Abstract
Stem cells of all types are characterized by a stable, heritable state permissive of multiple developmental pathways. The past five years have seen remarkable advances in understanding these heritable states and the ways that they are initiated or terminated. Transcription factors that bind directly to DNA and have sufficiency roles have been most easy to investigate and, perhaps for this reason, are most solidly implicated in pluripotency. In addition, large complexes of ATP-dependent chromatin-remodeling and histone-modification enzymes that have specialized functions have also been implicated by genetic studies in initiating and/or maintaining pluripotency or multipotency. Several of these ATP-dependent remodeling complexes play non-redundant roles, and the esBAF complex facilitates reprogramming of induced pluripotent stem cells. The recent finding that virtually all histone modifications can be rapidly reversed and are often highly dynamic has raised new questions about how histone modifications come to play a role in the steady state of pluripotency. Another surprise from genetic studies has been the frequency with which the global effects of mutations in chromatin regulators can be largely reversed by a single target gene. These genetic studies help define the arena for future mechanistic studies that might be helpful to harness pluripotency for therapeutic goals.
Keywords: epigenetics, chromatin remodeling, BAF complexes, stem cells, lineage specificity
GENERAL FEATURES OF CHROMATIN IN STEM CELLS
In eukaryotic cells, 146 base pairs (bp) of DNA wrap an octamer of core histones to form the nucleosome, the basic unit of chromatin (Kornberg 1974). In addition to conventional histones (H2A, H2B, H3, and H4), the incorporation of “variant histones” promotes nucleosome diversity and influences overall chromatin structure (Ahmad & Henikoff 2001). Throughout the genome, nucleosomes occur as repeating arrays, separated by linker DNA associated with a fifth histone, H1, which initiates higher-order chromatin structures. Local chromatin structure is specified by the positioning of nucleosomes, which are progressively folded into poorly characterized higher-order heterochromatin that shows visible differences between cell types and between closely related species (Le Douarin & Teillet 1974). In addition, heterochromatin occurs at sites of repetitive DNA and specific chromosomal regions such as centromeres.
Investigators have long suspected that stem cells maintain their stable, heritable state by epigenetic regulatory mechanisms. Only recently have some of the genes and mechanisms become defined. Embryonic stem (ES) cells, derived from the inner cell mass (ICM) of the blastocyst, possess self-renewal potential as well as the ability to generate all cell types other than the placenta within the body (pluripotency). These characteristics of ES cells, which distinguish them from tissue stem cells with more limited self-renewal and developmental potential (generally termed multipotent), are conferred by unique transcriptional regulation due in part to the specialized and dynamic nature of their chromatin. First, fewer and more diffuse transcriptionally inactive heterochromatic foci are detected in ES cell nuclei compared with their differentiated progeny (Meshorer & Misteli 2006, Meshorer et al. 2006). Upon differentiation, condensation of ES cell chromatin into a more repressive state is associated with increased global incorporation of specific histone variants (microH2A) and concentration of heterochromatin proteins (such as HP1) at discrete foci (Dai & Rasmussen 2007, Meshorer et al. 2006). Fluorescent recovery after photobleaching (FRAP) experiments revealed an increased fraction of loosely bound or soluble structural chromatin proteins in pluripotent ES cells, which become more stably associated with chromatin upon differentiation. Accordingly, the exchange of linker histone H1 by a more tightly chromatin-bound version inhibited ES cell differentiation, whereas replacement in chromatin of core histone H3 by its variant H3.3, a marker of active transcription, accelerated their differentiation (Meshorer et al. 2006). This suggests that reorganization of chromatin structure (more compact and repressive) during lineage specification is achieved, at least in part, through the dynamic exchange of structural proteins.
The status of histone modifications further indicates that the chromatin in ES cells is more transcriptionally permissive than in differentiated cells. Pluripotent chromatin displays properties of euchromatin, such as high levels of acetylated histones and increased nuclease accessibility. Lineage specification and differentiation is accompanied by a decrease in global levels of active histone marks (such as acetylated histone H3 and H4, including H3K4me3) and an increase in repressive histone marks (such as histone H3 lysine 9 methylation) (Azuara et al. 2006, Lee et al. 2004, Meshorer et al. 2006). However, proper histone methylation at H3K27 does not appear to be essential to maintain pluripotency, as loss of function of Polycomb repressive complex 2 (PRC2) components responsible for making the H3K27me3 repressive histone mark in ES cells does not abolish their self-renewal or their ability to produce all three germ layers (Montgomery et al. 2005, Pasini et al. 2007) but rather gives rise to later specific defects in the allocation and migration of mesoderm.
Consistent with the observation that the pluripotent chromatin is in an open conformation, ES cell chromatin is generally more permissive to the transcriptional machinery than that of differentiated cells, and tissue-specific genes that are expected to be silent in undifferentiated cells may be in a semipermissive transcriptional state in ES cells (Levings et al. 2006, Szutorisz et al. 2005). The proteosome is thought to be involved in this process by regulating the rapid turnover of transcription factors and Pol II binding at the promoters of developmentally regulated genes to restrict permissive transcriptional activity while keeping the genes in a potentiated state for later activation (Szutorisz et al. 2006). Altogether, these observations suggest that restriction of developmental potential is associated with a marked decrease in genome plasticity and the establishment of new heritable gene expression programs. As discussed below, the hyperdynamic nature of pluripotent chromatin may be essential to achieve rapid changes in transcriptional programs during lineage commitment and differentiation.
THE CORE PLURIPOTENCY CIRCUITRY
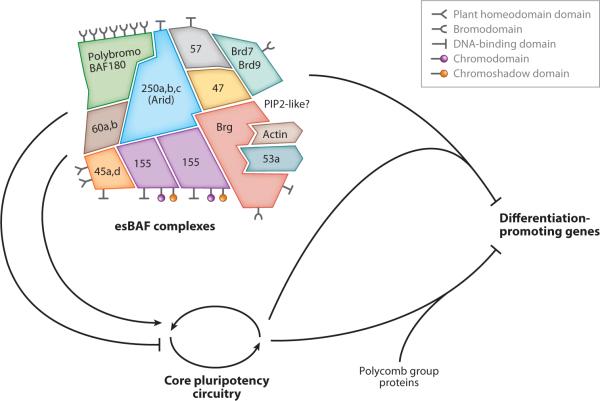
Recent studies have begun to uncover a transcriptional regulatory network in ES cells that provides insights into the molecular basis of how pluripotency is established and maintained. The genes essential or contributing to the pluripotent state are listed in Table 1 and in an extended form in Supplemental Table 1 (follow the Supplemental Material link from the Annual Reviews home page at http://www.annualreviews.org). To help the reader judge the quality of the data, null mutations that provide definitive evidence are given in bold, whereas RNAi studies are shown in plain type. Foremost among this list are the three key transcription factors, Oct4, Sox2, and Nanog, which form an intrinsic core-regulatory circuitry with positive feedback that maintains the pluripotent state of stem cells (Boiani & Scholer 2005; Boyer et al. 2005, 2006; Chew et al. 2005; Ivanova et al. 2006; Loh et al. 2006; Rao & Orkin 2006; Remenyi et al. 2003; Yeom et al. 1996). The POU family transcription factor Oct3/4 (encoded by Pou5f1) is a critical regulator of pluripotency. During mouse embryonic development, zygotic Oct4 expression begins at the four-cell stage of, and is subsequently restricted to, pluripotent stem cells (i.e., ICM, germ cells, and ES cells). Oct4 deficiency induces the differentiation of the ICM and ES cells into trophectoderm and later cell death, whereas its overexpression in ES cells promotes differentiation into the primitive endoderm and mesoderm lineages (highlighting the importance of negative feedback mechanisms) (Nichols et al. 1998; Niwa 2001, 2007; Niwa et al. 2000; Yeom et al. 1996). Nanog, a NK2-class homeobox transcription factor, is another component of the core pluripotency network that is required for the maintenance of pluripotency in both the ICM and ES cells (Mitsui et al. 2003). Nanog expression is restricted to pluripotent cells, and ES cells deficient for this gene spontaneously differentiate into the primitive endoderm lineage; yet, it is not essential for formation of ES cells (Chambers et al. 2003, Mitsui et al. 2003). Overexpression of Nanog in mouse ES cells can bypass the requirement for leukemia inhibitory factor in maintaining pluripotency in culture (Matsuda et al. 1999). Similarly, the SRY-related HMG-box transcription factor Sox2 is required for the maintenance of pluripotency (Avilion et al. 2003, Masui et al. 2007). Sox2 expression is not restricted to pluripotent cells in the embryo (in contrast to Oct4 and Nanog) and is maintained in early neural cells (Avilion et al. 2003). Sox2-null embryos die immediately after implantation (Avilion et al. 2003), and shRNA-mediated knockdown of Sox2 in ES cells promotes their differentiation into multiple lineages (Ivanova et al. 2006). Oct4, Sox2, and Nanog biochemically interact with each other and coregulate the expression of many target genes (Boyer et al. 2005, Kuroda et al. 2005, Loh et al. 2006, Masui et al. 2007, Rodda et al. 2005) including histone-modification enzymes (Loh et al. 2006, 2007; Matoba et al. 2006). Oct4, Sox2, and Nanog are also direct transcriptional targets of SWI/SNF-like BAF chromatin-remodeling complexes (Ho et al. 2009a,b) and are found associated with these complexes in pluripotent ES cells (Boyer et al. 2005; Ho et al. 2009a,b; Liang et al. 2008; Zhou et al. 2007) (Figure 1). As discussed below, biochemical and functional interactions between the core pluripotency network and chromatin-remodeling enzymes may promote a permissive chromatin structure that is essential to preserve genomic plasticity and pluripotency (Loh et al. 2007).
Table 1.
