Abstract
It has been generally assumed that product formation in DNA damaged by ionizing radiation is relatively independent of base sequence; i.e., that the yield of a given product depends primarily on the chemical properties of each DNA constituent and not on its base sequence context. We examined this assumption by comparing direct-type end products produced in films of d(CTCTCGAGAG)2 with those produced in films of d(GCACGCGTGC)2. Here we report the product yields in d(CTCTCGAGAG)2 hydrated to Γ = 2.5 and 15, where Γ is the hydration level given in mol H2O/mol nucleotide. Of the 16 products monitored by GC/MS, 7 exhibited statistically significant yields: 8-oxoGua, 8-oxoAde, 5-OHMeUra, 5,6-diHUra, 5,6-diHThy, 5-OHCyt, and 5-OHUra. These yields at Γ = 2.5 are compared with the yields from our previously reported study of d(GCACGCGTGC)2 (after projecting the yields to a CG/AT ratio of 1). The ratio of projected yields, d(CTCTCGAGAG)2 divided by d(GCACGCGTGC)2, are 1.3 ± 0.9, 1.8 ± 0.3, 1.6 ± 0.6, 11.4 ± 4.7, 0.2 ± 0.1, > 28, and 0.8 ± 1.1, respectively. Considering just d(CTCTCGAGAG)2, the ratio of yields at Γ = 2.5 divided by yields at Γ = 15, are 0.7 ± 0.2, 0.5 ± 0.1, 2.3 ± 4.0, 3.4 ± 1.2, 3.5 ± 3.3, 1.2 ± 0.2, and 0.4 ± 0.2, respectively. The effects of sequence and hydration on base product yields are explained by a working model emphasizing the difference between two distinctly different types of reaction: i) radical reactions that progress to non-radical intermediates and product prior to dissolution and ii) reactions that stem from radicals trapped in the solid state at room temperature that go on to yield non-radical product after sample dissolution. Based on these findings, insights into rates of hole and excess electron transfer relative to rates of proton transfer are discussed.
INTRODUCTION
Ionizing radiation damages DNA either indirectly, by creating water radical species that react with DNA, or directly, through ionizations that create sites of electron loss (radical cations), electron gain (radical anions), and excitations.1,2 Subsequent reactions of the radical anions/cations are believed to account for the majority of stable end products and these are referred to as either indirect-type or direct-type damage.3-5 Energy deposition via excited states appears to play a minor, perhaps negligible, role.6 It is well established that, for indirect-damage produced by low LET (linear energy transfer) radiation, each indirect-type product stems from an initial attack of either one OH• or one aqueous electron (e−aq). The reactions leading to an indirect-type product do not entail a subsequent reaction with OH• or e−aq. For direct-type damage by low LET, the analogous relationship between initial radical and final product is generally assumed. In other words, it is assumed that one, and only one, radical cation or radical anion is involved in the formation of a given direct-type product. Recent findings suggest this assumption is not entirely correct7,8.
In a study of strand break (sb) formation in DNA films, Purkayastha et al.9,10 discovered that the yields of deoxyribose-centered free radicals trapped in pUC 18 films at 4K were insufficient to account for the yield of sb in the same samples. This was the first reported case where product yield was shown to exceed the yield of the presumed free radical precursor. The difference between product yield and radical yield was called the “shortfall”. Following on this work, Sharma et al.7,11-13 measured deoxyribose free radical yields and free base release (fbr) in pUC18 films and in a series of oligodeoxynucleotide films. Free base release, just as in the case of strand breaks, correlate with oxidative damage to deoxyribose of the DNA backbone. Here too, a shortfall between assumed radical precursor and end product was observed. For example, the yield of free base release in pUC18 films at a hydration level (Γ) of 2.5 waters/nucleotide was 134 ± 5 nmol/J and the yield of radicals trapped on deoxyribose at 4K was 33 ± 4, giving a shortfall of 134 – 3 = 101 nmol/J.12 In order to explain these shortfalls, and other results such as the influence of base sequence on base release,8 we hypothesized that a significant fraction of sb and fbr is due to two-consecutive one-electron oxidations.
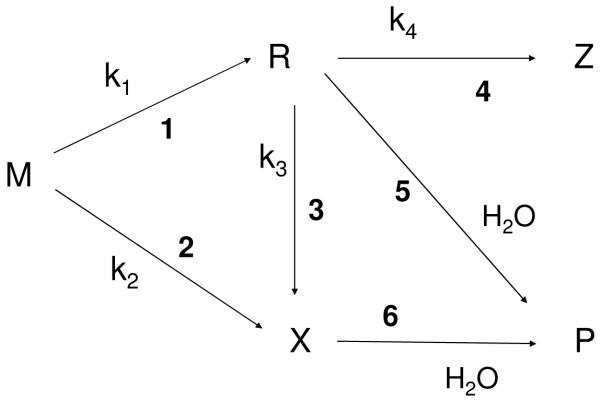
In a more comprehensive study, the yields of base damage, fbr, single strand breaks (ssb), and free radicals were measured in d(GCACGCGTGC)2 in both polycrystalline form and as Γ = 2.5 films.14 Statistically significant yields were obtained for 4 base damage products: 8-oxoGua, 8-oxoAde, 5,6-diHThy, and 5,6-diHUra. (See structures in Figure 1.) Although the dose response curves for fbr and ssb were linear over a wide dose range, 0 – 80 kGy, the dose response for the base products were not. Generally, the curves can be described by two straight lines, the larger slope from 0 Gy to ~10 kGy and the smaller slope from ~20 kGy to 80 kGy. These results were explained by a mechanistic model encapsulated by the reaction scheme shown in Figure 2. The scheme proposes three different pathways to product P, each initiated by ionizing molecule M: (1) trappable-radical single track (Reaction 1 gives the radical R and Reaction 5 leads to P), (2) trappable-radical multiple track (Reactions 1, 3, and 5), and (3) molecular (Reactions 2 and 6). Competing with these forward reactions are other reactions (Reaction 4), two important examples of which are recombination (Z is then ≡ M) and hole/excess electron transfer to another site (Z is then a radical on a nearby base). When this reaction scheme is applied to hole formation on M, the radical R is formed as a consequence of one-electron oxidation. A second one-electron oxidation (Reaction 3), gives X. If all of X were formed via a trappable radical R, there would be no shortfall. The fraction of X formed via the molecular pathway (Reaction 2) accounts for the shortfall.
