy. This makes the observation of the initial cell-cell contact and early transfer events exceedingly rare. Without a reliable method to control the induction of cell-cell virus transfer we cannot accurately or fully determine the quantitative parameters that affect this process in both donor and acceptor cells.
Manipulating microscopic objects by optical tweezers utilizes changes in the momentum of photons to drive a target with a high index of refraction (such as a cell in media) to the center of a tightly focused laser beam [31, 32]. Once a cell has been trapped in this fashion it will follow the light focus when moved. The use of near-infrared laser sources for optical tweezers enables the manipulation of non-adherent cells such as germ cells, red blood cells, and T lymphocytes (of interest here) because cells have no to little absorption at these wavelengths and the process is virtually non-invasive [33–38]. Another advantage is that optical tweezers do not require open access to the sample or the use of needle aspirators, both of which significantly escalate the risk of accidental exposure when handling infectious agents like HIV-1.
Here, we demonstrate how optical tweezers can be utilized to test contact between HIV-1 Gag-íGFP infected and uninfected primary CD4+ T cells and subsequently study the cellular response by rapid live cell 4D fluorescence microscopy. When taken beyond the proof-of-concept stage, this controlled initiation process has the potential to significantly improve the throughput of investigating and understanding HIV-1 cell-cell transmission as well as the natural immunological synapse.
2. Experimental
Spinning-disk confocal fluorescence microscopy
A spinning disk assembly (CSU-10, Yokogawa Industries, Japan) was attached to an inverted optical microscope (iX71, Olympus, Center Valley, PA) and provides confocal excitation and fluorescence collection capabilities. Laser light from a multiline argon-krypton (ArKr) ion gas laser (Innova 70C, Coherent, Santa Clara, CA) is sent through an acousto-optic tunable filter (AOTF), which serves as a narrow bandpass excitation filter (selecting both 488 and 568 nm), fast shutter, and laser power control (<1 mW at the microscope objective for both laser lines). The laser beam is then expanded and collimated through a 2-lens 8× telescope system. The collimated light is guided into the CSU-10, where a dichroic mirror (Quadband 405–488–568–647, Semrock, Rochester, NY) directs the laser beams to a spinning microlens disk. Approximately 800 microlenses in a Nipkow pattern simultaneously focus the laser light through a second disk with 50 μm pinholes. The microlens pattern evenly illuminates the sample when both disks are rotating (1,800 RPM in this case). The laser spots are projected onto the sample by a 60×, 1.42 NA oil immersion objective lens (PlanApo N, Olympus). Fluorescence emission from the sample is collected by the same microscope objective, sent through the pinhole disk, and projected onto a highly sensitive single photon counting electron-multiplying charge-coupled device (EM-CCD) camera (iXon+ DU897, Andor Technology, Belfast, Ireland). Rapid 3D sectioning of the sample is achieved by mounting the sample onto a closed-loop piezo-translation stage (PS-S1010, MadCity Labs, Madison) that is synchronized to the EM-CCD read-out rate by the data acquisition software (iQ, Andor Technology). An image splitter (Optosplit II, Cairn, Kent, UK) utilizing a dichroic mirror (570DCXR, Chroma, Rockingham, VT) simultaneously projects two image channels onto the EM-CCD. Both fluorescence channels are collected through bandpass filters with the green channel set up to image GFP (FF01-525/50-25, Semrock), while the red channel (FF01-609/54-25, Semrock) imaged cells labeled with CellTracker Red or light from a bandpass-filtered (HQ620/30m, Chroma) halogen lamp used for differential interference contrast.