Function of selected epigenetic regulators in mouse pluripotent ES cells
| Gene | Gene product | Mouse mutant | Function in ES cells | Phenotype of mouse germline mutation | Reference (s) |
|---|---|---|---|---|---|
| Brg1 | SWI/SNF subunit, ATPase | null and KD | Required for ES cell SR and pluripotency. Required for survival of the ICM and trophectoderm. KO ES cells cannot be derived from blastocysts* | KO embryos die during the pre-implantation stage | Bultman et al. (2000, 2006), Ho et al. (2009a,b), Kidder et al. (2009) |
| BAF250a/Arid1a | SWI/SNF subunit | null | Required for ES cell pluripotency, SR and differentiation. KO ES cells are impaired in their ability to differentiate into functional mesoderm-derived cardiomyocytes and adipocytes but are capable of differentiating into ectoderm-derived neurons. KO ES cells are prone to differentiate into primitive endoderm-like cells under normal feeder-free culture conditions | KO embryos arrest development at E6.5; they form the ICM but do not gastrulate or form mesoderm | Gao et al. (2008) |
| BAF250b/Arid1b | SWI/SNF subunit | null | Required for ES cell SR and proliferation. KO ES cells show a mild reduction in proliferation and more rapid differentiation | N/A; biallelic inactivation in ES cells | Yan et al. (2008) |
| BAF155/Srg3 | SWI/SNF subunit | null | Required for ICM outgrowth. KO ES cells cannot be derived from blastocysts* | KO embryos develop in the early implantation stage but undergo rapid degeneration thereafter | Kim et al. (2001) |
| BAF47/Snf5/ini1 | SWI/SNF subunit | null | Required for ICM outgrowth and formation of trophectoderm. KO ES cells cannot be derived from blastocysts* | KO embryos die between E3.5 and E5.5 at the periimplantation stage | Klochendler-Yeivin et al. (2000), Guidi et al. (2001) |
| Snf2h | ISWI subunit, ATPase | null | Required for survival and growth of trophectoderm and ICM | KO embryos die during the periimplantation stage | Stopka & Skoultchi (2003) |
| Bptf | ISWI subunit | null | Required for ES cell differentiation. KO ES cells are deficient in their ability to form the mesodermal, endodermal, and ectodermal lineages | KO embryos manifest growth defects at the postimplantation stage and are reabsorbed by E8.5 | Landry et al. (2008) |
| Mbd3 | NuRD subunit | null | Required for ES cell pluripotency. KO ES cells can be maintained in the absence of leukemia inhibitory factor (LIF) and initiate differentiation in embryoid bodies or chimeric embryos, but fail to commit to specific lineages. ICM of KO blastocysts fails to develop into mature epiblast after implantation | KO embryos die at around the time of implantation | Kaji et al. (2006, 2007) |
| Ring1b/Rnf2 | Polycomb group, PRC1, H2A E3 monoubiquitin ligase | null | Required to stably maintain undifferentiated state of mouse ES cells | KO embryos show gastrulation arrest | Voncken et al. (2003), van der Stoop et al. (2008), Roman-Trufero et al. (2009) |
| Ezh2/Enx1 | Polycomb group, PRC2, H3K27 HMTase | null | KO ES cells can be derived from blastocysts as well as self-renew | KO embryos stop developing after implantation or fail to complete gastrulation and die at around E8.5 | Shen et al. (2008) |
| Eed | Polycomb group, PRC2 | null | Eed null ES cells are pluripotent, even though they have a tendency to differentiate spontaneously in culture and display midly defective differentiation. Eed null chimeras have a paucity of mesoderm | KO embryos die at around E8.5 with all germ layers formed but defects in mesoderm formation | Faust et al. (1995), Montgomery et al. (2005) |
| Suz12 | Polycomb group, PRC2 | null | Required for ES cell differentiation in culture. KO ES cells cannot form neurons after in vitro differentiation and KO EBs fail to form a proper endodermal layer | KO embryos die during early postimplantation stages | Pasini et al. (2004, 2007) |
| Yy1 | PRC2/3 interaction | null | KO ES cells cannot be derived from blastocysts* | KO embryos die at around the time of implantation | Donohoe et al. (1999) |
| Jarid2/jumonji | Histone demethylase of jumonji family, PRC2 subunit | null | Required for ES cell differentiation. Modulates the balance between SR and differentiation. Lineage commitments are delayed in KO ES cells | KO embryos die before E15.5, required for neural tube formation | Takeuchi et al. (1995, 1999), Shen et al. (2009), Pasini et al. (2010) |
| Mll2/Wbp7 | H3K4 HMTase | null | Required for ES cell proliferation, proper differentiation and survival but dispensable for SR and pluripotency | KO embryos fail to develop beyond around E9.5 | Glaser et al. (2006), Lubitz et al. (2007) |
| G9a/Ehmt2 | H3K9 HMTase | null | KO ES cells exhibit growth defects upon induction of differentiation with all-trans retinoic acid (RA) | KO embryos die at around E8.5–E9.5 | Tachibana et al. (2002, 2005) |
| Glp/Ehmt1 | H3K9 HMTase | null | N/A | KO embryos die at around E9.5 | Tachibana et al. (2005) |
| Eset/Setdb1 | H3K9 HMTase | null | Required for ICM outgrowth. KO ES cells cannot be derived from blastocysts* | KO embryos die at around E3.5–E5.5 | Dodge et al. (2004), Bilodeau et al. (2009) |
| Dnmt1 | Dnmt (maintenance) | null | Required for ES cell differentiation. KO ES cells proliferate normally but die upon induction of differentiation and cannot form teratomas | Development of KO embryos is arrested prior to the eight-somite stage | Lei et al. (1996), Tucker et al. (1996), Gaudet et al. (1998) |
| Dnmt3a/3b | Dnmt (de novo) | null | Required for ES cell differentiation. Late-passage KO ES cells cannot form teratomas | Dnmt3a KO mice become runted and die at around 4 weeks of age; Dnmt3b KO mice die after E9.5; dKO mice die before E11.5 | Okano et al. (1999), Chen et al. (2003) |
| Dnmt1/3a/3b | Dnmt | null | Modest effect on ES cell proliferation. Triple KO ES cells grow robustly (although slightly slower than WT) and maintain their undifferentiated characteristics | N/A; triple-KO ES cells were studied | Tsumura et al. (2006) |
| p300 | HAT and coactivator | null | Required for ES cell differentiation but dispensable for SR | KO embryos die at or before E11.5 | Yao et al. (1998), Zhong & Jin (2009) |
| Thap11/Ronin | Thap and ZF-domain epigenetic regulator | null and OE | Promotes ES cell SR/proliferation, essential for pluripotency. Required for ICM outgrowth. KO ES cells cannot be derived from blastocysts.* OE inhibits ES cell differentiation | KO embryos die at periimplantation | Dejosez et al. (2008) |
Abbreviations: dKO, double knockout; Dnmt, DNA methyltransferase; EB, embryoid body; ES, embryonic stem; HAT, histone acetyltransferase; ICM, inner cell mass; KD, knockdown; KO, knockout; N/A, not available; OE, overexpression; SR, self-renewal; WT, wild type; ZF, zinc finger.
Deletion of these genes causes a failure of the ICM to give rise to ES cells in vitro, suggesting a direct role in the establishment or maintenance of pluripotency.
Figure 1.
A functionally and structurally specialized SWI/SNF-like complex, esBAF, cobinds across the genome with the factors of the pluripotency transcriptional circuit as well as those that initiate and maintain pluripotency. esBAF complexes are distinguished by containing a homodimer of BAF155 but not 170; Brg but not Brm; BAF45a and d, but not b and c; and BAF53a but not BAF53b. Proteomic studies of endogenous complexes have demonstrated biochemical interactions with Sox2, Oct4, and many of the proteins involved in induced pluripotent stem (IPS) cell formation or embryonic stem (ES) cell maintenance. Of particular note was the absence of binding to general transcription factors or proteins such as Sp-1 or Fos that are present at high levels in ES cells, indicating that the interactions of esBAF are functionally dedicated to pluripotency. In addition, esBAF complexes occupy the promoters of nearly all genes of the core pluripotency network, such as Oct4, Sox2, c-myc, KLF4, Sall4, TCF3, and Nanog. esBAF complexes also co-occupy target genes of Oct4, Sox2, and Nanog, suggesting a functional interaction between esBAF complexes and the core pluripotency circuitry. Recently, components of esBAF were shown also to facilitate pluripotency (Singhal et al. 2010). The subunits are shown as interlocking pieces to indicate that they must be partially denatured (2 M urea) to dissociate from the complex. The positions are not necessarily accurate.
EPIGENETIC MECHANISMS TO MAINTAIN PLURIPOTENCY
Chromatin-Remodeling Complexes and Pluripotency
Differentiation of ES cells or the cells of the ICM from pluripotent to developmentally more restricted states is accompanied by global epigenetic changes at the level of the chromatin structure and concomitant changes in gene expression. Stem-cell-specific genes are gradually silenced as differentiation occurs, whereas subsets of lineage-specific genes are turned on. This developmental transition occurs, at least in part, through chromatin regulatory mechanisms, which include covalent histone modification, DNA methylation of CpG dinucleotides, and ATP-dependent chromatin remodeling.
ATP-dependent chromatin-remodeling complexes and pluripotency
Perhaps the genetically most well-documented chromatin regulators of the pluripotent state are the ATP-dependent chromatin-remodeling enzymes. In mammalian cells, approximately 30 genes encode ATP-dependent chromatin regulators that can be roughly grouped into families based on the structural features of the ATPase domain. These include Brg, Brahma/Brm, SNF2H, SNF2L, CHD1, and Mi2-beta, all of which play genetically non-redundant roles. These characterized ATPases are assembled into complexes such as BAF (also called mSWI/SNF), NuRD, ISWI, CDH1, and Tip60 and interact with several other subunits, indicating that perhaps several hundred genes are involved in ATP-dependent chromatin regulation.
In mammalian cells, the Brm (Brahma) and Brg ATPases are assembled with 12 other subunits into BAF or mSWI/SNF complexes that share certain homologs with yeast SWI/SNF complexes, but have lost, gained, and shuffled subunits with other classes of ATPases. Highlighting fundamental mechanistic differences in the control of gene expression, mammalian BAF complexes often repress transcription from a distance, whereas the yeast SWI/SNF complex regulates all known targets by activation from promoters. Unlike the homologous complexes in yeast, flies, and worms, most subunits of mammalian BAF complexes are encoded by gene families and the complexes are combinatorially assembled (Ho et al. 2009b; Lemon et al. 2001; Lessard et al. 2007; Takeuchi & Bruneau 2009; Wang et al. 1996a,b; Wu et al. 2007, 2009). In certain cases (see below), complex composition confers functional specificity to these complexes.
