Abstract
DamIP is a new method for studying DNA-protein interaction in vivo. A mutant form of DNA adenine methyltransferase (DamK9A) from E. coli is fused to the protein of interest and expressed. The fusion protein will bind to target binding sites and introduce N-6-adenine methylation in nearby sites in the genomic DNA. Methylated DNA fragments are enriched with an antibody against N-6-methyladenine and used for further analysis, e.g. real-time PCR, microarray or high-throughput sequencing. This method is simple and does not require either protein-DNA crosslinking or a specific antibody to the protein of interest. This unit describes the application of this method for the identification of DNA binding sites in vivo.
Keywords: DNA adenine methyltransferase, transcription factor binding sites, DamIP, chromatin immunoprecipitation
INTRODUCTION
DNA-protein interactions are central to the regulation of gene expression. Historically, many methods have been developed to study these interactions (Moss and Leblanc, 2009). Currently, the most popular method for studying genome-wide DNA-protein interaction is chromatin immunoprecipitation (ChIP), which uses formaldehyde to cross-link protein and DNA and enriches target protein-DNA complexes with specific antibodies (Collas, 2009). When combined with DNA microarray or massively parallel sequencing technologies, ChIP makes it possible to profile the occupancy of DNA-interacting proteins at a genome wide scale (Ren et al., 2000; Robertson et al., 2007). However, the ChIP assay is rather cumbersome and is limited by its dependence on a highly specific antibody for each protein of interest.
van Steensel and colleagues developed an alternative strategy to study protein-DNA interactions (Orian et al., 2009; van Steensel et al., 2001; van Steensel and Henikoff, 2000). DNA adenine methyltransferase (Dam) from E. coli, which specifically methylates the adenine residue in a GATC recognition sequence, is fused to the protein of interest using a short linker. When expressed in cells, the fusion proteins bind genomic DNA via the protein of interest and introduce N-6-adenosine methylation (m6A) to nearby GATC tetramers. Locations of methylation can be identified with methylation-sensitive restriction enzymes Dpn I and Dpn II. This method, named Dam IDentification (DamID), has been successfully applied to several eukaryotic model systems, e.g. budding yeast, plant, fruit fly, and cultured mammalian cells (Bianchi-Frias et al., 2004; Filion et al., 2010; Orian et al., 2003; Venkatasubrahmanyam et al., 2007; Weber et al., 2005).
The high activity of E. coli Dam creates a signal to noise problem for the DamID approach. In addition, the tetrameric Dam recognition site occurs on average once in every 256 nucleotides in the genome and may not be present near specific DNA binding sites of interest, which limits its resolution. Dam has been extensively studied and its target sequence recognition is determined by several key amino acid residues in the catalytic pocket (Horton et al., 2006; Horton et al., 2005). Previously described mutations of these residues decrease both the activity of the enzyme and the specificity for the GATC tetramer, thereby increasing the frequency of potential methylation sites and addressing both concerns. We developed a new method, termed DamIP, using such a mutant form of Dam, combined with an antibody that specifically recognizes N-6-methylated DNA (Lopez et al., 2003). The mutant Dam is linked to the protein of interest, and the fusion protein introduces N-6-adenosine methylation to sequences adjacent to specific DNA binding sites. We have used human estrogen receptor alpha (hERα) in an initial test of this method, and have found that Dam-hER fusion protein can be used to specifically identify both direct and indirect hERα DNA binding sites with great resolution and sensitivity (Xiao et al., 2010).