Optical tweezers addition to the microscope
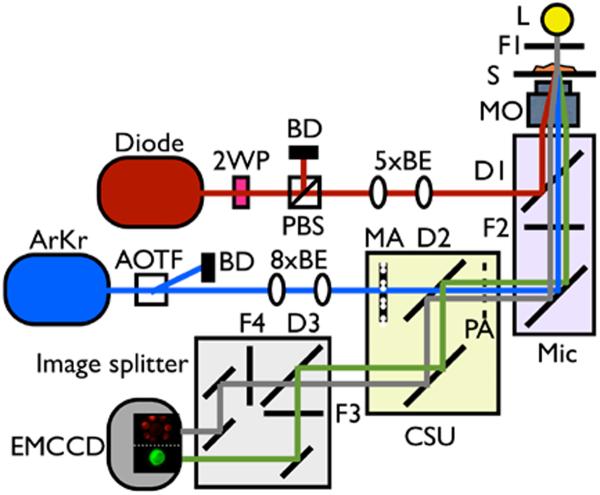
The optical tweezers system utilizes a 2W, 1064 nm CW laser (SDL 1064-2000T, Shanghai DreamLaser Tech, Shanghai, China) whose power was reduced to 5–15 mW using a λ/2 waveplate (NT43–705, Edmund Optics, Barrington, NJ) and polarizing beamsplitter (PBS-1064-100, CVI laser, Albuquerque, NM). After a 2-lens 5× beam expander the laser was directed into the back turret port of the microscope and combined with the ArKr laser light from the spinning disk assembly using a dichroic mirror (900dcsp, Chroma). After passing through the microscope objective and optically trapping the sample, backreflected or scattered 1064 nm light is blocked from the EM-CCD camera using a shortpass filter (FF01-750/SP-25, Semrock) underneath the dichroic mirror used for combining the laser beams. Manually moving the microscope stage allowed us to target and optically trap specific cells with the optical tweezers while the built-in turret shutter from the microscope enables us to turn the optical trap on or off by blocking the laser beam. The entire optical assembly is schematically depicted in Figure 1.
Figure 1.
Combined video-rate fluorescence confocal microscope and single beam laser tweezers system. The 1064 nm laser providing the ability to optically trap and manipulate cells is directly coupled into the back port of an inverted optical microscope and reflected towards the objective lens by an infrared-reflecting dichroic mirror. The spinning disk assembly is attached to the left side port of the microscope and the excitation light and fluorescence simply passes the dichroic mirror. 2WP is a half waveplate, 5×BE is a 5× beam expander, 8×BE is a 8× beam expander, AOTF is acoustic optical tuneable filter, ArKr is a multiline ArKr ion laser, BD is a beam dump, CSU is a confocal spinning unit, D1 is a 900 nm short pass dichroic mirror, D2 is a quad band dichroic mirror, D3 is a 560 nm long pass dichroic mirror, Diode is a 1064 nm CW diode laser, EMCCD a is electron multiplying charged coupled devise camera, F1 is a 620/30 nm bandpass filter, F2 is a 750 nm short pass dichroic, F4 is a 609/54 nm bandpass filter, F5 is a 525/50 nm bandpass filter, L is halogen lamp, Mic is the microscope, MA is a microlens array, MO is the microscope objective, PA is a pinhole array, PBS is a polarizing beam splitter, and S is sample.
Cell isolation
CD4+ transformed human T cells (Jurkat T cell line CE6.1) are obtained from American Type Culture Collection (ATCC, Rockville, MD) and maintained in culture with RPMI 1640 medium supplemented with 10% fetal calf serum. Human peripheral blood mononuclear cells (PBMC) from healthy HIV/HBV sero-negative donors are purified from leukocyte buffy coat by Ficoll gradient. T-lymphocytes are separated and enriched for CD4+ T cells by magnetic bead sorting according to manufacturer's instructions (Miltenyi Biotec). Cell preparations are routinely >99% viable as assessed by trypan blue dye exclusion. The purity of the cell sub-populations is defined by surface immunophenotype using flow cytometry and specific monoclonal antibodies to CD4 and checked for purity of >90% CD4+ T cells. If the purity is less than 90%, the magnetic bead separation step is repeated. The use of fresh human samples for this study is in accordance with the University of California Institutional Review Board practice.