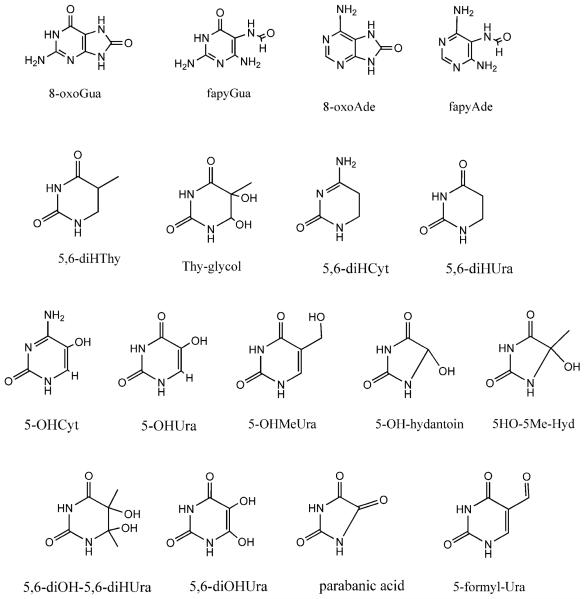
Figure 1.
Structural formulas and notation for products investigated by GC/MS of X-irradiated d(CTCTCGAGAG)2 films.
Figure 2.
A generalized scheme used to describe product formation by direct ionization of DNA. Reactions 1 & 5 describe the most commonly considered pathway in which parent molecule M forms a radical R that leads directly to product P (a trappable radical single-track pathway). Reactions 1,3, & 6 require a second one-electron reduction/oxidation, converting R to X (a trappable radical multi-track pathway). X is a non-radical intermediate that upon dissolving the film gives P. Reaction 2 is a molecular pathway in which X is produced directly with no trappable radical intermediate. Reaction 4 competes with the forward radical reactions either by going backward to parent (Z ≡ M) or by transferring the unpaired electron to a different site creating a new radical Z.
Under this reaction scheme, the dose response for product is described by
| [1] |
where the indexes of formal reaction rates kj match the reaction numbers in Figure 2.14 The chemical yield of product, G(P), is given by the slope of P vs. D at low dose
| [2] |
Equation 2 gave the following base damage yields: G(8-oxoGua) = 127 ± 160 nmol/J, G(8-oxoAde) = 4 ± 3 nmol/J, G(5,6-diHThy) = 39 ± 60 nmol/J, and G(5,6-diHUra) 111 ± 62 nmol/J. The yield for each of the products detected in d(GCACGCGTGC)2 was less than the yield of its presumed radical precursor; i.e., a shortfall was consistently observed.
The reaction model in Figure 2 predicts that base context should influence the distribution of base product yields. This is due to competition between Reaction 4 and product formation via Reactions 3 and 5, which depends on the rate of electron transfer to and from nearby bases. For example, if a hole is initially formed on Thy, transfer of that hole to a nearby Gua competes with forward reactions that give a damaged Thy. The goal of the work reported here was to determine whether or not base damage is influenced by base context. The sequence CTCTCGAGAG was chosen because it consists of a pyrimidine stretch and purine stretch; therefore, there is only one pyrimidine adjacent to a Gua. This contrasts with the previously studied GCACGCGTGC sequence in which all pyrimidines are adjacent to at least one Gua.
MATERIALS AND METHODS
Film preparation
The palindromic oligodeoxynucleotide, d(CpTpCpTpGpApGpApG), was purchased from Ransom Hill, where it was desalted by reverse phase chromatography. Without further purification, d(CTCTCGAGAG)2 was dissolved in Omnipur nuclease-free water (Merck) to give 5-7 mM of oligomer solution. The DNA concentration was determined by measuring the absorbance at 260 nm using a 20 μl aliquot diluted 50x. Films were prepared by drawing solution into silylated suprasil-quartz capillaries (1 mm o.d.) open at both ends.9 Evaporation inside a sealed container was governed by a much larger volume of saturated NaOH, presumed to maintain a relative humidity of 8% at room temperature. Clear pliable films reached steady state weights in 3-4 weeks. Initially, the oligomer solutions were dehydrated at 277 K by equilibrating at a relative humidity of 8% for 1 week and then further dehydrated at RT until a constant level of hydration was reached. The film weights, 200-250 μg, were measured with a Cahn C-30 Microbalance to an accuracy of ±1 μg. The DNA content of the films varied between 60-66 % across all sample sets; within any given data set (the data used for one dose response curve), the DNA content varied by 1-3%. Under this protocol, we assume that hydration level of DNA (Γ) is 2.5 mol water/mol nucleotide. 15 These ‘dry’ samples were weighed and rehydrated by equilibration, for a minimum of 3 weeks, against saturated solutions of KBr, which give a relative humidity of 84%. The samples were then reweighed to an accuracy of ±1 μg and Γ was calculated from the increase in weight. The film mass was 245-260 μg for the oligomers. The mass fraction of film consisting of DNA plus its solvation shell varies between 92 ± 3% at Γ of ~2.5 to 87 ± 4% at Γ of ~15; the remaining fraction is excess salt.
Irradiation
Films were X-irradiated using a Varian/Eimac OEG-76H tungsten-target tube operated at 70 kV at 20 mA through a 40 μm aluminum filter, giving a dose rate at the sample of 1.1 kGy/min.12 For electron paramagnetic resonance (EPR) spectroscopy, samples were irradiated at 4 K at a dose rate of 0.75 kGy/min and spectra recorded at 4 K.16 For GC/MS analysis, samples were irradiated at RT at a dose rate of 1.1 kGy/min, then prepared for GC/MS by dissolving in nuclease free water at a ratio of 1:1 (mass/volume), and stored at −20°C.