STRATEGIC PLANNING
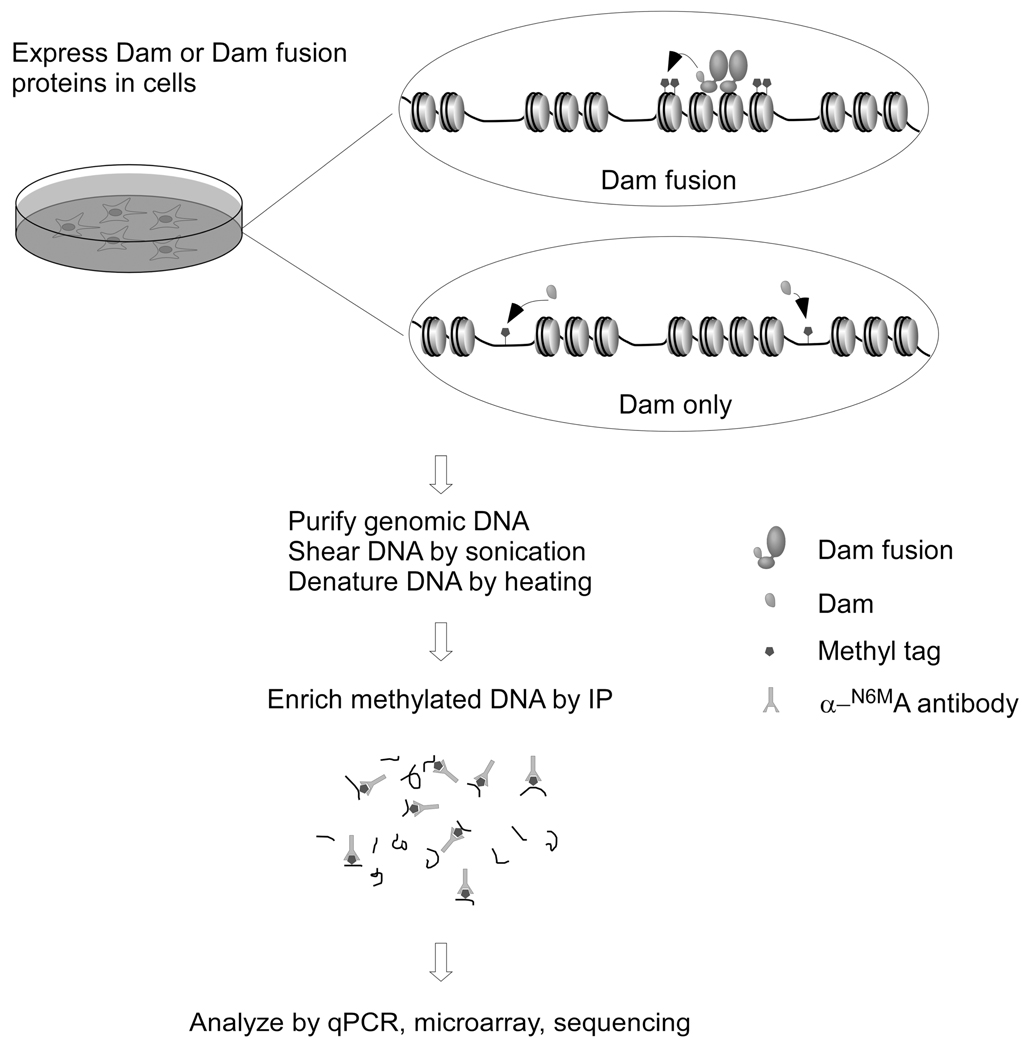
The first step in DamIP is to generate a fusion protein of DamK9A and the protein of interest. It is crucial to test whether the fusion protein maintains the expected functionality, and it may be necessary to fuse the DamK9A to either the N-terminus or C-terminus of the protein of interest. If the fusion protein functions well, the next step is to express the fusion protein in appropriate cells, either via transient or stable transfection or viral systems. When using a stable cell line strategy, it is necessary to use an inducible expression system, e.g. Tet-on/Tet-off Advanced Inducible Expression Systems from Clontech, because sustained expression of Dam or Dam fusion proteins will lead to excessive methylation of the genome. After fusion protein expression, typically for 24 to 48 hours, genomic DNA is harvested and fragmented and methylated DNA fragments are enriched by immunoprecipitation and detected by quantitative real time PCR (qPCR) or other methods (Figure 1).
Figure 1.
Illustration of the procedure of DamIP. DamK9A or DamK9A fusion proteins are expressed in cells. Genomic DNA is purified, sonicated and denatured before being mixed with anti-N-6-methyladenine antibodies. Methylated DNA recognized by the antibody is enriched and analyzed by various methods, e.g., qPCR, microarray and next-generation sequencing (adapted from Xiao et al., 2010).