Transduction of Jurkat T cells and sample preparation
HIV-1 proviral constructs are transfected into Jurkat cells using electroporation in a variable frequency electroporation device (Nucleofector II, Amaxa Biosystems, Germany). In brief, 3 ug of endotoxin-free HIV-1 proviral plasmids are nucleofected into 5×106 Jurkat cells, using Cell Line Nucleofector™ Kit V, program S-18. 24h after transfection, viable Jurkat cells are purified by centrifugation on a Ficoll Hypaque density gradient. 48h after transfection, cells are washed with complete buffer and enumerated for test. Transfected Jurkat and primary T cells labeled with CellTracker red (Invitrogen, Carlsbad) are loaded in a gas-permeable imaging chamber (IBIDI μ-slide VI, IBIDI GmbH, Germany) in a biosafety cabinet located in a biosafety level 2 facility, the chamber is sealed, and then transferred to the microscope system for live cell microscopy. Several hours before sample placement, the microscope and surrounding area are warmed to 37° C by heating them with an air stream incubator (ASI400, Nevtek, Williamsville).
Image acquisition and analysis
Transfected Jurkat T cells expressing HIV Gag-iGFP are identified by their strong cytoplasmic and plasma membrane localized GFP fluorescence. Once a cell is located, the optical tweezers are used to trap and move CellTracker Red labeled primary CD4+ T cells towards the Jurkat cell. Subsequently, the automated, continuous acquisition of digital images from the EM-CCD to optically dissect the cells is started. Rapid 3D time lapse images are taken at a rate of 30 frames per second using 0.5 μm z-steps for typically 20+ minutes. For processing, CCD images containing both color channels are separated, encoded by false color, overlaid, aligned and maximum intensity projected and analyzed using a variation of ImageJ (NIH, Bethesda, MD) called Fiji (Max Plank Institute of Molecular Cell Biology and Genetics, Dresden, Germany).
3. Results and discussion
After co-incubation of HIV-transfected and primary T cells, cells that are forming a virological synapse are identified as conjoined cells that move together indicating adhesive contact and might already have formed a bright synaptic button in the junction connecting the two cells. This process appears random and is uncontrolled therefore searching for such cells can take up considerable time on the microscope platform. Also, since the mechanisms of synapse formation are not yet fully understood, it is very difficult to determine the time that it takes from the initial engagement of the two cells until synapse formation and ultimately transfer of viral material. To solve these problems and attack these questions, we decided to integrate an optical tweezers system with a spinning disk confocal microscope system. This is motivated in particular by the fact that the chamber containing the infected and co-incubated cells is sealed, so that optical tweezers are currently the only way to allow for the controlled manipulation of select cells within the chamber.