GC/MS
DNA hydrolysis, derivatization, and data acquisition follow closely the methods described previously.14,17 Key steps are given here. DNA hydrolysis was performed on freeze-dried samples prepared with a known addition of internal standards (6-azathymine, 8-azaadenine, thymine-α,α,α,6-d4, diHUra-U-13C-U-15N, diHThy-α,α,α,5,6,6-d6, 5-OHMeUra-α,α-d2-1,3-15N, 5-OHCyt-2-13C-1,3-15N, Thy-glycol-α,α,α,6-d4, 8oxoAde-8-13C-6,9-15N2, 8oxoGua-4,5,6,8-13C4, fapyAde-formyl-13C-diamino-15N2, fapyGua-4,5,6-13C3-[5-formyl-15N]) to d(CTCTCGAGAG)2. Hydrolysis was by 88% formic acid for either 90 min or 30 min, the longer time was used to detect primarily 8-oxoGua, 8-oxoAde, diHUra, diHThy and the shorter time was used to detect primarily fapyGua and fapyAde. Subsequently, the samples were freeze-dried and then held under vacuum for 5 or more days. Hydrolyzed d(CTCTCGAGAG)2 was trimethylsilylated by adding 120 μl of degassed N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS)/5:1 acetonitrile+pyridine/butanethiol (10:4:1) to the freeze dried samples and then holding the samples under dry nitrogen gas (O2 < 100 ppm) in a sealed chamber at 22 °C for 60 minutes.
It is important to note that quantification of each product requires careful selection and testing of an internal standard specific to that product. In addition, there are most likely DNA lesions that cannot be revealed by GC/MS. These constraints limited the current study to the 16 products listed above.
GC/MS, working under selected ion monitoring (SIM), was used to quantify base products. The instrument was the same one as used previously, a Hewlett-Packard 5890B GC with a Hewlett-Packard 5970B MSD. See reference 17 for instrument settings and run conditions. Base damage products were quantified using isotope dilution methods. A careful calibration scheme minimized artifacts that arise during hydrolysis and derivatization.
In order to test the quantitative accuracy of the three major base products (8-oxoGua, diHUra, and diHThy) known amounts of the authentic product where added to d(CTCTCGAGAG)2 and then analyzed by the procedure described above in analyzing X-irradiated d(CTCTCGAGAG)2. The recovery of the authentic compounds was 97±9 % for 8-oxoGua , 117±14 % for diHUra, and 111±15 % for diHThy, where the standard deviations were based on 13 measurements. These recovery values were used to adjust the measured yields of product. For example, the yield of 8-oxoGua was reported in Table 1 as 1.03x (1.0/0.97) the measured yield.
Table 1.
Product yields for d(CTCTCGAGAG)2 films, with hydration levels of Γ = 2.5 and 15 waters/nucleotide, X-irradiated at RT. ki were calculated by a non-linear least squares fit, of the data in Figures 2-3, to Equation 1. G(P) = k1 + k2. G’(P) were calculated by a linear least squares fit to the data at doses < 13 kGy
| d(CTCTCGAGAG)2 | Γ=2.5/Γ=15 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Product (P) | k1 | k2 | k3 | k4 | R- value |
G(P) | G’(P) | sd | Ratioa | sd |
| Γ = 2.5 b | nmol/J | nmol/J | MGy−1 | MGy−1 | nmol/J | nmol/J | ||||
| 8-oxoGua | 43 | 4 | 82 | 100 | 0.93 | 47 | 36 | 2 | 0.7 | 0.2 |
| 8-oxoAde | 8 | 1 | 40 | 608 | 0.91 | 10 | 3.1 | 0.6 | 0.5 | 0.1 |
| 5-OHMeUra | 23 | 2 | 48 | 208 | 0.79 | 21 | 11 | 3 | 2.3 | 4.0 |
| 5,6-diHUra | 256 | 1 | 158 | 177 | 0.97 | 258 | 175 | 48 | 3.4 | 1.2 |
| 5,6-diHThy | 20 | 7 | 51 | 1000 | 0.77 | 28 | 6 | 4 | 3.5 | 3.3 |
| 5-OHCyt | 35 | 12 | 10 | 150 | 0.90 | 47 | 48 | 5 | 1.2 | 0.2 |
| 5-OHUra | 1 | 0 | 12 | 65 | 0.81 | 2 | 0.8 | 0.2 | 0.4 | 0.2 |
| Γ = 15 c | ||||||||||
| 8-oxoGua | 45 | 8 | 10 | 101 | 0.90 | 54 | 53.1 | 17.5 | ||
| 8-oxoAde | 14 | 1 | 22 | 303 | 0.96 | 15 | 6.1 | 0.8 | ||
| 5-OHMeUra | 6 | 7 | 1000 | 0 | 0.96 | 13 | 4.5 | 7.7 | ||
| 5,6-diHUra | 37 | 35 | 844 | 0 | 0.98 | 72 | 51.8 | 11.9 | ||
| 5,6-diHThy | 3 | 3 | 0 | 3 | 0.93 | 6 | 1.8 | 1.3 | ||
| 5-OHCyt | 82 | 7 | 9 | 274 | 0.93 | 89 | 40.7 | 2.5 | ||
| 5-OHUra | 2 | 0 | 15 | 154 | 0.96 | 2 | 1.7 | 0.3 | ||
Standard deviations (±1 sd) for the ratios were calculated using (A ±a)/(B ±b) = (D ±d), where A/B = D, then d = D * [(a/A)2 + (b/B)2]1/2.
Standard deviations were based on three independent data sets fit simultaneously.
Standard deviations were based on one data set.
Calculation of Yields
The chemical yields were based on a target mass consisting of the DNA and one counter ion plus the number of water molecules/nucleotide. The counter ion was NH4+ and the remainder of the film mass was assumed to be excess salt and, as such, was assumed not to be part of the target mass.
RESULTS
Applying GC/MS to damage formation in irradiated films of d(CTCTCGAGAG)2, 16 products were monitored: 8oxoGua, 8oxoAde, diHUra, diHThy, 5HO-Cyt, 5HO-Ura, 5HOMeUra, fapyGua, fapyAde, Thy-gly, 5HO-5Me-Hyd, 5HO-5,6diHThy, 5HOMe-Cyt, parabanic acid, 5HO-Hyd, 5OH-5Met-Hyd, and 5-formyl-Ura (see Figure 1). Of these, the chemical yields for the first 7 were found to be statistically significant. For these 7 products, the dose response curves are given in Figure 2 for Γ 2.5 films and in Figure 3 for Γ 15 films. The yields were calculated by two approaches.