Genetic studies in mice have demonstrated that BAF complexes are essential for early embryonic development and pluripotency. In mice, inactivation of most BAF subunits including the ATPase Brg as well as the BAF47, BAF57, BAF60, BAF155, BAF180, and BAF250a subunits results in early embryonic lethality, and in the case of Brg, BAF47, and BAF155, a failure of formation of pluripotent cells (Bultman et al. 2006, Doan et al. 2004, Gao et al. 2008, Guidi et al. 2001, Kim et al. 2001, Klochendler-Yeivin et al. 2000, Lickert et al. 2004, Roberts et al. 2000). Conversely, mice with deletion of the alternative ATPase Brm are viable and approximately 15% larger than controls (Reyes et al. 1998). Maternally derived Brg is required for zygotic genome activation, a nuclear reprogramming event that establishes totipotency in the cleavage-stage embryo and is required for embryonic development (Bultman et al. 2000). Consistent with this, nuclear reprogramming of permeabilized somatic human cells using extracts from Xenopus laevis eggs and early embryos requires Brg, demonstrating the importance of these complexes in the establishment of pluripotency (Hansis et al. 2004). Brg, BAF155, and other components of the complex were also identified in a large-scale RNAi screen targeted against chromatin regulatory factors as being required for the maintenance of ES cell colony morphology (Fazzio et al. 2008) and in a screen for genes required for Nanog expression (Schaniel et al. 2009). Interestingly, in these screens, components not characteristic of esBAF were not detected. Recently, components of esBAF were found to facilitate pluripotency (Singhal et al. 2010).
BAF or mSWI/SNF complexes have been considered to be general regulators of transcription, suggesting that the essential roles of this complex could simply reflect a general role in transcription. However, several observations argue strongly against a general role, but rather for a specific and programmatic role. First, recent proteomics studies by Ho et al. (2009b) revealed that pluripotent ES cells express distinctive complexes (termed esBAF) defined by the presence of Brg, BAF155, and BAF60a and the absence of Brm, BAF170, and BAF60c subunits (Figure 1). These studies indicated that the ATPase Brg is essential for the self-renewal ability of pluripotent ES cells. shRNA-mediated depletion of Brg in ES cells generated small colonies with flattened morphology indicative of spontaneous differentiation. These studies also showed that ES cells require a specific esBAF composition with respect to BAF155 and BAF170 subunits. BAF155 depletion in ES cells diminished ES cell proliferation and increased cell death, whereas enforced expression of BAF170 decreased ES cell competitive self-renewal ability and teratoma formation in immunocompromised mice (Ho et al. 2009b). Similarly, combinatorial assembly of subunits of the BAF250 family regulates esBAF function. BAF250a and BAF250b subunits are both required to maintain ES cell pluripotency and self-renewal, but they differentially regulate the potential of ES cells to develop into specific lineages (Gao et al. 2008, Yan et al. 2008). BAF250a and b are alternative subunits and esBAF complexes contain either one or the other, which imply that these subtypes of complexes are dedicated to different, non-redundant pluripotency programs. Mouse embryos lacking BAF250a (ARID1a) form the ICM but do not gastrulate or form mesoderm. ES cells deficient for BAF250a are capable of differentiating into primitive endoderm- and ectoderm-like cells but cannot generate mesoderm-derived cardiomyocytes (Gao et al. 2008). Conversely, disruption of BAF250b in ES cells results in downregulation of pluripotency genes, reduced proliferation, and increased expression of lineage-specific genes, including markers of mesodermal differentiation. Interestingly, deletion of components of the related PBAF complex, defined by the signature subunit BAF180 or polybromo, leads not to a reduction in pluripotency, but instead to specific late developmental effects (see below). Confirming the importance of the specific subunit composition of esBAF complexes, only esBAF subunits have been detected in RNAi screens for pluripotency of ES cells (Fazzio et al. 2008, Schaniel et al. 2009).
An important question regarding the role of esBAF complexes is whether their function is simply to act in a general way, promoting the transcription of whatever genes are active in a given cell type, or whether they function in a programmatic way as an essential component of the core pluripotency circuit. Genome-wide studies of direct targets also strongly support a programmatic and unexpected function. High-resolution genome-wide analysis of Brg-containing esBAF occupancy in ES cells revealed that these complexes bind approximately 3% of the murine genome with an average footprint of approximately 2.1 kb. Transcriptional start sites show a clear peak; however, most peaks are not at the transcriptional start site and many enhancers and silencers are also sites of Brg binding (Ho et al. 2009a). Although repression at a distance had been previously demonstrated for the CD4 gene in T cells (Figure 2), this finding was a surprise because the yeast SWI/SNF complex activates all its genomic targets by binding to promoters. This reinforces the apparent mechanistic difference between SWI/SNF and BAF complexes and suggests caution when generalizing between the two complexes. Biochemical and genetic studies indicated that Brg-containing esBAF complexes directly interact with Oct4 and Sox2 and are required for ES self-renewal and pluripotency (Ho et al. 2009a,b). esBAF complexes occupy the enhancers and promoters of nearly all genes of the core pluripotency network, such as Oct4, Sox2, c-myc, KLF4, Sall4, TCF3, and Nanog. In addition, esBAF complexes co-occupy target genes of Oct4, Sox2, and Nanog, suggesting a functional interaction between esBAF complexes and the core pluripotency circuitry (Figure 1). Microarray analysis of the genes acutely affected by conditional deletion of Brg in ES cells revealed that Brg-containing esBAF complexes function mainly as transcriptional repressors in pluripotent ES cells. Consistent with a role for these complexes in maintaining the expression of stem-cell-specific genes within the correct range for ES cell function, Brg represses a significant number of differentiation-specific genes as well as many targets of the core pluripotency network in these cells (Ho et al. 2009a,b). Altogether, these studies suggest that esBAF functionally interacts with Sox2 and Oct4 to refine the expression of pluripotency genes, while repressing the transcription of differentiation-specific genes. This suggests a revision of the conventional view that Trithorax genes maintain the expression of developmental genes, whereas Polycomb group (PcG) genes repress them, and it implies that in the case of stem cells these regulatory circuitries may be more complex.
Figure 2.
BAF complexes commonly repress their targets at a distance (indicated here for the CD4 gene). In developing T lymphocytes, BAF complexes bind to the CD4 silencer and repress transcription of the CD4 gene at a distance. Deletion of Brg or the silencer itself by homologous recombination results in similar phenotypes with derepression of the CD4 gene in common lymphoid progenitors. This mode of function is probably the norm for BAF complexes as shown from genome-wide studies of embryonic stem cells. Highlighting fundamental mechanistic differences in the control of gene expression, mammalian BAF complexes primarily repress transcription from a distance, whereas the yeast SWI/SNF complex regulates all known targets by activation from promoters.
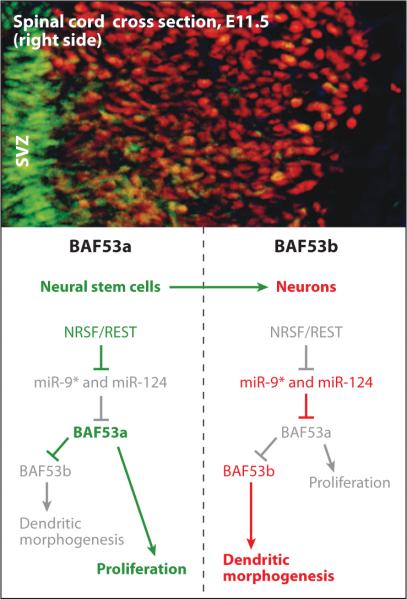
Combinatorial assembly of ATP-dependent BAF chromatin-remodeling complexes also orchestrates the development of the nervous system. A switch in subunit composition of neural, SWI/SNF-like BAF chromatin-remodeling complexes underlies the transition from proliferating neural stem/progenitors to postmitotic differentiated neurons (Lessard et al. 2007). Most compellingly, the self-renewal and proliferative activities of neural stem/progenitor cells require a specialized npBAF complex containing the double–plant-homeodomain (PHD) domain BAF45a/d subunit and the actin-related protein BAF53a assembled on the Brg/Brm ATPases. The dynamic exchange of these progenitor-specific subunits for the homologous BAF45b, BAF45c, and BAF53b subunits in postmitotic neurons orchestrates cell-cycle withdrawal and the acquisition of neuronal properties. The subunits of the npBAF complex are essential for neural-progenitor proliferation, and mice with reduced dosage for the genes encoding its subunits have defects in neural-tube closure similar to those in human spina bifida. BAF45a expression appears sufficient for inducing proliferation of neural progenitors, implying an instructive role of npBAF complexes. In contrast, the BAF45b/BAF53b-containing neuron-specific BAF (nBAF) complex is essential for postmitotic neuronal function, including activity-dependent dendritic outgrowth, via its association with the Ca2+-responsive dendritic regulator CREST (Wu et al. 2007). Remarkably, these studies indicated that the highly homologous BAF53a protein, which is a component of neural-progenitor and non-neural BAF complexes, cannot replace BAF53b's role in dendritic development and that this functional specificity of BAF53b is conferred by its actin fold subdomain 2. More recent studies have found that microRNA-mediated regulation of specific subunits of BAF chromatin-remodeling complexes is essential for mitotic exit and the onset of dendritic morphogenesis in the vertebrate nervous system (Yoo et al. 2009) (Figure 3). In postmitotic neurons, BAF53a repression is mediated by sequences in the 3′ untranslated region corresponding to the recognition sites for microRNAs miR-9* and miR-124, which are selectively expressed in these cells. Mutation of these sites leads to persistent expression of BAF53a and defective activity-dependent dendritic outgrowth in neurons, whereas overexpression of miR-9* and miR-124 in neural stem/progenitor cells impaired cellular proliferation. Altogether, these studies indicate that functional specificity to ATP-dependent chromatin-remodeling complexes is achieved, at least in part, by miRNA-mediated switching of specific subunits, allowing differential interaction with specific factors that promote cell-lineage commitment and terminal differentiation.
Figure 3.
Genetic/epigenetic circuitry controlling mitotic exit of neural stem cells. (left) In neural stem cells in the subventricular zone (SVZ), NRSF/REST represses the microRNAs miR-9* and miR-124, allowing constitutive expression of BAF53a (green) and proliferation. npBAF complexes containing BAF53a repress BAF53b, preventing dendritic morphogenesis. Inactive paths are gray. (right) In postmitotic neurons, REST is repressed, leading to expression of miR-9* and miR-124, repression of BAF53a, and derepression of BAF53b (red). BAF53b is necessary for dendritic development in both mice and Drosophila. Photograph by Brett Staahl.