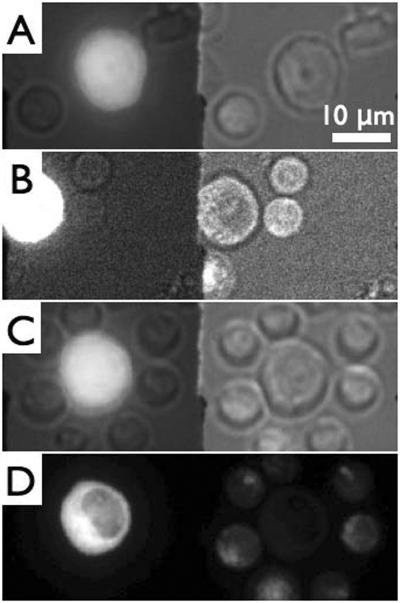
Figure 2 illustrates the typical process of “loading” an infected cell with primary T cells. Gag-iGFP expressing Jurkat cells are identified by their larger size and the strong green fluorescence emitted by the fluorescently labelled Gag protein in the cytoplasm and partially at the plasma membrane (Figure 2A). Figure 2A–C shows an unmodified sequence of CCD images as they are being collected during the experiment. The spinning disk system is running continuously, while images are being streamed to the computer display at a cropped CCD frame size. Here, the left hand image within each frame is the green (GFP) channel, whereas the right-hand channel is the red channel, which, in this case shows white light images of all the cells within the field of view after illumination with a spectrally filtered white light source. The image in the green channel also shows a weak white light background due to some white light bleed-through. Once a suitable infected Jurkat T cell has been identified (”suitability” being judged mostly by a cell's healthy morphology and strong fluorescence intensity, indicating a highly HIV Gag-iGFP expressing cell), the optical tweezers are then used to move primary T cells in the vicinity of the infected cell. As can be seen in Fig. 2A, initially no primary CD4+ T cell (identifiable by their smaller size and CellTracker red fluorescence (see Fig. 2D)) were touching this Jurkat T cell. The nearest eight CD4+ T cells (up to several fields of view away) were then selected and moved in close vicinity of or brought in direct contact with the Jurkat T cell. Loading the Jurkat T cell with eight potential acceptor cells was accomplished in approximately 4 minutes by repeated manual stage movement and laser shuttering. Figures 2B–C show images demonstrating the search for primary cells and their manipulation towards the Jurkat T cell. Upon completion of loading the infected cell with the primary T cells a single z-stack image of the GFP and CellTracker red fluorescence channels was acquired with the white light source turned off. An intensity-projected image of the Jurkat cell decorated with primary T cells is shown in Figure 2D. This “loading” process enables us to exercise some control over the timing of the formation of virological synapses as well as the subsequent transfer of viral material from the infected cell to one or more uninfected cells. It also allows us to potentially greatly increase the efficiency of observing cell-cell transfer of HIV. This process, however, likely also depends on a number of other parameters that are still unknown and are thus uncontrolled.
Figure 2.
Sequence of video micrographs demonstrating the “loading” of an HIV-iGFP-infected Jurkat T cell with uninfected primary CD4+ T cells. Images taken through an image splitter with the Gag-iGFP fluorescence image (left) with partial transmission of white light (right). A) Initial image identifying the infected Jurkat cell by its size and fluorescence (bright cell in the middle). B) 2 primary cells have been moved to upper right corner near the infected cell. C) The Jurkat cell surrounded with eight primary cells. D) Simultaneous 2-color fluorescence image of C) showing GFP in the Jurkat cell (left) and CellTracker red in primary CD4+ T cells. All images are maximum intensity projections.
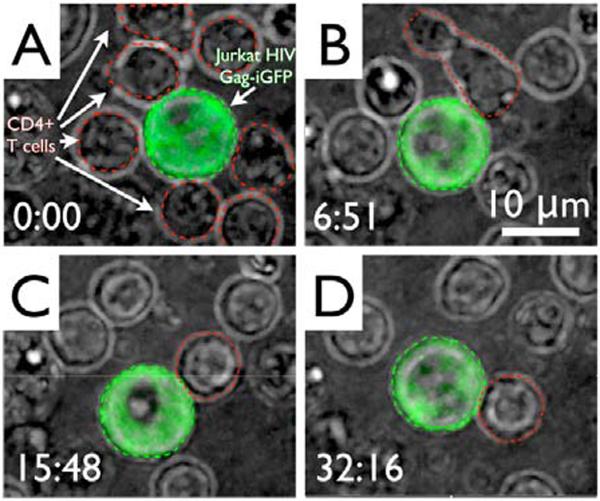
After surrounding the Jurkat T cell with primary T cells by optical tweezers, rapid 3D imaging was conducted on the group of cells for over 60 minutes total by acquiring 30 frames per second per 0.5 μm z-step through the cells, resulting in complete optical sectioning of the cells every ~ 1.5 seconds. To permit long-term viewing of the cells, the red fluorescence channel was replaced with the white transmitted light image of the cells. Figure 3 shows a sequence of images with the green fluorescence channel overlaid as an intensity-projected image onto the white light channel to highlight the Jurkat T cell. The primary T cells are highlighted by red outlines in the first image and a particular cell of interest in the subsequent images to improve visibility. The time in minutes:seconds after the start of the initial image acquisition at which each image in the sequence was obtained is shown in the lower left corner of each image. Out of all eight cells that were relocated to decorate the infected cell, each of the eight cells were observed to transiently touch the infected cell, some on multiple occasions, but without formation of a stable adhesion. Only one primary CD4+ T cell created a stable adhesion with the infected cell approximately 7 minutes into the recording. These cells then remained connected until the end of image acquisition. The formation of a stable adhesion junction is evidenced by the fact that the two cells maintained a large contact zone and moved together despite Brownian motion, drift, and rotation of the two cells.