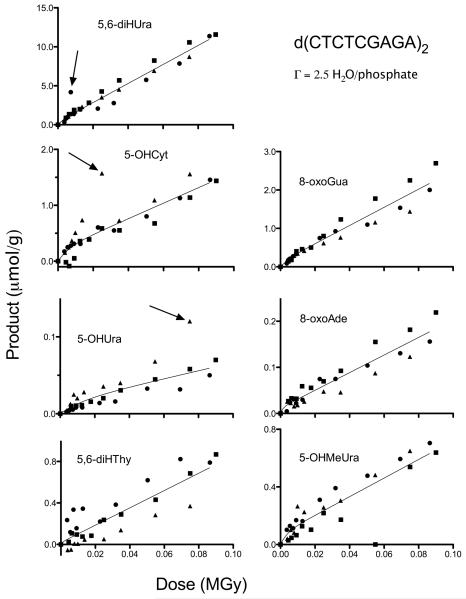
Figure 3.
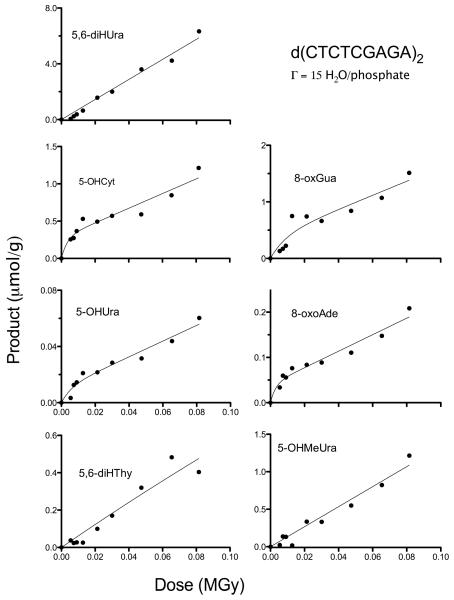
Dose response data for 7 products measured in d(CTCTCGAGAG)2 films hydrated to Γ = 2.5, X-irradiated at RT, dissolved in water, and analysed by GC/MS. Circles, squares, and triangles show data from three separate experiments. The curves were generated by a non-linear least squares fit to Equation 2 with the four parameters (k1-k4 values given in Table 1) shared by the three data sets, constrained to > 0, and in one case (5,6-diHThy) k4 was constrained to ≤ 1000 MGy−1. Three data points, indicated by arrows, were determined to be outliers and excluded from the respective fits.
The first approach used a non-linear least squares fit of the data to Equation 1, giving parameters k1, k2, k3, and k4. The ki values are given in Table 1 along with the R-values for the fit. As can be seen from the R-values, the fit is reasonable. The covariance between the parameters, however, is very large. Therefore, the fit shows only that the model is consistent with the data. One way to resolve the covariance problem would be to obtain independent measures of k2 and k3 by determining the yields of R using EPR spectroscopy. While this is feasible, it was beyond the scope of this work.
The second approach was to determine the initial slope of the dose response curves using linear least-squares fit to the low dose data (<13 kGy). We call the yields measured by this approach G’(P). While not the true chemical yield, G’(P) provides the minimum value with a standard error. The degree to which G’(P) underestimates the product yield can be gauged by a comparison with the values of G(P) calculated using Equation 2 and given in Table 1.
The reproducibility the GC/MS data was surprisingly good over 3 replicate experiments at Γ 2.5. Only one experiment was completed at Γ 15 (a consequence of irreparable GC/MS equipment failure). Under a cautionary note that the standard errors for the Γ 15 data were based on one set of dose response curves, the ratios of yields for Γ 2.5/Γ 15 are given in Table 1.
In order to compare the yields from d(CTCTCGAGAG)2 with those from d(GCACGCGTGC)2, the yields need to be normalized to the same GC content. For both oligomers, the yields projected to a CG/AT ratio of 1 are given in Table 2. For d(GCACGCGTGC)2, it was necessary to revisit our data in order to select data from just the experiments conducted under air and calculate values of G’(P) from the initial response at low dose (as done here for d(CTCTCGAGAG)2). In addition, we report here the yields of 5-OHMeUra, 5-OHCyt, and 5-OHUra for d(GCACGCGTGC)2, which given the low yields and large percent errors in G(P) were not reported earlier. In Table 2, it can be seen from the ratio, G’(P) for d(CTCTCGAGAG)2 divided by G’(P) for d(GCACGCGTGC)2, that the distribution of base damage changes substantially. The yields increase for 8-oxoAde, 5-OHMeUra, 5,6-diHUra, and 5-OHCyt, they decrease for 5,6-diHThy, and they are about the same for 8-oxoGua and 5-OHUra.
Table 2.
Product yields (nmol/J) projected to a CG/AT ratio of 1
| d(CTCTCGAGAG)2 Γ=2.5, under air |
d(GCACGCGTGC)2 Γ=2.5, under air |
d(CTCTCGAGAG)2/ d(GCACGCGTGC)2 |
||||
|---|---|---|---|---|---|---|
| Product (P) | G’(P) a | sd | G’(P) b | sd | Ratio c | sd |
| 8-oxoGua | 30.4 | 1.8 | 24.3 | 18.0 | 1.3 | 0.9 |
| 8-oxoAde | 3.9 | 0.7 | 2.2 | 0.1 | 1.8 | 0.3 |
| 5-OHMeUra | 13.1 | 3.6 | 8.5 | 2.3 | 1.6 | 0.6 |
| 5,6-diHUra | 145.5 | 40.0 | 12.8 | 3.9 | 11.4 | 4.7 |
| 5,6-diHThy | 8.0 | 4.9 | 43.7 | 4.6 | 0.2 | 0.1 |
| 5-OHCyt | 39.6 | 4.5 | <0.1 | 1.4 | >28d | NAe |
| 5-OHUra | 0.6 | 0.2 | 0.8 | 1.1 | 0.8 | 1.1 |
Calculated from values given in Table 1.
Calculated using data from our earlier study17 using a linear least squares fit to the data at doses < 33 kGy and confined to experiments conducted under air; there were 1 to 3 experiments per G’(P).