DamIP FOR STUDYING DNA PROTEIN INTERACTIONS IN VIVO
This protocol uses adherent cultured cells that express DamK9A alone as control, and the protein of interest fused to DamK9A. Expression of these proteins results in non-specific and specific introduction of m6A modifications, which are enriched by immunoprecipitation with a specific antibody against m6A and analyzed by PCR, microarray, etc.
Support Protocol
Prepare genomic DNA for DamIP
To perform DamIP, we typically use 5 to 10µg genomic DNA. If not mentioned specifically, all of the following steps are performed at room temperature (20~25°C).
Materials
Cells expressing DamK9A fusion protein or the control DamK9A only. We have observed good signal to noise ratios for specific fusion protein binding with 24 to 48 hours of fusion protein expression.
Phosphate-buffered saline (PBS; APPENDIX 2)
Lysis buffer (see recipe)
10x DamIP buffer (see recipe)
TE buffer (see recipe)
Digestion buffer (see recipe)
RNase A, DNase free
Proteinase K, DNase free
25:24:1 phenol/chloroform/isoamyl alcohol, pH 7.9 (Invitrogen)
Isopropanol
70% ethanol
Branson Sonifier 250 with microtip
Collect cells that express DamK9A fusion protein or DamK9A only by trypsinization or using cell scrapers. Wash once with 1xPBS and pellet with centrifugation at 800×g for 2 minutes at room temperature. Estimate cell number by hemocytometer or automated cell counter.
Add lysis buffer to cell pellet. Use 0.6ml lysis buffer for up to 107 cells. If the mixture becomes too viscous as the genomic DNA is released, a brief sonication of 10 seconds can be applied.
Add RNAse A to a final concentration of 10µg/ml and incubate at 37°C for 30 minutes.
Add proteinase K to a final concentration of 250µg/ml and incubate at 55°C for more than 3 hours.
Extract DNA with phenol/chloroform/isoamyl alcohol and precipitate with isopropanol (UNIT 2.1A).
Resuspend genomic DNA with 500µL TE buffer and quantify DNA concentration by measuring absorption at 260nm with UV spectrometer (APPENDIX 3D).
- Sonicate DNA on ice to produce approximately 500-bp fragments. Use 15 second pulses with 1 minute intervals on ice.Sonication conditions are highly dependent on the instrument and need to be optimized and checked by agarose gel electrophoresis. With a Branson Sonifer 250 we find that 4 rounds of 15-second sonication with duty cycle 50% and output 4 yields ~500-bp fragments. Since DNA concentration affects the efficiency of sonication, it is preferable to keep the DNA concentrations of various samples at similar levels.
Support Protocol
Antibody Pretreatment
DamIP depends on an antibody that specifically recognizes N-6 methylated adenosine. The antibody we use was raised as a polyclonal and it retains very weak affinity against non-modified adenosine, which will introduce some background. To optimize DamIP, we tried various immunoprecipitation conditions and found the best is to pre-treat the antibody with agarose conjugated with single-stranded DNA (ssDNA).
Prepare ssDNA-agarose
Materials
CNBr-activated agarose (Sigma, C9210)
1mM HCl: store at 4°C
Buffer A: 10mM potassium phosphate buffer, pH 8.0
0.2M glycine-NaOH, pH 8.0
Eukaryotic genomic DNA: purified from cells or salmon sperm DNA, sheared by sonication to ~500bp. Proteinase K treated and extracted twice with phenol/chloroform/isoamyl alcohol (see Support Protocol above: Prepare genomic DNA for DamIP)
Dissolve 5 mg gDNA in 5ml Buffer A to final concentration of 1mg/ml in 15ml falcon tube. Boil for 10 minutes and then place in ice-water bath for 10 minutes.
Wash and swell 2g CNBr-activated agarose resin in cold 1mM HCl for at least 30 minute. Use 200ml 1mM HCl per gram dry resin.