Finally, deletion of Brg, BAF180, and BAF60c subunits in the mouse has been associated with distinct cardiac developmental outcomes. Mice lacking BAF180 or polybromo have specific defects in formation of the ventricular chambers of the heart that are consistent with a role for this subunit in response to retinoic acid. Interestingly, earlier retinoic acid–dependent processes do not seem to be affected (Wang et al. 2004). Conditional mutation of Brg in the heart indicated that Brg maintains cardiomyocytes in an embryonic state (promotes their proliferation and preserves differentiation) by interacting with histone deacetylases (HDACs) and poly (ADP ribose) polymerase (PARP) and controlling developmental gene expression. In adult cardiomyocytes, Brg is turned off but can be reactivated by cardiac stress to induce a pathological program of gene expression by interacting with HDAC and PARP (Hang et al. 2010). Similarly, RNAi interference of BAF60c in the early mouse embryo revealed a specific requirement in skeletal and cardiac development (Lickert et al. 2004). More recent studies have shown that BAF60c is critical to establish the regions of the embryo that give rise to the heart, a function quite different from that of BAF180 in cardiac development. Remarkably, BAF60c appears to have an instructive role in heart development, because its injection into noncardiogenic regions of the embryo can result in the generation of beating cardiomyocytes (Takeuchi & Bruneau 2009). These studies suggest the existence of a specialized cBAF complex. However, purification of this putative cardiogenic complex has not yet been reported.
NuRD complexes
Mammalian nucleosome remodeling deacetylase (NuRD) complexes contain at least six subunits that are encoded by gene families (Bowen et al. 2004). These complexes possess both ATP-dependent chromatin-remodeling and HDAC activities (Denslow & Wade 2007). The activity of Hdac1 and Hdac2 within the complexes requires the presence of the chromodomain-containing Mi2a and Mi2b, which are SNF2/SWI2-like ATPase subunits. Other subunits of these complexes include the methyl-CpG-binding proteins Mbd 1/2/3, the metastasis-associated Mta1/2/3 proteins, the WD40-containing RbAP46 and RbAP48 proteins, and two zinc fingers proteins, p66a and p66b. Mi2b-containing NuRD complexes, which possess both transcriptional repressive and activating functions, are required for hematopoietic stem cell self-renewal and multilineage differentiation (Wade et al. 1999, Williams et al. 2004, Yoshida et al. 2008). Several subunits of these complexes are also important for ES cell pluripotency and differentiation. ES cells lacking Mbd3 are viable but fail to form a stable NuRD complex and display a profound defect in differentiation that results in persistent self-renewal. Mbd3-deficient ES cells can be maintained in the absence of leukemia inhibitory factor and can initiate differentiation in embryoid bodies or chimeric embryos, but they fail to commit to developmental lineages, except when induced with retinoic acid (Kaji et al. 2006). Recent studies indicated that Mbd3 is required for the ICM of blastocysts to develop into mature epiblast after implantation. Expression of the pluripotency factors Oct4, Nanog, or Sox2 and their targets did not seem to be affected in the absence of MBD3 (methyl-binding domain 3), but transcription of genes that are normally expressed at the preimplantation stage and then silenced failed to be repressed. Unlike Mbd3-null ES cells, Mbd3-deficient ICMs grown ex vivo fail to expand Oct4-positive pluripotent cells despite producing robust endoderm outgrowth (Kaji et al. 2007). Together, these findings define a role for MBD3 in cell-fate commitment of pluripotent ES cells and epiblast formation after implantation.
Interestingly, a subfamily of NuRD complexes (termed NODE for Nanog and Oct4 associated deacetylase) containing Hdac1/2- and Mta1/2- and near absence (or substoichiometric levels) of Mbd3 and Rbbp7 interacts with the pluripotency factors Nanog and Oct4 (Liang et al. 2008). NODE HDAC activity seems to be comparable to NuRD, and NODE is recruited to Nanog/Oct4 target genes independently of Mbd3 in ES cells. In contrast to Mbd3 loss-of-function, knockdown of NODE subunits in ES cells increased expression of developmentally regulated genes and promoted differentiation. shRNA-mediated depletion of Mta1 also has different effects than MBD3 depletion on target genes. In contrast to Mbd3, which is required to repress preimplantation genes, Mta1 is required to repress lineage-specific factors, such as Gata6 and Foxa2. Thus, a subfamily of NuRD complexes containing Hdac1/2- and Mta1/2 is essential to maintain pluripotency by interacting with components of the core pluripotency circuitry. The question remains whether different NuRD-related complexes possess distinct enzymatic activities and play generic or specialized roles in the regulation of stem cell self-renewal, proliferation, and differentiation.
ISWI complexes
The ISWI family of chromatin remodelers contains two to four subunits based on the alternative ATPases SNF2L and SNF2H, the mammalian homologs of the Drosophila ISWI ATPase (Eberharter & Becker 2004). ISWI subunits differ in their expression pattern and assemble into at least seven distinct complexes. SNF2L is a component of the NURF complex, together with BPTF and RbpAp46/48. The PHD-domain-containing BPTF subunit appears to mediate the selective recruitment of ISWI complexes to target genes with transcriptionally active histone marks such as H3K4me3 (Wysocka et al. 2006), but genetic studies on mice lacking the BPTF PHD domain will be essential to confirm this result. BPTF null embryos have growth defects leading to their death by E8.5 (Goller et al. 2008), and BPTF deletion in ES cells impairs their ability to form the mesodermal, endodermal, and ectodermal lineages (Landry et al. 2008).
The chromatin-remodeling activity of at least six subfamilies of ISWI complexes, namely hACF, hCHRAC, hWICH, RSF, NoRC, and SNF2H/cohesin, is regulated by the presence of the alternative ATPase subunit SNF2H (Eberharter & Becker 2004). Snf2h−/− embryos die during the periimplantation stage, and Snf2h is required for the survival and proliferation of both the trophectoderm and ICM (Stopka & Skoultchi 2003). As genetic analyses indicate that ISWI complexes play important roles in diverse biological processes (such as transcriptional regulation, heterochromatin replication, chromatin assembly, and the formation of higher-order chromatin structure), it will be interesting to investigate whether combinatorial assembly of ISWI subunits assembled on SNF2H and SNF2L generates a family of heterogeneous complexes with distinct and specialized functions in embryonic and adult stem cells (Bozhenok et al. 2002, Eberharter et al. 2001, Hamiche et al. 1999, Ito et al. 1999, Langst et al. 1999, Poot et al. 2004, Strohner et al. 2001).
Tip60-p400 complexes
The Tip60-p400 family of complexes [whose subunits, on the basis of tagging overexpressed proteins, appear to be composed of Ruvbl1, Ruvbl2, Dmap1, Ep400 (p400), Htatip (tip60), Trrap, Tip49 (TAP54α), Tip48 (TAP54β), BAF53a, β-actin, E(Pc), and MRGBP] possesses both histone acetyltransferase and chromatin-remodeling activities and can act either as positive or negative regulators of transcription (Ikura et al. 2000, Cai et al. 2003). Tip60-p400 transcriptional activity seems to be mediated, at least in part, by the incorporation of the histone variant H2AZ into nucleosomes and by the catalysis of histone acetylation at target genes (Sapountzi et al. 2006, Squatrito et al. 2006). Embryos lacking Tip60 and Trrap, two components of the Tip60-p400 complexes, also die before implantation (Gorrini et al. 2007, Herceg et al. 2001), suggesting a role in early development. Interestingly, Tip60-p400 was recently identified in a large-scale RNAi screen for chromatin-remodeling proteins involved in ES cell function (Fazzio et al. 2008). Depletion of several subunits of Tip60-p400 complexes inhibited the self-renewal ability of ES cells, impaired their ability to differentiate, and/or generated ES cell colonies with altered morphology without affecting the expression of the pluripotency transcription factors. Chromatin immunoprecipitation experiments indicated that Tip60-p400 colocalizes with the pluripotency factor Nanog and the transcriptionally active histone mark H3K4me3 in ES cells. Interestingly, the authors observed a significant overlap between Tip60-p400 target genes and that of Nanog and further demonstrated that both Nanog and H3K4me3 are required for Tip600-p400 binding at target promoters in ES cells, whereas binding of Tip60-p400 is required to mediate histone H4 acetylation at both activated and repressed target genes in ES cells.
CHD1 complexes
Although there is a strong correlation between open chromatin and the undifferentiated state of stem cells, it has long been debated whether open chromatin is necessary for stem cell potential. In support of this idea, RNAi knockdown of the chromatin remodeler Chd1 reduced chromatin decondensation and pluripotency of ES cells (Gaspar-Maia et al. 2009). Chd1 contains an ATPase SNF2-like helicase domain and belongs to the chromodomain family of proteins (Woodage et al. 1997). The two chromodomains in Chd1 are essential for recognition of H3K4me2/3 (Sims et al. 2005) and Chd1 is involved in transcriptional activation in several organisms (Simic et al. 2003, Sims et al. 2007, Stokes et al. 1996). Chromatin immunoprecipitation studies in mouse ES cells indicated that the Chd1 promoter is bound by several pluripotency-associated factors such as Oct4, Nanog, Sox2, and Zfx (Chen et al. 2008), highlighting a potential mechanism by which CHD1 complexes function downstream of the pluripotency factors to maintain open chromatin of mouse ES cells and regulate their pluripotency.
Polycomb group genes regulate pluripotency by suppressing developmental as well as metabolic pathways
PcG proteins are an evolutionarily conserved family of chromatin regulators known best for their role in establishing and maintaining the silent state of homeotic gene expression during embryonic development (Ringrose & Paro 2004). Mammalian PcG proteins assemble into at least three biochemically distinct complexes: PRC1, PRC2, and PhoRC. The four core subunits (PHC, CBX, Bmi1, and RING1) of mammalian PRC1 complexes are homologs of Drosophila Ph, Pc, Psc, and dRing, respectively. Mammalian PRC2 complexes contain EED, SUZ12, and either EZH1 or EZH2. The SET-domain-containing proteins EZH2 and potentially EZH1 of PRC2 are required for the initiation of silencing through the di- and tri-methylation of the K27 residue of histone H3. This modification forms the recruiting mark for the PRC1 complex, which is implicated in the maintenance of gene repression through the formation of higher-order chromatin structures (Valk-Lingbeek et al. 2004). This process appears to involve Ring1b-mediated monoubiquitination of H2AK119, an activity that is stimulated by the Bmi1 and Mel18 PRC1 subunits (Elderkin et al. 2007). Although this simple relationship between the two biochemical activities of PRC2 and PRC1 is appealing, genetic evidence in mammals indicates that this sequential action is not used broadly (see below).