Standard deviations (±1 sd) for the ratios were calculated using (A ±a)/(B ±b) = (D ±d), where A/B = D, then d = D * [(a/A)2 + (b/B)2]1/2.
Calculated using + 1 sd: 39.6/1.4 = 28.
Not applicable because the formula for sd approaches infinity as either value of G’ approaches zero.
DISCUSSION
We discuss here how the general scheme presented in Figure 2 can be used to explain the formation of each of the observed products, their relative yields, and the influence of sequence on their relative yields. But before doing so it is important to draw a clear distinction between reactions occurring in the solid state as opposed to those that occur upon sample solvation.
Reactions before and after sample solvation
In the solid state prior to dissolving the DNA films in water, the majority of the free radical reactions have progressed to non-radical intermediates or end product. This is supported by the observation that the total yield of products exceeded the yield of free radicals that remain in the samples at room temperature prior to dissolution. The total yields of product in films of d(CTCTCGAGAG)2 and d(GCACGCGTGC)2 are given in the penultimate row of Table 3. Included in these totals are the previously measured yields of free base release,13,14 a consequence of electron loss from the DNA backbone. The yield of total measured product ranged from 191 to 366 nmol/J. While we have not measured the free radical yields for these specific films at RT, the yield of radicals in crystalline GCACGCGTGC were reported as 0 nmol/J.18 Typically yields of free radicals in crystals and films of DNA irradiated at RT have been found to be around 100 nmol/J and not greater than 200 nmol/J. The fact that product yield exceeds the yield of radicals trapped at room means that non-radical intermediates (or end product) must be present in the solid state prior to dissolution. In the solid phase, therefore, some free radical reactions must have progressed to give non-radical intermediates. Only the relatively small concentration of radicals, remaining in the solid phase, goes forward to create products via radical reactions promoted by dissolution.
Table 3.
Balance between products initiated by one-electron oxidation and one-electron reduction. The sums of base product yields (nmol/J) are calculated from the G’(P) in Table 2
| d(CTCTCGAGAG)2 | d(GCACGCGTGC)2 | ||||||
|---|---|---|---|---|---|---|---|
| Line # | Ionization event | Γ=2.5 | Γ=15 | Γ=2.5 | |||
| G’(P) | ±1 sd | G’(P) | ±1 sd | G’(P) | ±1 sd | ||
| 1 | base reduction a | 229 | 48 | 96 | 12 | 57 | 6 |
| 2 | base oxidation b | 50 | 4 | 64 | 19 | 5 | 18 |
| 3 | free base release c | 87 c | 5 | 152 d | 27 | 93 e | 15 |
| 4 | total oxidation f | 137 | 6 | 216 | 33 | 98 | 24 |
| 5 | total ox + red g | 366 | 49 | 312 | 36 | 191 | 24 |
| 6 | % oxidation h | 37 | 5 | 69 | 13 | 51 | 14 |
Sum of G’(P) for 5,6-diHUra, 5,6-diHThy, 5-OHCyt, and 5-OHUra.
Sum of G’(P) for 8-oxoGua, 8-oxoAde, and 5-OHMeUra.
Ref 13
Calculated by multiplying G’(P) (line 3 under Γ = 2.5) by 1.75 ± 0.30. This enhancement factor, which is due to increasing Γ to 15, is estimated from studies on 4 different DNA samples 8 and unpublished data.
Ref 14
Sum of lines 2 and 3
Sum of lines 1 and 4
Line 4 divided by line 5 times 100
Damage pathways, whereby radical reactions are complete in the solid phase, have received little consideration for chemistry initiated by low LET radiation. The prevailing assumption is that each end product can be assigned to a single radical event within the solid state. But that assumption proves problematic when considering how to explain, for example, the dependence of free base release on base sequence.8 If fbr is assumed to be only due to one-electron oxidation of the deoxyribose-phosphate backbone of DNA, it is hard to find a mechanism that adequately explains the magnitude and variation in yields for release of each of the four bases. As mentioned in the introduction, results on sb and fbr yields lead us to propose the model encapsulated by the scheme in Figure 2. This working model is extended beyond backbone damage to include the base damage products observed in d(CTCTCGAGAG)2 and d(GCACGCGTGC)2.
8-oxoGua
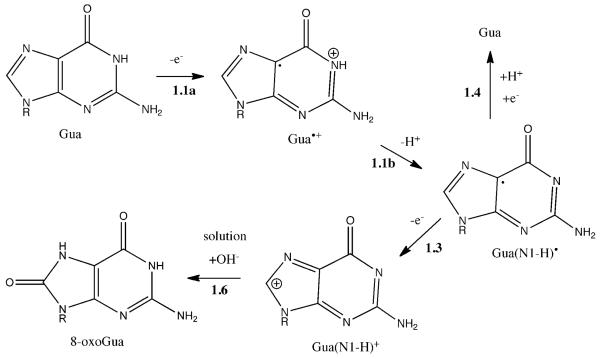
Of the four bases, Gua is considered to be the most probable site for one-electron oxidation. This is based on the order of the base oxidation potentials, Gua < Ade < Thy < Cyt, particularly when the bases are incorporated within duplex DNA.19-21 Contributing to the smaller oxidation potential of Gua is the driving force provided by deprotonation of the radical cation Gua•+ giving Gua(N1-H)•.22 Indeed, this neutral radical is trapped when DNA constituents containing Gua are irradiated at 10K.23,24 Gua(N1-H)• is unstable at higher temperatures, decaying in the 120-230K range. 25,26 At RT, no radicals assignable to the one-electron oxidation of Gua have been observed. Therefore, either 8-oxoGua is already formed in the solid state or, what we believe is occurring, a non-radical precursor is formed that yields these products upon dissolution.
At low hydration levels, ~2.5 waters per nucleotide, the water is tightly bound to phosphate. It is improbable that this water has the mobility at < 200K for OH− to react with Gua•+ or Gua(N1-H)•. But if it did so, it would result in radicals of the type Gua(C8+OH)• or a rearrangement thereof.27 And the yield of Gua(C8+OH)• would have to be sufficiently large to account for the yield of 8-oxoGua. Given 8-oxoGua yields of 40-120 nmol/J, such a radical should be readily discernable. Yet there is no evidence that this type of radical is formed in DNA films at the hydration levels employed here or in other types of samples related to DNA constituents with low water content. The 8-oxoGua precursor, trapped in the solid state at RT must be primarily or exclusively a non-radical.