Wash resin with at least 10 packed resin volume (p.r.v.) of distilled water extensively.
- Wash once with 5ml buffer A and aspirate buffer A. Combine with DNA solution prepared in step 1 IMMEDIATELY.It is important to perform this step quickly to get the best conjugation efficiency.
Rotate overnight at 4°C.
- Centrifuge at 800×g for 5 minutes at room temperature and transfer the supernatant to a new tube. Add 5 p.r.v. of Buffer A and mix by inverting the tube. Centrifuge 800×g for 5 minutes at room temperature and discard the supernatant.Save first supernatant to estimate coupling efficiency. Quantify DNA concentration of the supernatant by absorption at 260nm with Buffer A as reference (APPENDIX 3D). The efficiency can be estimated as (total starting DNA – DNA in first supernatant)/volume of the resin. Usually more than 1mg DNA can be coupled to 1ml wet resin.
Block unreacted groups by incubating with 0.2M glycine-NaOH, pH8 for 2 hours at room temperature.
Wash with 10 p.r.v. Buffer A containing 1M KCl and then 10 p.r.v. buffer A.
Wash with distilled water extensively and wash once with TE.
Resuspend resin in TE buffer containing 0.05% NaN3 and store up to 12 months at 4°C.
Support Protocol
Pretreat antibody before DamIP
To reduce nonspecific binding, pre-treat antibody with DNA-agarose beads before immunoprecipitation. Use DNA-agarose with more than 20ug conjugated DNA per 10ug antibody. Antibody pretreatment can be set up several hours or the day before conducting DamIP.
Materials
DNA-agarose beads: describe in protocol 2
Affinity purified antibody against m6A (Megabase Research or Synaptic Systems)
0.2M NaOH
1xDamIP buffer: diluted from 10x DamIP buffer (see recipe)
Benchtop centrifuge, refrigerated
Wash DNA-agarose with 0.2M NaOH for 10 minutes at room temperature to denature DNA on agarose.
Wash the agarose beads 3 times with cold water, 3 times with TE buffer and once with 1xDamIP buffer.
Add 100ul 1xDamIP buffer and 10µg antibody to DNA-agarose, so the final antibody concentration will be about 0.1ug/ul in 1xDamIP buffer.
Rotate at room temperature for 2 hours or at 4°C overnight. Centrifuge beads 5min at 800×g. Carefully transfer supernatant (containing antibody) to a new tube and use for DamIP immediately.
Basic Protocol
DamIP
Materials
Sonicated genomic DNA (from protocol 1)
Pretreated antibody (from protocol 3)
10x DamIP buffer (see recipe)
TE buffer (see recipe)
Digestion buffer (see recipe)
Proteinase K, DNase free
25:24:1 phenol/chloroform/isoamyl alcohol, pH 7.9 (Invitrogen)
Isopropanol
70% ethanol
QIAquick PCR purification kit (Qiagen)
Benchtop centrifuge
Water bath, 37°C and 55°C
Dilute 10 µg of sonicated DNA in 400 µl TE buffer in a 1.7 ml eppendorf tube.
Denature for 10 minutes in boiling water and immediately cool on ice/water bath for at least 5 minutes. Save 40µl as 10% input.
Centrifuge briefly to bring all liquid to the bottom of the tube. Add 40 µl of 10x DamIP buffer and 10 µg (~100ul) of pretreated 6mA antibody or control antibody to the remaining 360 µl DNA.
Rotate 1–2 hours at room temperature
Mix 40µl A/G plus agarose beads (Santa Cruz, SC-2003) for each sample with 1ml 1xDamIP buffer by inverting the tube. Centrifuge at 800×g for 2 minutes and carefully pipet off the supernatant. Add 1ml 1xDamIP buffer and incubate at room temperature for 30 minutes. Centrifuge again and remove the supernatant. Resuspend in 1x DamIP buffer to the original volume.
Add 40µl 50% agarose slurry to each sample and rotate at room temperature for 1 hour.