A role for PcG proteins in maintaining ES cell identity and pluripotency was first suggested on the basis that most PcG components are required for early embryonic development (mainly PRC2 subunits, see below) (Pasini et al. 2004, Shumacher et al. 1996, Voncken et al. 2003), the self-renewal/maintenance of different types of adult stem cells (Molofsky et al. 2003, Park et al. 2003), and the formation of the bivalent chromatin state of stem cells (Bernstein et al. 2006). EED is required for PRC2 activity and early embryonic development in mice (Faust et al. 1995, Shumacher et al. 1996). Eednull embryos, which lack all detectable H3K27 methylation, display disrupted A/P patterning of the primitive streak during gastrulation and contain excess extraembryonic mesoderm but reduced embryonic mesoderm. Despite the absence of the repressive H3K27me3 mark, Eednull ES cells can be derived from blastocysts, and chimeric embryo analyses indicated that they are pluripotent, even though they have a tendency to express differentiation-promoting genes (and differentiate spontaneously) in culture (Boyer et al. 2006, Chamberlain et al. 2008). Primordial germ cells are specified in Eednull embryos, suggesting that they can contribute to the germline (Faust et al. 1995). However, high-contribution Eednull chimeras have a paucity of mesoderm, suggesting that Eed is required for the specification of embryonic mesoderm (Faust et al. 1995) and/or for the differentiation or maintenance of multipotent progenitors (Chamberlain et al. 2008). Similarly, Suz12 is essential for PRC2 activity and its inactivation results in early lethality of mouse embryos (Pasini et al. 2004). ES cells and the ICM form in the absence of Suz12, and embryos lacking Suz12 produce all three germ layers. Suz12−/− ES cells are also characterized by global loss of H3K27 tri-methylation (H3K27me3) and higher expression levels of differentiation-specific genes. However, in contrast to Eed, Suz12 is apparently required for differentiation of ES cells in culture, as Suz12−/− ES cells cannot form neurons after in vitro differentiation, and Suz12−/− Embryoid bodies fail to form a proper endodermal layer (Pasini et al. 2007). A molecular explanation for this apparent paradox is not clear, but it may be related to a role of Suz12 in other complexes. Despite the crucial role of EZH2 in the di- and tri-methylation of H3K27 in ES cells, a recent study by Orkin and colleagues showed that EZH2-deficient ES cells can be derived from blastocysts as well as self-renew (Shen et al. 2008). Surprisingly, known PcG targets (derepressed in EED-deficient ES cells) remained unaffected in EZH2-deficient ES cells and still contained the H3K27me3 repressive mark. This work also revealed that EZH1 exhibits histone methyltransferase activity in vitro and colocalizes with EED at PcG targets. Depletion of EZH1 in EZH2−/− ES cells was sufficient to remove the repressive H3K27me3 mark from these important developmental targets, demonstrating functional complementation between these two PRC2 subunits. The PRC2-associated PCL2 (Polycomb-like 2) protein was identified in a genome-wide screen for regulators of ES cell self-renewal and pluripotency. Knockdown of Pcl2 in mouse ES cells resulted in enhanced self-renewal, differentiation defects, and altered patterns of histone methylation (Walker et al. 2010). Although these studies suggest that PcG proteins may be dispensable for the establishment of pluripotency in ES cells, they suggest that at least some components of PRC2 complexes are required for the maintenance of pluripotency in its strictest meaning (i.e., potential of ES cells to generate all differentiated cell types in a cell-autonomous fashion as well as chimeras with germline potential). At present, it is still not clear why PRC2 mutant embryos die, but it may relate to a failure to assemble mesodermally derived tissues such as blood vessels or withdrawal of essential cytokines and growth factors.
How might PcG genes be involved in regulating aspects of ES cell identity? Genome-wide studies indicated that PcG targets are preferentially activated upon ES cell differentiation, suggesting that they regulate pluripotency by repressing the premature expression of lineagespecific genes (Bernstein et al. 2006, Boyer et al. 2006, Buszczak & Spradling 2006, Lee et al. 2006) (Figure 1). Consistently, PRC1 and PRC2 targets in ES cells were enriched in genes involved in developmental patterning, signaling, morphogenesis, and organogenesis (Boyer et al. 2006, Lee et al. 2006). A significant subset of PcG target genes was co-occupied by Oct4, Sox2, and Nanog (Bernstein et al. 2006, Boyer et al. 2006, Lee et al. 2006), suggesting functional interaction between PcG proteins and the core pluripotency network (Figure 1). However, a much larger fraction of combined Oct4/Sox2/Nanog targets are co-occupied by Brg (Ho et al. 2009a). Finally, recent studies revealed that one of the founding members of the Jumonji C (JmjC) domain protein family, JARID2, forms a stable complex with PRC2 in pluripotent ES cells and promotes its recruitment to target genes while inhibiting its histone methyltransferase activity (Pasini et al. 2010, Peng et al. 2009, Shen et al. 2009). Jarid2-deficient mice form all germ layers and die with defects in the organization of the cardiovascular system at approximately E10.5. In other genetic backgrounds, the mice survive until birth and are fully formed, indicating that pluripotency in the early embryo is not significantly compromised. Surprisingly, Jarid2 is required for the differentiation of mouse ES cells, and activation of genes marked by H3K27me3 and lineage commitments are delayed in JARID2−/− ES cells. However, one group of investigators found the opposite result, i.e., that Jmjd1a or Jmjd2c depletion leads to enhanced ES cell differentiation (Loh et al. 2007). One interpretation is that the dynamic regulation of PRC2 activity by JARID2 fine-tunes the relative balance between self-renewal and differentiation decisions in pluripotent ES cells. Why these defects in pluripotency are not seen or are dramatically blunted in the embryo is not clear, but this may become apparent upon a focused analysis of the Jarid2 embryonic phenotype.
One curious feature of the phenotype of PRC2-deficient mice is that the embryos die significantly after gastrulation and slightly before or at the time that an organized vasculature becomes essential for viability (the vascular/oxygenation checkpoint). For example, VEGF-, VEGF receptor-, and calcineurindeficient mice die at about the same time with a similar appearance (Carmeliet et al. 1996, Graef et al. 2001, Fong et al. 1995, Shalaby et al. 1995). Because cells that simply fail to differentiate properly do not necessarily die, this suggests a fundamental defect in either the metabolism of PRC2-deficient cells or the initiation of a checkpoint-induced cell death. For these reasons, reanalysis of PRC2-deficient embryos may be quite informative and provide a framework for possible mechanisms underlying PRC2 action.
Whereas deletion of any of the PRC2 subunits in mice is embryonic lethal (embryos die with defects in gastrulation 7 to 9 days post-fertilization), mice with deletion of PRC1 subunits, with the exception of Ring1b, are viable, suggesting that the PRC1 complex may be redundant with another mechanism in early development (Faust et al. 1995, Pasini et al. 2007). In any case, these genetic observations indicate that it is unlikely that PRC2 functions only to set up later repression by PRC1 (Figure 4), because this sequential mechanism would lead to similar phenotypes for PRC1 and PRC2 complex family members. However, several PRC1 components are required for the self-renewal/maintenance of different types of multipotent adult stem cells. For example, Bmi1 is required for the maintenance of hematopoietic stem cells (Lessard & Sauvageau 2003, Park et al. 2003); leukemic hematopoietic stem cells (Lessard & Sauvageau 2003); and neural, mammary, lung, and intestinal stem cells (Dovey et al. 2008, Liu et al. 2006, Molofsky et al. 2003, Pietersen et al. 2008, Sangiorgi & Capecchi 2008). In addition to Bmi1, several other subunits of PRC1 (Mel18, Phc1/Rae28, Ring1b) and PRC2 (EZH2) complexes are required for hemopoietic stem cell function (Kajiume et al. 2004, Kamminga et al. 2006, Kim et al. 2004, Ohta et al. 2002). Even though the targets of Polycomb complexes are commonly thought to be developmental genes, a recent study demonstrated that Bmi1 mutant mice show defects in mitochondrial function resulting in the release of reactive oxygen species with subsequent DNA damage. Remarkably, the Bmi defect in many stem cell populations could be repressed with a second mutation in the DNA damage checkpoint gene, CHK2 (Liu et al. 2009), indicating that a substantial role of Bmi1 in stem cell populations is to control the generation of reactive oxygen species in mitochondria (Figure 4). If indeed PRC2 functions upstream of PRC1, then there should also be defective mitochondrial function in Suz12, Eed, and Ezh2 mutant mice, possibly explaining early embryonic death. Altogether, these findings support a model in which Polycomb repression could act not only in pluripotent stem cells to ensure proper lineage choice, but also in progenitor cells to guide their further developmental potential by ensuring proper regulation of subtype-specific genes (Figure 4).
Figure 4.
Potential roles of the Polycomb repressive complex 1 (PRC1) and PRC2 complexes in the maintenance of multipotent and pluripotent cells. A/P, anterior/posterior; HSC, hemopoietic stem cell; NSC, neural stem cell.
DNA Methylation and Pluripotency
DNA methylation is a covalent modification of cytosine at position C5 in CpG dinucleotides. In mammals, DNA methylation has been implicated in processes as diverse as tissue-specific gene expression, cell-fate determination, cellular differentiation, X chromosome inactivation, and imprinting (Farthing et al. 2008). In the genome of mammalian cells, nearly all DNA methylation occurs on CpG dinucleotides, more than 70%–80% of which are methylated predominantly in areas of repetitive sequences (Bird 2002). This epigenetic modification is catalyzed by several DNA methyltransferases (Dnmts). Dnmt3a and Dnmt3b are de novo methyltransferases responsible for remethylating the genome in postimplantation mouse embryos and primordial germ cells (Okano et al. 1999), whereas the maintenance of methylation relies on Dnmt1, which favors hemimethylated DNA and methylates the complementary strand (Bestor 2000). Dnmt3l lacks enzymatic activity but may act as a cofactor for the de novo Dnmts (Dnmt3a and Dnmt3b). Recent studies indicated that unmethylated H3K4 is specifically recognized by Dnmt3l (Ooi et al. 2007). Dnmt2 does not have methyltransferase activity and its function remains obscure (Okano et al. 1998). Recent studies suggest that, in addition to Dnmts, the epigenetic regulator Hells (Lsh, lymphoid-specific helicase) is directly involved in the control of de novo methylation of DNA (Zhu et al. 2006). Silencing of gene expression upon DNA methylation could occur through the recruitment of methyl-CpG binding proteins (such as MBD1, MBD2, MBD3, MBD4, MECP2, and Kaiso) or, alternatively, by blocking the binding of transcription factors to their cognate response elements. The maintenance DNA methylation enzyme (Dnmt1) can act as transcriptional repressor and associate with HDACs to silence gene expression (Robertson et al. 2000). Although DNA demethylase activity has been reported for MBD2 (Bhattacharya et al. 1999), whether DNA demethylation is a reversible process remains to be determined (see below).