Figure 3.
Formation of an adhesion junction between an HIV Gag-iGFP transfected Jurkat cell (larger, green with green dashed outline) and primary CD4+ T cells (smaller, with red dashed outline) that were loaded around it using optical tweezers. Images are maximum intensity projections of a false color overlay of GFP and red filtered white light image. A) First image after loading. B) Initial contact between the cell pair. C) & D) Stable adhesion between the cell pair over time.
The primary T cell that ended up adhering to the infected cell in this instance started out at the top of the screen and was one of a few that were not initially in direct contact with the Jurkat T cell. As the primary cell moved closer to the infected cell, actively changing morphology along the way, all seven other primary cells just drifted randomly in solution while preserving a round shape. As can be seen in the video sequence from which the screen shots in Figure 3 were taken (see supplemental information), three of these cells appeared to make contact with the infected cell during this time, but did not engage in the formation of an adhesion junction. The act of “crawling” towards the infected cell and the creation of an adhesion junction is shown in Figures 3B–D, and the accompanying movie in the supplemental materials. While adhering to the infected cell the primary cell continued to actively change its shape from round to elongated to round again. Although we imaged this pair of cells for over one hour we did not observe the formation of a bright fluorescent button, but nevertheless found a discrete accumulation of Gag-iGFP signal in the junction between the cells (see Figure 4). This is indicative of an early virological synapse and precedes transfer of viral material. The long time that it took these cells to accumulate Gag-iGFP in the synapse is consistent with previous observations that viral transfer can occur over a wide time span ranging from minutes to over an hour - after a stable virological synapse has already formed. Figure 4 further demonstrates the versatility of our experimental system, which enables the acquisition of full 3D data of these cells. Here, we took advantage of this fact and show a cross-section of just the plane in which the virological synapse is located, which highlights the accumulation of Gag-iGFP and creates much better contrast then the intensity-projected image shown in Figure 3D.
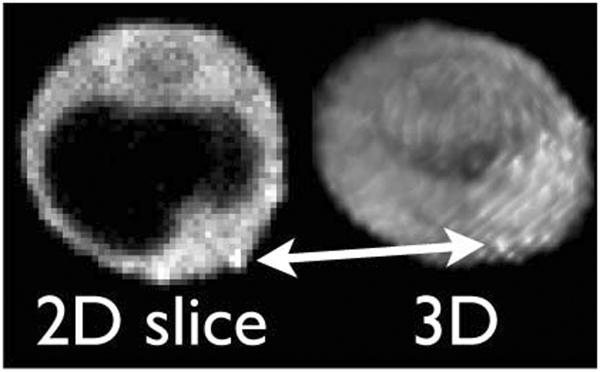
Figure 4.
Single frame of a sequence of 3D confocal fluorescence microscope data obtained from the conjoined cells shown in Figure 3 after 65 minutes of imaging. The left image is a z-slice through the middle of the cell while the image on the right is the corresponding 3D rendering. For orientation, the red arrow highlights the same feature by pointing to one of several bright Gag-iGFP spots in the Gag-iGFP enriched synapse. The acceptor cell (not labelled) is to the lower right side of the cell, just as shown in Figure 3D.