Centrifuge at 800×g for 2 minutes at room temperature. Carefully remove supernatant by pipetting without disturbing the beads. Add 800µl 1xDamIP buffer and rotate for 10 minutes at room temperature.
Repeat step 7 for 4 more times (wash 5 times in total).
Resuspend the beads in 150 µl digestion buffer containing 300 µg/ml proteinase K and incubate 3 hours at 50°C with shaking or rotating.
Purify DNA by phenol/chloroform/isoamyl alcohol extraction (UNIT 2.1A) or Qiagen Qiaquick columns. Elute DNA in 100 µl Qiagen EB buffer.
Use 2 µl of purified DNA for quantitative real-time PCR analysis of specific binding sites.
REAGENTS AND SOLUTIONS
Use deionized, distilled water in all recipes and protocol steps. For common stock solutions, see APPENDIX 2; for suppliers, see APPENDIX 4.
Lysis buffer
10mM Tris, pH 8.0
25mM EDTA, pH 8.0
150mM NaCl
0.5% SDS
Store at room temperature for up to several months
1M Na-Phosphate buffer, pH 7.0
Make solution A containing monobasic sodium phosphate monohydrate, 138g/L and solution B containing dibasic sodium phosphate, 142g/L.
Mix 39.0 ml solution A and 61.0 ml solution B to get 1M Na-phosphate buffer, pH 7.0
10x DamIP buffer
100mM Na-Phosphate, pH 7.0 (from 1M Na-Phosphate buffer)
3M NaCl
0.5% Triton X-100
Store at room temperature for up to several months
Digestion buffer
50mM Tris, pH 8.0
10mM EDTA
0.5% SDS
Store at room temperature for up to several months
TE buffer
10mM Tris, pH 8.0
1mM EDTA
Store at room temperature for up to several months
COMMENTARY
Background Information
Methylation at the N6 position of adenosine in DNA is thought to be specific for prokaryotes and absent in eukaryotes except some lower eukaryotic organisms (Ratel et al., 2006a; Wion and Casadesus, 2006). In mouse, it is estimated that each genome has less than one thousand N6-methylated adenosines, if any (Ratel et al., 2006b). This allows the use of Dam to introduce m6A into eukaryotic genomes as a unique mark. Although m6A is essentially absent in eukaryotic DNA, it is present in mRNA (Rottman et al., 1994). Therefore, it is necessary to completely remove RNA before DamIP.
The enzyme used in DamIP to introduce m6A in DNA is from Escherichia coli. This enzyme has been extensively studied and the crystal structure has been resolved (Horton et al., 2006; Horton et al., 2005). Based on the crystal structures and mutagenesis studies, several amino acid residues of E. coli Dam mediate the specific recognition of the target methylation sequence GATC. Mutation of these residues changes both specificity and activity of E. coli Dam. For example, mutation of lysine 124 to alanine reduces the catalytic activity by more than 100-fold, and also strongly decreases the specificity against the fourth position of GATC. We mutated several residues individually or combinatorially and found that the K9A (lysine 9 to alanine) mutant shows the best combination of activity and specificity. This mutation loses the recognition of the first position of GATC on either strand, but retains reasonable activity. This reduction in Dam specificity increases the number of potential potential methylation sites near the binding site for the DamK9A fusion protein and hence increases the ability of DamIP to introduce appropriate tags.
However, since the mutant form of Dam methylates more than GATC sequences, effective detection of methylated adenosines in the genome cannot be achieved by Dpn I/II enzymes as in DamID. Instead, we use antibodies that specifically recognize m6A (Lopez et al., 2003).
Compared with the conventional ChIP assay, which uses formaldehyde to crosslink protein and DNA and provides a snap-shot of protein-DNA interaction, DamIP generates a very different kinetic profile of protein-DNA interaction. Dam-introduced m6A modifications in genomic DNA are apparently very stable and we did not see any loss of m6A during our cell culture or DNA storage. We believe that m6A modification accumulates as the Dam fusion protein is present in the cell and interacting with DNA dynamically. Therefore, DamIP generates a cumulative binding profile over the time of fusion protein expression.