Several studies suggest that DNA methylation may play a key role in cell-fate determination and pluripotency (Reik et al. 2001). Dnmt1 and Dnmt3b knockout mice die by E10.5, whereas Dnmt3a-deficient mice, which are born occasionally, suffer from serious malformations and die within weeks after birth (Li et al. 1992, Okano et al. 1999). Dnmt1-deficient ES cells are viable but undergo cell death upon induction of differentiation (Panning & Jaenisch 1996). Dnmt3a and Dnmt3b inactivation in ES cells results in progressive loss of DNA methylation patterns at both single-copy genes and repetitive sequences. In mouse ES cells, both of these enzymes directly interact (Li et al. 2007) and function synergistically to methylate the promoters of pluripotency genes such as Oct4 and Nanog. Hypomethylation of the Oct4 promoter region in ES cells allows cells to maintain high levels of Oct4 expression, thus keeping them in a pluripotent state, whereas hypermethylation of its promoter in differentiating cells correlates with its silencing. Together, these studies indicate that DNA methylation/demethylation may regulate the expression of master developmental regulators in ES cells. Interestingly, recent genome-wide studies revealed that DNA methylation at CpG-rich sequences is very low in stem cells and that methylation can occur at CpG island promoters and at CpG-rich sequences outside of promoter regions during lineage determination (Farthing et al. 2008, Fouse et al. 2008, Illingworth et al. 2008, Meissner et al. 2008, Mohn et al. 2008). Interestingly, many of the genes that are de novo methylated upon cellular differentiation are stem-cell- and germline-specific genes (Farthing et al. 2008, Mohn et al. 2008, Weber et al. 2007). These studies collectively suggest that DNA methylation is involved (either causally or as a result of) in shutting down the pluripotency program upon lineage specification and in preventing its aberrant reactivation under physiological conditions.
Recent studies have highlighted a critical role for DNA methylation in regulating adult stem cell function. For example, de novo Dnmts Dnmt3a and Dnmt3b are required to promote hemopoietic stem cell self-renewal (but not differentiation) (Tadokoro et al. 2007). Similarly, Dnmt1, MBD1, and MeCP2 are essential for fetal or adult neural stem cell function (Fan et al. 2005, Kishi & Macklis 2004, Zhao et al. 2003). However, how DNA methylation specifically contributes to pluripotency, commitment, and phenotypic maturation of specific differentiated cells is not well understood.
As mentioned above, DNA methylation has been generally considered to be irreversible, raising the following question: What removes the methylation during the induction of pluripotency? Recently, the work of Blau and colleagues has indicated that the cytosine deaminase AID (activation-induced cytidine deaminase) is required for active DNA methylation and nuclear reprogramming of somatic cell nuclei toward pluripotency (Bhutani et al. 2009). The mechanism proposed involves AID-mediated promoter demethylation and induction of OCT4 and NANOG gene expression. Base-excision repair mechanisms seem a risky way of removing methylation because mutations may result from the extensive removal of methyl marks at thousands of sites over the genome. If this is indeed the case, such mutations may reduce the therapeutic potential for induced pluripotency.
Chromatin Modifications: The Generation of Histone Marks
The diversity and complexity of histone modifications, which together act as "marks" that can signal transcriptional activation or repression, are being studied intensively. The core histones (H2A, H2B, H3, and H4) are subject to dozens of different modifications (including acetylation, methylation, phosphorylation, and ubiquitination) that can be epigenetically inherited. Lysine acetylation, the most studied modification, is generally associated with gene expression, whereas lysine methylation can lead to either gene activation or repression, depending on the residue involved. The level of methylation of a particular lysine residue (i.e., mono-, di-, and tri-methylation) influences the levels of gene expression or repression by recruiting different effector proteins. Each histone modification can induce or inhibit subsequent modification, and this cross-talk can operate both in cis, on the same histone, or in trans, between histones. As discussed below, histone modifications can impinge on transcription by promoting the binding of transcriptional regulators and by directly altering chromatin structure. Understanding of histone modifications is undergoing revision owing to the finding that these modifications are reversible by specific demethylases. In addition, results of genome-wide studies have demonstrated remarkable lability of acetylation marks (Wang et al. 2009).
At active promoters (H3K4me3 and H3/H4Ac)
Recent studies have highlighted the molecular mechanisms responsible for generating, removing, and recognizing the histone marks located at active promoters. H3/H4Ac, H3K4me3, or H4K4me2 marks are generally associated with accessible chromatin structures and gene activation (Santos-Rosa et al. 2002, Schubeler et al. 2004). These active marks are found in the promoters of nearly all transcribed genes, whereas H3K36me3 and H3K79me3 appear to be located along the actively transcribed regions (Edmunds et al. 2008). In mammals, the trimethylation of H3K4 is catalyzed by SET-domain-containing proteins of the Trithorax group, which are encoded by at least six genes in the mouse (MLL1–4, SET1a, and SET1b). The recent discovery of histone demethylases revealed that this modification is more dynamic than previously thought (Klose & Zhang 2007). Several histone demethylases belonging to the Jumonji domain-containing (Jmjd) protein family [such as lysine-specific demethylase 1 (LSD1), JHDM1A, JHDM2A, JHDM3/JMJD2] catalyze the demethylation of H3K4me2/3, H3K27me2/3, or H3k9me2/3 marks and play important roles in promoting ES cell self-renewal, pluripotency, and differentiation (Christensen et al. 2007; Cloos et al. 2008; Iwase et al. 2007; Klose & Zhang 2007; Loh et al. 2007; Pasini et al. 2008, 2010; Peng et al. 2009; Shen et al. 2009; Tsukada et al. 2006; Yamane et al. 2006, 2007). PRC2 and Rbp2 are both displaced from promoters that are activated during ES cell differentiation, resulting in removal of the H3K27me3 mark and deposition of the H3K4me3 mark (Pasini et al. 2008). The H3K4me3 mark seems to be specifically recognized by PHD-domain-containing proteins. For example, the BPTF subunit of NURF complexes is specifically recruited to H3K4me3 at the HOXC8 promoter leading to its activation (Wysocka et al. 2006). In ES cells, removal of the H3K4me3 mark by the RBP2 demethylase leads to the silencing of HOX gene expression (Christensen et al. 2007). The PHD-domain-containing TAF3 subunit of the general transcription factor TFIID also recognizes the H3K4me3 mark and may contribute to the assembly of the polymerase II initiation complex at active or poised promoters (Vermeulen et al. 2007). Notably, a role for this mark in protecting inactive CG-rich promoters from de novo DNA methylation by Dnmt3L has been proposed (Ooi et al. 2007, Weber et al. 2007).
Recent genome-wide studies in ES cells have indicated that the abundance of the H3K36me3 mark better correlates with levels of gene expression than does the H3K4me3 mark. In these studies, H3K4 tri-methylation in ES cells was found at more than 80% of the annotated promoters (Guenther et al. 2007). Similarly, RNA polymerase II was detected at more than 50% of the annotated promoters in ES cells, including many silent genes. The discrepancy between RNA pol II binding, H3K4me3 levels, and gene activation may be explained by the recent observation that short abortive transcripts are synthesized at these promoters (Guenther et al. 2007). Although the underlying mechanisms are still obscure, H3K27 methylation by PcG proteins may be responsible for blocking elongation at these promoters (Bernstein et al. 2006, Boyer et al. 2006, Lee et al. 2006). As the presence of a “poised polymerase” at silent promoters was also observed in B and T lymphocytes and Drosophila (Barski et al. 2007, Guenther et al. 2007), inhibition of gene elongation may represent a general mechanism to keep inactive genes “poised” for activation. In agreement with a role for histone methyltransferases (HMTases) in regulating adult stem cell populations, MLL1, MLL2, and MLL5 are required for some aspects of hemopoietic (MLL1 and MLL5) (Ernst et al. 2004, Heuser et al. 2009, Lim et al. 2009, McMahon et al. 2007), neural (MLL1) (Lim et al. 2009), and ES cell function (MLL2) (Lubitz et al. 2007).
The acetylation of histones H3 and H4, which is catalyzed by interplay between histone acetyltransferase (HAT) and HDAC enzymes (Lee & Workman 2007, Xu et al. 2007), is also associated with gene activation. Many active transcription factors either recruit HATs or utilize their own internal HAT domains (e.g., CREB binding protein) to catalyze H3 and H4 acetylation and lead to accessible chromatin structure and transcriptional activation. Bromodomain-containing proteins (such as Brg and the BAF180 subunit of BAF chromatin-remodeling complexes) are generally targeted to acetylated histone residues and may be involved in opening the chromatin structure at these sites. Interestingly, the HAT p300 is required for proper ES cell differentiation and Nanog expression (Zhong & Jin 2009), and a role for the Querkopf (Qkf) (Merson et al. 2006, Thomas et al. 2000), Moz, and CBP HATs in regulating neural and hemopoietic stem cell function has been reported (Katsumoto et al. 2006, Rebel et al. 2002, Thomas et al. 2006).
At silenced promoters (H3K27me3 and H3K9me3)
Methylated H3K9, H3K27, or H4K20 residues are mainly associated with transposons, repetitive sequences, and pericentromeres and usually link to gene repression (Mikkelsen et al. 2007). The enzymes responsible for making these repressive chromatin marks are currently being elucidated (Swigut & Wysocka 2007). The best studied of these marks, H3K9me3, is catalyzed by SUV39h (mouse Suv39H1, Suv39H2), SetDB (mouse ESET), and G9a. These HMTases are likely recruited to methylated DNA by MBD proteins. H3K9 methylation allows the recruitment of heterochromatin protein-1 (HP1) and the formation of higher-order chromatin structures (Agarwal et al. 2007, Fujita et al. 2003). Heterochromatin-mediated gene silencing is propagated through cell division by an interaction between HP1, HDACs, and Dnmts (Lachner & Jenuwein 2002). Several H3K9me3 demethylases have been discovered including LSD1, Jmjd1a, and Jmjd2c (Klose et al. 2006, Loh et al. 2007, Whetstine et al. 2006). Removal of the H3K9me3 marks at the promoter of the pluripotency factor Nanog by Jmjd2c is required to prevent HP1 and KAP1 repressor binding (Loh et al. 2007).