The images shown in Figures 3 and 4 indicate that the ability of an infected T cell to engage in a stable adhesion with another T cell is likely dependent upon activation of local adhesion that may be influenced greatly by cell polarity induced by cell migration. As is apparent by its rapid change of morphology and the active crawling towards the infected cell, this particular cell is obviously in a different state than the others. The further use of optical tweezers will be quite beneficial in preselecting target cells with particular morphologies to determine the importance of pre-existing cell polarity in inducing virological synapse-mediated adhesion. When combined with the fluorescent labeling of cell markers, the importance of cell states (e.g. activation, health, etc) can be addressed.
4. Conclusion
Here we describe the combination of a high speed spinning disk confocal imaging platform with an optical tweezers system, which we demonstrate to be useful in probing cell-cell interactions under controlled in vitro conditions. The utility of the platform is demonstrated in probing the initial interactions of HIV infected cells with uninfected cells as the cells form virological synapses.
HIV-1 cell-cell transfer was previously shown to be a highly efficient process when considered in bulk, but our ability to capture the initial adhesion event between cells and the events that immediately follow has been rather inefficient. Optical tweezers are useful in probing this process by moving putative cells to touch HIV Gag-iGFP infected cells allowing us to test the receptiveness of the cell to synapse formation. We expect this to enable us to consistently and quantifiably probe this complex interaction. The combination of optical tweezers with rapid spinning disk confocal fluorescence microscopy into a single instrument enables not only the manipulation of infected cells in an enclosed sample chamber, but also their rapid characterization in all 3 dimensions with diffraction-limited spatial resolution. Additional modifications that further improve the performance of such a hybrid instrument can be envisioned. For example, the future use of spatial light modulators will enable us to dynamically create holographic optical tweezers across the entire field of view, which further improves the speed of cell manipulation, or enables us to actively maintain contact between several cells simultaneously. Also, as exemplified by what has been observed in this paper, such a system will allow us to explore CD4+ T cell states of cell activation or cell subtypes and their effect on the propensity to bind to HIV infected cells. All avenues of improving cell selection warrant exploration and some have even been implicated in affecting infection and transfer efficiency. Further attempts, not shown here, used 52 different cells that lacked a dynamic morphology to make 74 contacts with HIV Gag-iGFP expressing Jurkat cells all failed to create adhesion. This fact in particular is another strong motivation for multiplexing the capability to establish contacts between cells in the near future. It is particularly interesting to note that the adhesion between infected and uninfected T cells appears to require a cell in a certain morphological state, which may be difficult to probe without methods which rely on direct visualization. The combination of optical tweezers and fast time-lapse microscopy has potential in aiding our understanding in HIV-1 cell-cell adhesion by systematically initiating and tracking the process from beginning to end, one cell pair at a time, without physically breaching a closed infectious environment.
Finally, this approach of optical manipulation to initiate T lymphocyte cell contacts can help to enable systematic studies of the related immunological synapse, a crucial aspect in mounting immune responses.
Acknowledgements
We thank Drs. F. Chuang, D. Asmuth, X.-D. Li, P. Chen and B. Dale for critiques and stimulating discussions. Work was supported by NIH AI074420-02, Burroughs Wellcome Fund Investigator Award, and Hirschl Weill-Caulier Career Scientist Award to B.K.C.. This work was also supported by the NSF Center for Biophotonics Science and Technology (Cooperative Agreement PHY012099), a UC Davis Health System Research award to T.H., and the UCD CTSC (NCRR grant ULRR024146 (T.H.).
Biographies
Gregory McNerney is a Biophysics PhD graduate student at the University of California Davis, where he also received his B.S. in Optical Science and Engineering in 2005. He is currently working on the applying advanced optical tools for studying infectious diseases, including HIV-1 virological synapse mediated cell-cell transfer, at the NSF Center for Biophotonics Science and Technology in Sacramento, California.