By our model the radical cation, Gua•+, deprotonates (even at 4 K) to give the neutral radical Gua(N1-H)•. Subsequent one-electron oxidation Gua(N1-H)• gives the carbocation, Gua(N1-H)+. These three reactions, labeled 1.1a, 1.1b, and 1.3, respectively, are shown in Scheme 1. Because these reactions have gone to completion at RT in the solid phase, there are no radical precursors present in the films at room temperature. When the film is dissolved Reaction 1.6 gives 8-oxoGua by OH− addition to Gua(N1-H)+.
Scheme 1.
Proposed reaction pathway for 8-oxoGua formation by the direct effect. The reactions are label, e.g. 1.3, such that the 1 refers to Scheme 1 and the 3 refers to Reaction 3 of Figure 2. In the cases where more steps or pathways exist than in the general scheme shown in Figure 2, a lower case letter is added, e.g., 1.1a and 1.1b refer to two steps contained within Reaction 1 of Figure 2. In the Schemes that follow this same reaction numbering system is used.
Possible oxidants, for oxidation the Gua(N1-H)• radical, are radical cations formed on nearby phosphate or deoxyribose. While thermodynamically less favorable the pyrimidine radical cations are also worth considering. Because the redox properties of these radicals would be very difficult to measure, this is an area ripe for the application of computation chemistry. On the other hand, if the second oxidation occurs within the ionization spur, simple equilibrium thermodynamics will not apply and formation of the carbocations would be controlled by kinetics.
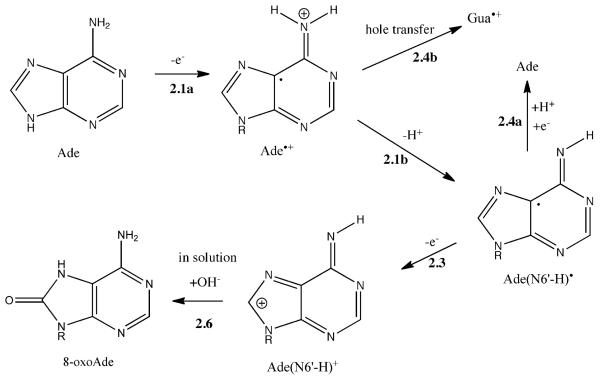
8-oxoAde
One-electron oxidation of Ade gives Ade•+, subsequent deprotonation yields Ade(N4-H)•,28,29 and a second one-electron oxidation yields the carbocation Ade(N4-H)+. In Scheme 2, these reactions are 2.1a, 2.1b, 2.3, respectively. Dissolution makes reaction 2.6 possible, giving 8-oxoAde by OH− addition to the carbocation. The reaction sequence is analogous with that for Gua except for two important differences. One is that deprotonation of Gua•+ is facilitated by the proton accepting power of its conjugate base, Cyt, where proton transfers across the N1-H…N3 hydrogen bond gives the Cyt(N3+H)+ cation.22 In the case of Ade•+, Thy is a relatively poor proton acceptor;22 therefore, the proton lost via Reaction 2.1b may be by transfer to either water in a Γ = 15 film or whatever proton acceptor is coordinated with the NH2 group in “dry” (Γ = 2.5) DNA. This suggests that hydration of DNA would be expected to increase the yield of 8-oxoAde relative to 8-oxoGua. As can be seen from the ratio in Table 1, it increased about 2 fold. The other difference is that hole transfer from Ade to Gua is thermodynamically favorable, Reaction 2.4b in Scheme 2. But the CTCTCGAGAG sequence, which should favor transfer of holes from Ade to Gua, gave higher yields of 8-oxoAde (Table 2). This suggests that the formation rate of Ade(N4-H)• is significantly faster than that of the rate of hole transfer to an adjacent Gua. As pointed out by Steenken, rates of deprotonation, ~1013 s−1, are fast enough to compete with rates of hole transfer, ~109-1014 s−1.30
Scheme 2.
Proposed reaction pathway for 8-oxoAde formation by the direct effect. See Scheme 1 for the reaction numbering system.
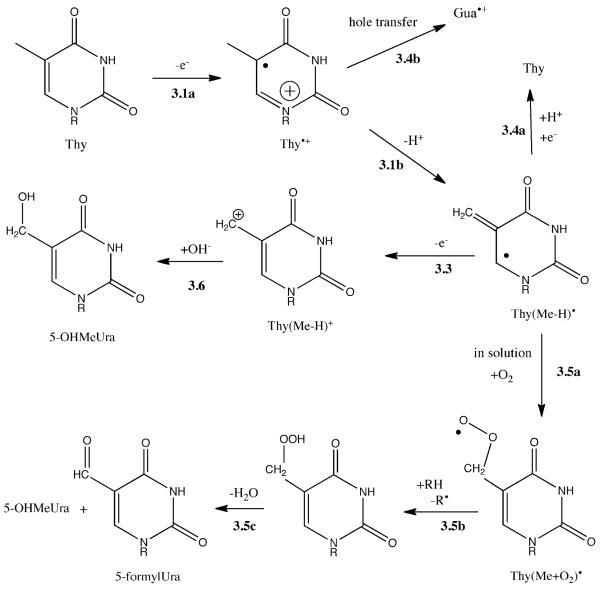
5-OHMeUra
From earlier work,31,32 it is known that the radical cation of thymine, Thy•+, is trapped at 4K in poly(AT) and poly(A):poly(T). When the samples were annealed from 77K to RT, the radical cation decayed, and the EPR signal of Thy(Me-H)• appeared. This conversion is consistent with deprotonation at the methyl group as shown by Reaction 3.1b in Scheme 3. Evidence of this conversion was first observed by Sevilla and Engelhardt33 and formation of Thy(Me-H)• in DNA was reported by Weiland and Hüttermann.34 This allylic radical is known to be extraordinarily stable, persisting in 1MeThy crystals up to 200°C.35 Given its distinctive five-line EPR signature,32 this radical is not difficult to detect but it has not been observed at room temperature in DNAs containing a mix of all four bases. By inference, this means that at temperatures approaching room temperature a significant fraction of the Thy(Me-H)• radical has either gained or lost an electron. If it gains an electron, subsequent protonation reverses the damage, reforming Thy (Reaction 3.4a). If it looses an electron, a carbocation is formed, Reaction 3.3. This non-radical precursor, having accumulated in the solid phase, would react in water to give the observed end product, 5-OHMeUra.