H3K27me3 is another repressive histone mark, which is catalyzed by the SET-domain-containing EZH2 subunit of the PRC2 (Barski et al. 2007, Mikkelsen et al. 2007). Subsequent recognition of this mark by the PRC1 at the silenced promoters ensures the formation of higher-order chromatin structures and its propagation through mitosis (Cao & Zhang 2004). In ES cells, several PRC2 subunits are essential for lineage specification, suggesting an important role for H3K27 tri-methylation (Lee et al. 2006). Jmjd proteins, notably UTX1, UTY1, and JMJD3, have been identified as H3K27 demethylases (Agger et al. 2007, De et al. 2007, Lan et al. 2007, Lee et al. 2007). Interplay between histone demethylases and methyltransferases in gene activation is suggested by the recent observation that UTX1 and MLL2 (an H3K4 HMT) biochemically interact (Agger et al. 2007, Issaeva et al. 2007, Lee et al. 2007). Interestingly, a role for the HMTases Carm1, Mll2/Wbp7, G9a/Ehmt2, Glp/Ehmt1, and Setdb1 (mouse Eset) has recently been demonstrated in pluripotent ES cells, and several of those HMTases are required for ICM outgrowth (Dodge et al. 2004; Lubitz et al. 2007; Tachibana et al. 2002, 2005; Wu et al. 2009) (see Table 1 and Supplemental Table 1).
Bivalent domains and pluripotency
The ES cell genome has a specific epigenetic profile characterized by a general abundance of transcriptionally active chromatin marks, such as H3K4me3, H3K9ac3, and H4Ac, and a more localized distribution of histone marks associated with gene silencing, such as H3K27me3 (Azuara et al. 2006, Mikkelsen et al. 2007, Bernstein et al. 2006). These short active and long silent clusters of histone marks are associated with highly conserved noncoding elements termed bivalent domains. As bivalent domains frequently overlap the binding sites of the core pluripotency factors Oct3/4, Sox2, and Nanog, it has been proposed that they promote pluripotency in undifferentiated cells by maintaining the expression of lineage-specific factors in a silent state, but poised for transcription. Consistently, the “primed” gene loci replicate earlier in S phase than in their differentiated progeny (Azuara et al. 2006, Perry et al. 2004) and can be enriched for key developmental regulators that are silenced in pluripotent ES cells but activated upon differentiation (Bernstein et al. 2006). Upon ES cell differentiation, repressive marks (H3K27me3) are removed from the promoters of activated genes, whereas activating marks (H3K4me3) are erased from genes that remain silent (Bernstein et al. 2006). Several subunits of the PcG PRC2 complexes, such as Eed and Suz12, are detected at these bivalent domains, and repression of developmentally regulated genes at bivalent domains is dependent on Eed (Boyer et al. 2006, Loh et al. 2006).
The enrichment for bivalent marks at conserved elements in pluripotent mouse ES cells (versus adult tissues) initially suggested a functional relationship between bivalent domains and pluripotency (Bernstein et al. 2006). However, it was recently shown that bivalent domains are not a unique feature of pluripotent cells but are also present in differentiated cell types and can even form de novo during cellular differentiation (Azuara et al. 2006, Barski et al. 2007, Mikkelsen et al. 2007, Pan et al. 2007, Roh et al. 2006, Zhao et al. 2007). In addition, the genetic studies of Magnuson and colleagues has shown that ES cells can be formed in the absence of H3K27me3 (Chamberlain et al. 2008), indicating that bivalent marks are not essential for pluripotency, but rather mark genes that will become activated during differentiation. Based on these observations and the fact that the number of promoters with a bivalent domain configuration gradually decreases during ES cell differentiation, Bernstein and colleagues recently proposed an alternative model whereby the relative abundance of bivalent domains in a given cell type corresponds to its degree of pluripotency (Mikkelsen et al. 2007). It is important to keep in mind that current studies have examined only a small fraction of the known histone modifications in the human genome (for which we know the relationship to gene expression) and only in a small number of cell types. More comprehensive genome-wide maps of histone modifications in ES cells and their differentiated progeny as well as their impact on gene expression may help decipher the molecular mechanisms underlying stem cell pluripotency and lineage specification.
CONCLUSIONS AND PERSPECTIVES
During the past five years, genome-wide analysis combined with proteomic studies and genetics in mice have provided important advances in our understanding of the molecular basis of the stable heritable state of pluripotency. A more dynamic picture of chromatin has emerged from the discovery of demethylases and deacetylases, prompting investigations into the mechanisms stabilizing competing activities that control histone modifications. In addition, specialized assemblies of ATP-dependent chromatin-remodeling complexes, such as esBAF, appear to give robustness and stability to the pluripotent state by interacting directly with pluripotency proteins, interacting with their regulatory regions, and binding across the genome with pluripotent factors such as Oct4, Nanog, and Sox2. These ATP-dependent chromatin-remodeling complexes undergo sequential changes in subunit composition in the development of the vertebrate nervous system to coordinate mitotic exit and the onset of postmitotic neural functions. Whether such changes occur in the development of other tissues remains to be determined. Genome-wide studies have also challenged the traditional view of the opposing action of Polycomb and Trithorax genes, revealing that the Trithorax gene Brg and esBAF complexes repress most of their targets, including many developmentally regulated genes, a function that was thought to be largely due to Polycomb action. The view that ATP-dependent chromatin remodeling is a permissive mechanism is being challenged by the observation that specific subunits, such as BAF45a and BAF53a, play instructive roles in directing progenitor division in the vertebrate nervous system, whereas subunits such as BAF60c appear to play instructive roles in the initiation of cardiac development. Finally, subunits of esBAF complexes facilitate reprogramming of induced pluripotent stem cells. Although chromatin regulation has generally been considered to be global and to affect vast numbers of genes, the recent discovery that most phenotypes of PRC1 mutations can be repressed by mutation of a single gene indicates that a few critical targets may mediate most of the actions of these chromatin regulators. Similar observations for the neural nBAF complex indicate that this may be a general feature of chromatin regulators. A final area of future investigations must be directed at understanding the mechanisms used by ATP-dependent chromatin-remodeling complexes. Although genetic studies strongly implicate several ATP-dependent chromatin-remodeling complexes in pluripotency, the biochemical mechanisms involved remain a mystery. Could it really be that these complexes, which in the case of the esBAF complex are 12 times the mass of a nucleosome and contain two highly active ATPases, function in vivo to move nucleosomes, a task that can be produced by the binding of a transcription factor? The development of better assays to explore the mechanisms of chromatin regulatory complexes will be critical to understanding their role in stem cells and as potential therapeutic targets.
Supplementary Material
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review, although they may be hugely biased by their egos.
LITERATURE CITED
- Agarwal N, Hardt T, Brero A, Nowak D, Rothbauer U, et al. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35(16):5402–8. doi: 10.1093/nar/gkm599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–34. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 2001;153(1):101–10. doi: 10.1083/jcb.153.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17(1):126–40. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8(5):532–38. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9(16):2395–402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397(6720):579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2009;463:1042–47. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau S, Kagey MH, Frampton GM, Rahl PB, Young RA. SetDB1 contributes to repression of genes encoding developmental regulators and maintenance of ES cell state. Genes Dev. 2009;23:2484–89. doi: 10.1101/gad.1837309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6(11):872–84. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim. Biophys. Acta. 2004;1677(1–3):52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21(9):2231–41. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La MC, Nicholson J, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6(6):1287–95. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20(13):1744–54. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M, Spradling AC. Searching chromatin for stem cell identity. Cell. 2006;125(2):233–36. doi: 10.1016/j.cell.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, et al. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 2003;278:42733–36. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004;14(2):155–64. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380(6573):435–39. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26(6):1496–505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113(5):643–55. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell Biol. 2003;23:5594–605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133(6):1106–17. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell Biol. 2005;25(14):6031–46. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128(6):1063–76. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–40. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai B, Rasmussen TP. Global epiproteomic signatures distinguish embryonic stem cells from differentiated cells. Stem Cells. 2007;25(10):2567–74. doi: 10.1634/stemcells.2007-0131. [DOI] [PubMed] [Google Scholar]
- De SF, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–94. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Dejosez M, Krumenacker JS, Zitur LJ, Passeri M, Chu LF, et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell. 2008;133:1162–74. doi: 10.1016/j.cell.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26(37):5433–38. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Doan DN, Veal TM, Yan Z, Wang W, Jones SN, Imbalzano AN. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene. 2004;23(19):3462–73. doi: 10.1038/sj.onc.1207472. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell Biol. 2004;24(6):2478–86. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, et al. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol. Cell Biol. 1999;19:7237–44. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey JS, Zacharek SJ, Kim CF, Lees JA. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc. Natl. Acad. Sci. USA. 2008;105(33):11857–62. doi: 10.1073/pnas.0803574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB. ATP-dependent nucleosome remodelling: factors and functions. J. Cell Sci. 2004;117(Pt 17):3707–11. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, et al. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 2001;20(14):3781–88. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–20. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderkin S, Maertens GN, Endoh M, Mallery DL, Morrice N, et al. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell. 2007;28(1):107–20. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev. Cell. 2004;6(3):437–43. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- Fan G, Martinowich K, Chin MH, He F, Fouse SD, et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development. 2005;132(15):3345–56. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4(6):e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121(2):273–85. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134(1):162–74. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong GH, Rossant J, Gertsenstein M, Breitman ML. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature. 1995;376(6535):66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2(2):160–69. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J. Biol. Chem. 2003;278(26):24132–38. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. USA. 2008;105(18):6656–61. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460(7257):863–68. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Talbot D, Leonhardt H, Jaenisch R. A short DNA methyltransferase isoform restores methylation in vivo. J. Biol. Chem. 1998;273:32725–29. doi: 10.1074/jbc.273.49.32725. [DOI] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, Van Der Hoeven F, et al. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–32. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- Goller T, Vauti F, Ramasamy S, Arnold HH. Transcriptional regulator BPTF/FAC1 is essential for trophoblast differentiation during early mouse development. Mol. Cell Biol. 2008;28(22):6819–27. doi: 10.1128/MCB.01058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, et al. Tip60 is a haplo-insufficient tumor suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448(7157):1063–67. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca2+/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell. 2001;105(7):863–75. doi: 10.1016/s0092-8674(01)00396-8. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130(1):77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell Biol. 2001;21(10):3598–603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97(7):833–42. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng HL, Shang C, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr. Biol. 2004;14(16):1475–80. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, et al. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 2001;29(2):206–11. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- Heuser M, Yap DB, Leung M, de Algara TR, Tafech A, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113(7):1432–43. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. USA. 2009a;106(13):5187–91. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA. 2009b;106(13):5181–86. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102(4):463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Illingworth R, Kerr A, Desousa D, Jorgensen H, Ellis P, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6(1):e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell Biol. 2007;27(5):1889–903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13(12):1529–39. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533–38. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, Torre-Ubieta L, Huarte M, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128(6):1077–88. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006;8(3):285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD corepressor complex, is required for development of pluripotent cells. Development. 2007;134(6):1123–32. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Kajiume T, Ninomiya Y, Ishihara H, Kanno R, Kanno M. Polycomb group gene mel-18 modulates the self-renewal activity and cell cycle status of hematopoietic stem cells. Exp. Hematol. 2004;32(6):571–78. doi: 10.1016/j.exphem.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–79. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumoto T, Aikawa Y, Iwama A, Ueda S, Ichikawa H, et al. MOZ is essential for maintenance of hematopoietic stem cells. Genes Dev. 2006;20(10):1321–30. doi: 10.1101/gad.1393106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–28. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- Kim JK, Huh SO, Choi H, Lee KS, Shin D, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell Biol. 2001;21(22):7787–95. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Sawada A, Tokimasa S, Endo H, Ozono K, et al. Defective long-term repopulating ability in hematopoietic stem cells lacking the Polycomb-group gene rae28. Eur. J. Haematol. 2004;73(2):75–84. doi: 10.1111/j.1600-0609.2004.00268.x. [DOI] [PubMed] [Google Scholar]