Wolfgang Hübner, Ph.D., is scientific officer at EMBL Heidelberg (Germany) responsible for advanced light microscopy in P-CUBE (Infrastructure for Protein Production Platforms) within the Seventh Framework Program (FP7) of the European Commission. He worked until 2009 as a postdoctoral fellow on HIV-1 assembly mechanisms and transfer through the T cell virological synapse in Benjamin Chen's laboratory in the Immunology Institute in Mount Sinai School of Medicine, New York.
Benjamin Chen, M.D., Ph.D., is an Assistant Professor in the Division of Infectious Diseases, Department of Medicine at Mount Sinai School of Medicine. He is a molecular virologist interested in understanding how Human Immunodeficiency Virus assembly is regulated during infection of T cells. He has developed a number of important virological tools that facilitate quantitative studies of the virus life cycle. The development of intrinsically fluorescent HIV variants in his laboratory has enabled high-resolution live microscopy studies in T cells, which are among the first to characterize viral assembly during cell-cell transmission.
Thomas Huser is an Associate Professor in the Department of Internal Medicine at the University of California, Davis. He also serves as Chief Scientist for the Center for Biophotonics Science and Technology, and Co-Director of the Translational Working Group for the Clinical Translational Science Center at UC Davis. Until November 2005, Dr. Huser was a Group leader for Biophotonics and Nanospectroscopy at Lawrence Livermore National Laboratory in Livermore, CA, where he developed and applied novel nano-biophotonics tools to characterize the nanosystems biology of individual cells. He worked at Lawrence Livermore National Laboratory since joining LLNL in 1998 as a postdoctoral researcher and became a staff scientist in 2000. Dr. Huser obtained his Ph.D. in Physics from the University of Basel, Switzerland, where he worked primarily on near-field optical microscopy. At UC Davis he applies Raman spectroscopy and imaging, and single molecule fluorescence microscopy to biological and medical problems at the single cell level.
References
- 1.Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Nature. 2005;435(7044):964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- 2.Bazzoni G, Dejana E. Physiol. Rev. 2004;84(3):869–901. doi: 10.1152/physrev.00035.2003. [DOI] [PubMed] [Google Scholar]
- 3.Schneeberger E, Lynch R. Am. J. Physiol. - Cell Physiol. 2004;286(6):C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- 4.Kumar N, Gilula N. Cell. 1996;84(3):381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 5.Murthy VN, De Camilli P. Annu. Rev. Neurosci. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]
- 6.Sudhof T. Annu. Rev. Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 7.Araque A, Carmignoto G, Haydon P. Annu. Rev. Physiol. 2001;63:795–813. doi: 10.1146/annurev.physiol.63.1.795. [DOI] [PubMed] [Google Scholar]
- 8.Çakur T, Alsan S, Saybaşulu H, Akun A. Theor. Biol. Med. Model. 2007;4(48) doi: 10.1186/1742-4682-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vernadakis A. Prog. Neurobiol. 1996;49(3):185–214. doi: 10.1016/s0301-0082(96)00012-3. [DOI] [PubMed] [Google Scholar]
- 10.Erickson J, Tooker A, Tai Y-C, Pine J. J. Neurosci. Meth. 2008;175(1):1–16. doi: 10.1016/j.jneumeth.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izzo AD, Walsh JT, Ralph H, Webb J, Bendett M, Wells J, Richter C-P. Biophys. J. 2008;94(8):3159–3166. doi: 10.1529/biophysj.107.117150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colicos MA, Syed NI. J. Exp. Biol. 2006;209(12):2312–2319. doi: 10.1242/jeb.02163. [DOI] [PubMed] [Google Scholar]