Scheme 3.
Proposed reaction pathway for 5-OHMeUra formation by the direct effect. See Scheme 1 for the reaction numbering system.
If a fraction of the Thy(Me-H)• radical persists at room temperature upon dissolving the films, the most likely reaction is oxygen addition, Reaction 3.5a, to form the peroxy radical Thy(Me+O2)•. Subsequent decomposition would lead to 5-formylUra and 5-OHMeUra.
Competing with deprotonation of Thy•+ is hole transfer to Gua (Reaction 3.4b), forming the energetically more favorable Gua•+ radical cation. When Thy is more distant from Gua, the rate of hole transfer to Gua will be slower making deprotonation at the methyl more competitive. This is consistent with previously observed yields of Thy(Me-H)• and Thy•+ in the AT polymers.31,32 It also explains the greater yield of 5-OHMeUra in d(CTCTCGAGAG)2 compared to d(GCACGCGTGC)2 (see ratio in Table 2) by the changes in distance between Thy and Gua.
5-OHCyt
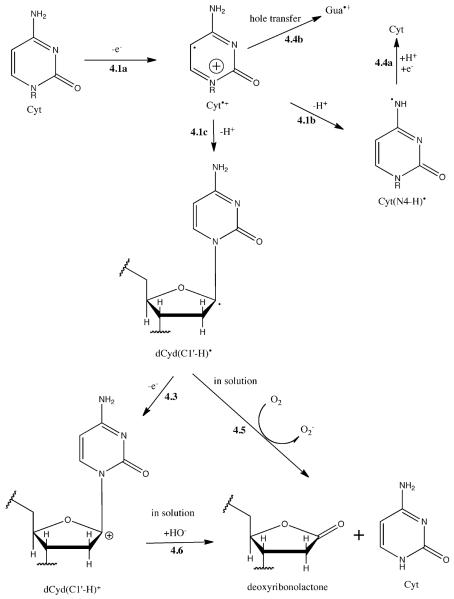
One might consider the Cyt•+ radical as a precursor to formation of the products 5-OHCyt and 5-OHUra. For the following reasons, we suggest that this is not the case.
In order to fix oxidative damage at Cyt, deprotonation of Cyt•+ must compete with hole transfer (Reaction 4.4b) from Cyt to Gua. The two sites of deprotonation are shown in Scheme 4, Reaction 4.1b giving Cyt(N4-H)• and Reaction 4.1c giving the neutral deoxyribose radical dCyd(C1′-H)•. The dCyd(C1′-H)• radical was proposed as a key intermediate in the release of free cytosine.8 The Cyt(N4-H)• radical was observed in crystalline cytosine at 10K36 but has not been observed in solid state DNA. A second oxidation of dCyd(C1′-H)• (Reaction 4.3) is predicted to be favorable,37 which would result in a carbocation precursor that upon sample dissolution would lead to the release of free Cyt (Reaction 4.6). If a fraction of the dCyd(C1′-H)• radicals persist at room temperature, oxidation by O2 will occur following dissolution and subsequent hydration of the carbocation will lead to the same products (Reaction 4.5).
Scheme 4.
Reaction pathway proposed to explain why one-electron oxidized Cyt gives rise to deoxyribonolactone53 and free Cyt but not 5-OHCyt. See Scheme 1 for the reaction numbering system.
Further, if either Cyt•+ or Cyt(N4-H)• are precursors to 5-OHCyt, then the yield of 5-OHCyt should have increased when the hydration of the d(CTCTCGAGAG)2 films was increased from Γ = 2.5 to 15. The lack of change (ratio in Table 1) supports the proposal that 5OH-Cyt and 5OH-Ura were not derived from the cytosine radical cation.
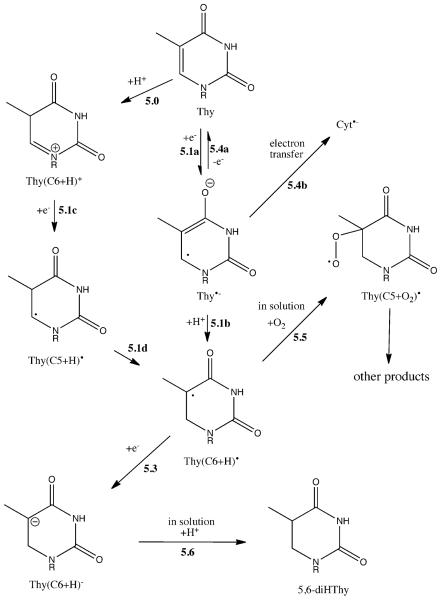
5,6-diHThy
One-electron reduction of Thy yields Thy•− (Reaction 5.1a), a radical anion that is readily trapped in DNA at temperatures < 90K.38-40 At temperatures above ~140K, Thy•− protonates at C6 (Reaction 5.1b) to give Thy(C6+H)•.41,42 Upon warming to RT, or when irradiation occurs at RT, we propose that a fraction of this neutral radical is one-electron reduced, Reaction 5.3. The carboanion, Thy(C6+H)−, would then be trapped in the solid state. Upon dissolution, protonation of Thy(C6+H)− , Reaction 5.6, yields the observed end product 5,6-diHThy. The fraction of Thy(C6+H)• that escapes one-electron reduction would be stable even at room temperature.43 Upon dissolution, this fraction would react with oxygen, Reaction 5.5, giving products that were not targeted in our study. To the degree that such products were formed, products assignable to the reduction side of the oxidation-reduction balance sheet would be missing.