- Kishi N, Macklis JD. MECP2 is progressively expressed in postmigratory neurons and is involved in neuronal maturation rather than cell fate decisions. Mol. Cell Neurosci. 2004;27(3):306–21. doi: 10.1016/j.mcn.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1(6):500–6. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442(7100):312–16. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007;8(4):307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184(139):868–71. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, et al. Octamer and Sox elements are required for transcriptional cis-regulation of Nanog gene expression. Mol. Cell Biol. 2005;25(6):2475–85. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14(3):286–98. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(7163):689–94. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4(10):e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97(7):843–52. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM, Teillet MA. Experimental analysis of the migration and differentiation of neuroblasts of the autonomic nervous system and of neurectodermal mesenchymal derivatives, using a biological cell marking technique. Dev. Biol. 1974;41(1):162–84. doi: 10.1016/0012-1606(74)90291-7. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38(1):32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Lee KK, Workman JL. Histone acetyltransferase complexes: One size doesn't fit all. Nat. Rev. Mol. Cell Biol. 2007;8(4):284–95. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125(2):301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–28. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423(6937):255–60. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–15. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings PP, Zhou Z, Vieira KF, Crusselle-Davis VJ, Bungert J. Recruitment of transcription complexes to the beta-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. FEBS J. 2006;273(4):746–55. doi: 10.1111/j.1742-4658.2005.05107.x. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li JY, Pu MT, Hirasawa R, Li BZ, Huang YN, et al. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell Biol. 2007;27(24):8748–59. doi: 10.1128/MCB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Wan M, Zhang Y, Gu P, Xin H, et al. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10(6):731–39. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, von Both I, Walls JR, McAuliffe F, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458(7237):529–33. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459(7245):387–92. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66(12):6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38(4):431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21(20):2545–57. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz S, Glaser S, Schaft J, Stewart AF, Anastassiadis K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol. Biol. Cell. 2007;18(6):2356–66. doi: 10.1091/mbc.E06-11-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007;9(6):625–35. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Matoba R, Niwa H, Masui S, Ohtsuka S, Carter MG, et al. Dissecting Oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS One. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Nakamura T, Nakao K, Arai T, Katsuki M, et al. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18(15):4261–69. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, et al. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1(3):338–45. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454(7205):766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merson TD, Dixon MP, Collin C, Rietze RL, Bartlett PF, et al. The transcriptional coactivator Querkopf controls adult neurogenesis. J. Neurosci. 2006;26(44):11359–70. doi: 10.1523/JNEUROSCI.2247-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7(7):540–46. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006;10(1):105–16. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113(5):631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell. 2008;30(6):755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425(6961):962–67. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 2005;15(10):942–47. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–91. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct. Funct. 2001;26(3):137–48. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134(4):635–46. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24(4):372–76. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Ohta H, Sawada A, Kim JY, Tokimasa S, Nishiguchi S, et al. Polycomb group gene rae28 is required for sustaining activity of hematopoietic stem cells. J. Exp. Med. 2002;195(6):759–70. doi: 10.1084/jem.20011911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 1998;26(11):2536–40. doi: 10.1093/nar/26.11.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–17. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1(3):299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10(16):1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–5. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell Biol. 2007;27(10):3769–79. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini DE, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23(20):4061–71. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–10. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22(10):1345–55. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry P, Sauer S, Billon N, Richardson WD, Spivakov M, et al. A dynamic switch in the replication timing of key regulator genes in embryonic stem cells upon neural induction. Cell Cycle. 2004;3(12):1645–50. [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, et al. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr. Biol. 2008;18(14):1094–99. doi: 10.1016/j.cub.2008.06.070. [DOI] [PubMed] [Google Scholar]
- Poot RA, Bozhenok L, Van Den Berg DL, Steffensen S, Ferreira F, et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat. Cell Biol. 2004;6(12):1236–44. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- Rao S, Orkin SH. Unraveling the transcriptional network controlling ES cell pluripotency. Genome Biol. 2006;7(8):230. doi: 10.1186/gb-2006-7-8-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc. Natl. Acad. Sci. USA. 2002;99(23):14789–94. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293(5532):1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17(16):2048–59. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α) EMBO J. 1998;17(23):6979–91. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–43. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA. 2000;97(25):13796–800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25(3):338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, et al. Transcriptional regulation of Nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280(26):24731–37. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. USA. 2006;103(43):15782–87. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman-Trufero M, Mendez-Gomez HR, Perez C, Hijikata A, Fujimura Y, et al. Maintenance of undifferentiated state and self-renewal of embryonic neural stem cells by Polycomb protein Ring1B. Stem Cells. 2009;27:1559–70. doi: 10.1002/stem.82. [DOI] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40(7):915–20. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419(6905):407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 2006;38(9):1496–509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27(12):2979–91. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18(11):1263–71. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376(6535):62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–14. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383(6597):250–53. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22(8):1846–56. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J. Biol. Chem. 2005;280(51):41789–92. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, et al. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and premRNA splicing. Mol. Cell. 2007;28(4):665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, et al. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell. 2010;141(6):943–55. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16(9):433–42. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Stokes DG, Tartof KD, Perry RP. CHD1 is concentrated in interbands and puffed regions of Drosophila polytene chromosomes. Proc. Natl. Acad. Sci. USA. 1996;93(14):7137–42. doi: 10.1073/pnas.93.14.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopka T, Skoultchi AI. The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. USA. 2003;100(24):14097–102. doi: 10.1073/pnas.2336105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, et al. NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20(17):4892–900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131(1):29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N. Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol. Cell Biol. 2005;25(5):1804–20. doi: 10.1128/MCB.25.5.1804-1820.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127(7):1375–88. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16(14):1779–91. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19(7):815–26. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J. Exp. Med. 2007;204(4):715–22. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459(7247):708–11. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Kojima M, Nakajima K, Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech. Dev. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, et al. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–22. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Thomas T, Corcoran LM, Gugasyan R, Dixon MP, Brodnicki T, et al. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Genes Dev. 2006;20(9):1175–86. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T, Voss AK, Chowdhury K, Gruss P. Querkopf, a MYST family histone acetyltransferase, is required for normal cerebral cortex development. Development. 2000;127(12):2537–48. doi: 10.1242/dev.127.12.2537. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–16. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–14. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Tucker KL, Talbot D, Lee MA, Leonhardt H, Jaenisch R. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc. Natl. Acad. Sci. USA. 1996;93:12920–25. doi: 10.1073/pnas.93.23.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SWM, van Lohuizen M. Stem cells and cancer: the polycomb connection. Cell. 2004;118(4):409–18. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, et al. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131(1):58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Roelen BAJ, Roefs M, de Vries S, Verhoeven E, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA. 2003;100(5):2468–73. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 1999;23(1):62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Walker E, Chang WY, Hunkapiller J, Cagney G, Garcha K, et al. Polycomb-like 2 associates with PRC2 and regulates transcriptional networks during mouse embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2010;6(2):153–66. doi: 10.1016/j.stem.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996a;15(19):5370–82. [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996b;10(17):2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138(5):1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhai W, Richardson JA, Olson EN, Meneses JJ, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18(24):3106–16. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39(4):457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125(3):467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Williams CJ, Naito T, Arco PG, Seavitt JR, Cashman SM, et al. The chromatin remodeler Mi-2β is required for CD4 expression and T cell development. Immunity. 2004;20(6):719–33. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. USA. 1997;94(21):11472–77. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136(2):200–6. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56(1):94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Wu Q, Bruce AW, Jedrusik A, Ellis PD, Andrews RM, et al. CARM1 is required in embryonic stem cells to maintain pluripotency and resist differentiation. Stem Cells. 2009;27(11):2637–45. doi: 10.1002/stem.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442(7098):86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26(37):5541–52. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25(6):801–12. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–95. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26(5):1155–65. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122(3):881–94. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460(7255):642–46. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hazan I, Zhang J, Ng SY, Naito T, et al. The role of the chromatin remodeler Mi-2β in hematopoietic stem cell self-renewal and multilineage differentiation. Genes Dev. 2008;22(9):1174–89. doi: 10.1101/gad.1642808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, et al. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. USA. 2003;100(11):6777–82. doi: 10.1073/pnas.1131928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1(3):286–98. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zhong X, Jin Y. Critical roles of coactivator p300 in mouse embryonic stem cell differentiation and Nanog expression. J. Biol. Chem. 2009;284(14):9168–75. doi: 10.1074/jbc.M805562200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104(42):16438–43. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, et al. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25(2):335–45. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.