- 13.Brigl M, Brenner M. Annu. Rev. Immunol. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 14.Bromley S, Burack W, Johnson K, Somersalo K, Sims T, Sumen C, Davis M, Shaw A, Allen P, Dustin M. Annu. Rev. Immunol. 2001;19:375–396. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- 15.Davis S, van der Merwe P. Curr. Biol. 2001;11(8):R289–R290. doi: 10.1016/s0960-9822(01)00165-8. [DOI] [PubMed] [Google Scholar]
- 16.Grakoui A, Bromley S, Sumen C, Davis M, Shaw A, Allen P, Dustin M. Science. 1999;285(5425):221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 17.Gunzer M, Schafer A, Borgmann S, Grabbe S, Zanker K, Brocker E, Kampgen E, Friedl P. Immunity. 2000;13(3):323–332. doi: 10.1016/s1074-7613(00)00032-7. [DOI] [PubMed] [Google Scholar]
- 18.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. Science. 1994;263(5148):802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Campbell R, Ting A, Tsien R. Nat. Rev. Mol. Cell Biol. 2002;3(12):906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 20.Lippincott-Schwartz J, Snapp E, Kenworthy A. Nat. Rev. Mol. Cell Biol. 2001;2(6):444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 21.Cubitt A, Heim R, Adams S, Boyd A, Gross L, Tsien R. Trends Biochem. Sci. 1995;20(11):448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 22.Giepmans B, Adams S, Ellisman M, Tsien R. Science. 2006;312(5771):217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 23.Hübner W, Chen P, Del Portillo A, Liu Y, Gordon RE, Chen BK. J. Virol. 2007;81:12596–12607. doi: 10.1128/JVI.01088-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hübner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li X-D, Asmuth DM, Huser TR, Chen BK. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P, Huebner W, Spinelli MA, Chen BK. J. Virol. 2007;81(22):12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolly C, Kashefi K, Hollinshead M, Sattentau Q. J. Exp. Med. 2004;199(2):283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jolly C, Sattentau Q. J. Virol. 2005;79:12088–12094. doi: 10.1128/JVI.79.18.12088-12094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jolly C, Sattentau Q. Traffic. 2004;5:643–650. doi: 10.1111/j.1600-0854.2004.00209.x. [DOI] [PubMed] [Google Scholar]
- 29.Jolly C, Sattentau QJ. J. Virol. 2007;81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fackler O, Alcover A, Schwartz O. Nat. Rev. Immunol. 2007;7(4):310–317. doi: 10.1038/nri2041. [DOI] [PubMed] [Google Scholar]
- 31.Ashkin A. Phys. Rev. Lett. 1970;24(4):156–159. [Google Scholar]
- 32.Ashkin A, Dziedzic J, Bjorkholm J, Chu S. Opt. Lett. 1986;11(5):288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 33.Chan JW, Taylor DS, Lane SM, Zwerdling T, Tuscano J, Huser TR. Anal. Chem. 2008;80(6):2180–2187. doi: 10.1021/ac7022348. [DOI] [PubMed] [Google Scholar]
- 34.Ozkan M, Wang M, Ozkan C, Flynn R, Birkbeck A, Esener S. Biomed. Microdev. 2003;5(1):61–67. [Google Scholar]
- 35.Nilsson J, Evander M, Hammarstrom B, Laurell T. Anal. Chim. Acta. 2009;649(2):141–157. doi: 10.1016/j.aca.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 36.Anvari B, Torres J, McIntyre B. J. Biomed. Opt. 2004;9(5):865–872. doi: 10.1117/1.1778178. [DOI] [PubMed] [Google Scholar]
- 37.Rao S, Balint S, Cossins B, Guallar V, Petrov D. Biophys. J. 2009;96(1):209–216. doi: 10.1529/biophysj.108.139097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Shao B, Chen T, Berns M. J. Biophot. 2009;2(3):167–177. doi: 10.1002/jbio.200810053. [DOI] [PubMed] [Google Scholar]