Critical Parameters and Troubleshooting
Antibody
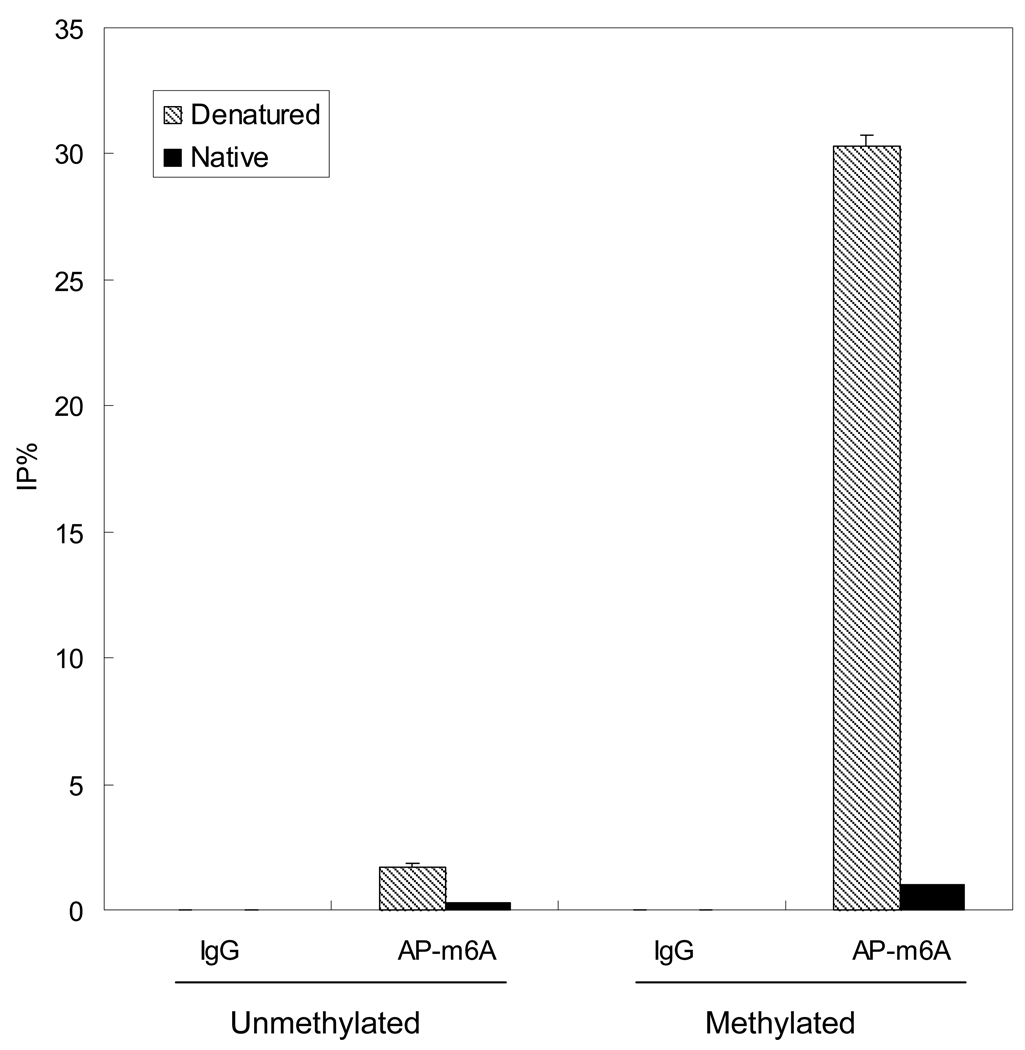
We have tested antibodies from different commercial providers (Megabase Research and Synaptic Systems) and both worked well for DamIP. We also successfully generated our own antibody against m6A for DamIP. All available m6A antibodies are polyclonal and they retain weak affinity against unmethylated adenosine even after affinity purification. This affinity causes elevated background signals. To reduce background, antibodies can be pretreated with ssDNA-conjugated agarose as described in Support Protocol (Figure 2).
Figure 2.
Denaturation is important for recognition of m6A by antibodies. Unmethylated or methylated plasmids were mixed with affinity purified m6A antibody (AP-m6A) and enriched fragments were quantified by real-time PCR. Denaturation of DNA by heating greatly increases the affinity of m6A antibody against methylated DNA.
Expression of Dam fusion proteins
Like other tagging strategies, generating and verifying the appropriate function of the Dam fusion proteins is important. We recommend generating both N-terminal and C-terminal fusions of the target protein. Particularly as we have observed significantly decreased activity with some nuclear receptor fusions, it is essential to test whether the fusion protein is functional. In addition, it may be necessary to optimize the time and level of Dam fusion protein expression before harvesting cells, since m6A modification of genomic DNA is stable and the total amount will be time-dependent. We usually express the fusion protein 24 to 48 hours and obtain satisfactory signal-noise ratios for positive control binding sites.
Denature DNA before DamIP
The methyl mark of m6A is located in the major groove of a DNA double helix (Guarne et al., 2002). Antibody recognition of m6A in double stranded DNA is very inefficient and denaturation of DNA before adding antibodies is essential for successful DamIP. Consequently, the enriched DNA fragment after DamIP will be essentially single stranded DNA and conversion to double stranded DNA may be necessary for some analyses, e.g. microarray. However, PCR analysis of DamIP samples does not require any modification of the protocol.
Methylation of plasmids
Many common E. coli strains, e.g. DH5α, used for plasmid amplification are Dam positive (dam+), which means plasmids grown in these strains are fully methylated at GATC N6-adenosine sites. Although DamIP uses mutant Dam and the expression of fusion proteins is driven by eukaryotic promoters, leaky expression from the plasmid can cause significant m6A modification of plasmids even when grown in dam−/dcm− strains. If such plasmids are introduced by transient transfection, they will be recovered by DamIP and may introduce background for some assays, e.g. microarrays. However, this will not be a problem if one uses PCR or next generation sequencing for analysis, or if stable inducible cell clones or viral expression systems are used for fusion protein expression.
Internal control for DamIP
A convenient method to normalize the variations introduced during the immunoprecipitation procedure is to take advantage of the E. coli-introduced m6A by adding a constant amount of an unrelated methylated DNA sequence into each sample before starting DamIP. We use a plasmid that contains a fragment from an organism unrelated to the target genome. Since this plasmid is propagated in normal Dam-positive E. coli strains, e.g. DH5α, it will be fully N6-adenosine methylated at GATC sites. This internal control plasmid is added at a concentration of one plasmid per genome (about 10 pg plasmid per 10µg genomic DNA). The level of detection of the control plasmid signal by PCR after DamIP is used to normalize variations between samples.
Anticipated Results
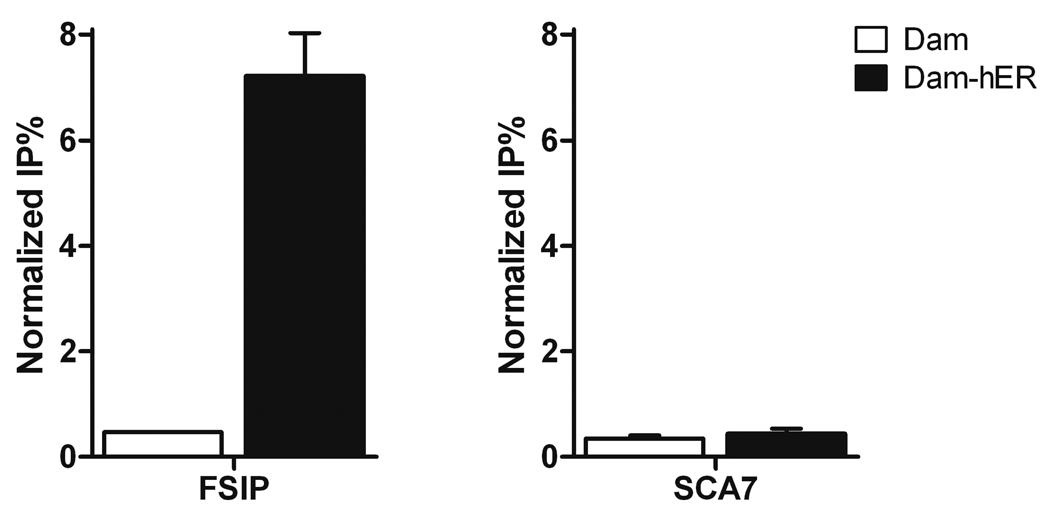
When using real-time PCR to quantify DamIP samples, we usually see significant enrichment of specific binding sites for DamK9A fusion protein in expressing cells compared with cells expressing DamK9A alone, as determined by quantitative real time PCR (Figure 3). DamIP can also be combined with either tiling arrays or high throughput sequencing for genome wide binding studies. Depending on the analysis strategy, it may be necessary to convert the single stranded enriched DNA produced by DamIP into double stranded. We have observed better results with random hexamers and the Klenow fragment of DNA polymerase than with adaptor based strategies.
Figure 3.
Representative DamIP result. Primers were designed to amplify genomic regions with (FSIP) or without (SCA7) known hER binding. DNA enriched with DamIP from MCF-7 cells transfected with DamK9A-hER (Dam-ER) or Dam alone was analyzed by qPCR. Data are presented as mean +/− SEM.
Time Considerations
Typically it takes one to two weeks to generate and verify the fusion protein. If one chooses to set up stable cell clones for DamIP, it will take about 3 weeks. If using viral, e.g. lentiviral systems, it takes one to two weeks to generate the virus for DamIP. Generation of DNA-agarose takes one day. Starting from genomic DNA, it takes about 2 days to complete a DamIP experiment.
Literature Cited
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collas P. The State-of-the-Art of Chromatin Immunoprecipitation. Methods Mol Biol. 2009;567:1–25. doi: 10.1007/978-1-60327-414-2_1. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarne A, Zhao Q, Ghirlando R, Yang W. Insights into negative modulation of E. coli replication initiation from the structure of SeqA-hemimethylated DNA complex. Nat Struct Biol. 2002;9:839–843. doi: 10.1038/nsb857. [DOI] [PubMed] [Google Scholar]
- Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J Mol Biol. 2006;358:559–570. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JR, Liebert K, Hattman S, Jeltsch A, Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121:349–361. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez OJ, Quintanar A, Padhye NV, Nelson M. Genotyping of DNA using sequence-specific methyltransferases followed by immunochemical detection. J Immunoassay Immunochem. 2003;24:11–28. doi: 10.1081/IAS-120018466. [DOI] [PubMed] [Google Scholar]
- Moss T, Leblanc B. DNA-Protein Interactions: Principles and Protocols. 3 edn. Vol 543. New York: Humana Press; 2009. [Google Scholar]
- Orian A, Abed M, Kenyagin-Karsenti D, Boico O. DamID: A Methylation-Based Chromatin Profiling Approach. Methods Mol Biol. 2009;567:155–169. doi: 10.1007/978-1-60327-414-2_11. [DOI] [PubMed] [Google Scholar]
- Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006a;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratel D, Ravanat JL, Charles MP, Platet N, Breuillaud L, Lunardi J, Berger F, Wion D. Undetectable levels of N6-methyl adenine in mouse DNA: Cloning and analysis of PRED28, a gene coding for a putative mammalian DNA adenine methyltransferase. FEBS Lett. 2006b;580:3179–3184. doi: 10.1016/j.febslet.2006.04.074. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Rottman FM, Bokar JA, Narayan P, Shambaugh ME, Ludwiczak R. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76:1109–1114. doi: 10.1016/0300-9084(94)90038-8. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S, Hwang WW, Meneghini MD, Tong AH, Madhani HD. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–16614. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Roman-Sanchez R, Moore DD. DamIP: a novel method to identify DNA binding sites in vivo. Nucl Recept Signal. 2010;8 doi: 10.1621/nrs.08003. e003. [DOI] [PMC free article] [PubMed] [Google Scholar]