Generally, it is important to realize that, for every excess electron generated by ionization, a radical cation deprotonates. This must be the case because all of the radical cations produced in DNA deprotonate44 and, with the possible exception of Thy,32 do so even at 4K. When the deprotonation is from a carbon site, as it is for radical cations located on the DNA backbone, cytosine and thymine, the radiation-released proton must attach to a nearby site that favors proton addition. We illustrate this point with the possibility of proton addition to C5 of Thy, giving the Thy(C5+H)+ carbocation shown in Scheme 5. (This Reaction is labeled 5.0, using 0, because an analogous reaction is not shown in Figure 2.) Thy(C5+H)+ would be a highly favorable electron trapping site; when reduced, the Thy(C5+H)• radical would be formed (Reaction 5.1c). Thy(C5+H)• is not thermally stable, converting to Thy(C6+H)• (Reaction 5.1d) with an activation energy of 96.7 ± 2.4 kJ/mol.45 While there is no evidence of the Thy(C5+H)• radical trapped in DNA at low temperatures, this does not rule out this pathway when DNA is irradiated at room temperature. As discussed below for the case of Cyt, there is evidence that a “protonation first” pathway plays a role.
Scheme 5.
Proposed reaction pathway for 5,6-diHThy formation by the direct effect. See Scheme 1 for the reaction numbering system.
Because Cyt is an energetically more favorable electron trapping site than Thy 22,40, one should expect competition between the rate of electron transfer from Thy•− to Cyt (Reaction 5.4b) and the rate of protonation at C6 (Reaction 5.1b). This competition offers an explanation for the lower yield of 5,6-diHThy in d(CTCTCGAGAG)2 relative to d(GCACGCGTGC)2 as seen from the ratio of 0.2 ± 0.1 in Table 2. In d(CTCTCGAGAG)2, Cyt blocks electron transfer to Thy from both the 3′ to 5′ direction and 5′ to 3′ direction.
It is also important to note that 5,6-diHThy is a major product reported for TpTpT exposed to low energy electrons (LEE)46. The suggested reaction pathway is by hydride addition to the 5,6 double bond of thymine followed by protonation. The hydride ion arises from dissociative electron attachment (DEA). In order for this reaction to account for a significant fraction of the 5,6-diHThy yield observed in our system, a comparable yield of radicals created from the same DEA event is required. To date, there is no EPR evidence of such residual radicals in either amorphous or crystalline DNA; this suggests that DEA by LEE is not likely to account for a major fraction of the 5,6-diHThy yield observed under our conditions.
diHUra, 5OH-Cyt, and 5OH-Ura
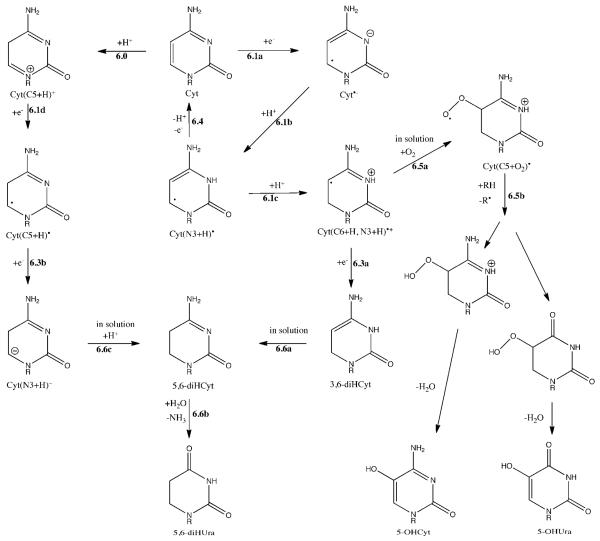
The Cyt radical anion, Cyt•−, behaves quite differently than Thy•−. In crystalline derivatives of Cyt and duplex DNA, Cyt•− has not been observed. Even at 10K it is protonated at N3,47, shown by Reaction 6.1b in Scheme 6. The resulting neutral radical Cyt(N3+H)•, as best explained by Steenken,22 is the most favorable electron trapping site in DNA because of proton transfer from N1 of Gua to N3 Cyt. Another difference between Thy and Cyt is that protonation at C6 occurs in addition to protonation at N3 (Reaction 6.1c), giving the radical cation Cyt(C6+H, N3+H)•+. This radical was formed at 4K and the dose response was linear at low dose, indicating that it is formed by single-track chemistry.48,49 The formation of Cyt(C6+H, N3+H)•+ is thus explained by Reactions 6.1a, 6.1b, and 6.1c. If Cyt(C6+H, N3+H)•+ captures an electron, Reaction 6.3a, 3,6-diHCyt forms. Upon dissolution, rearrangement (6.6a) would give 5,6-diHCyt. Subsequently, deamination would give 5,6-diHUra (6.5a and 6.5b).
Scheme 6.
Proposed reaction pathway for 5,6-diUra, 5-OHCyt, and 5-OHUra formation by the direct effect. As for 6-OHCyt, it was not one of the 16 products monitored in this work. See Scheme 1 for the reaction numbering system.
The concentration of Cyt(C6+H, N3+H)•+ does not increase upon annealing and it is known to be relatively stable at room temperature. 48,49 Therefore, some fraction of this radical may persist until the DNA is dissolved. That fraction would react with O2 to give the peroxyl radical (Reaction 6.5a), which would abstract hydrogen (Reaction 6.5b). Subsequent loss of water yields the observed products 5-OHCyt and 5-OHUra. It was suggested by Swarts, et al., in their study products in lyophilized herring sperm DNA, that this type of reaction could account for the unexpected low yields of reduction products such as dihydropyrimidines.17 Deamination during the course of Reactions 6.5a and 6.5b could explain the small amount of 5-OHUra. We think it likely, therefore, that these two products, 5-OHCyt and 5-OHUra are “crossover” products, i.e., they are initiated by one-electron reduction but terminated by oxidation.
Debije et al. also observed formation of a radical due to a net gain of hydrogen at C5 of Cyt.48,49 In this case, the EPR spectra could not distinguish between a structure of Cyt(C5+H)• or Cyt(C5+H, N3+H)•+. We favor the former neutral radical because it nicely fits our model. If a radiation released proton attaches to C5 (Reaction 6.0), subsequent electron capture yields the Cyt(C5+H)• radical (Reaction 6.1d). The concentration of Cyt(C5+H)• did not increase upon annealing. If conversion of Cyt(N3+H)• to either Cyt(C5+H)• or Cyt(C6+H, N3+H)• occurred, it must have been offset by a comparable level of loss. Loss would occur, e.g., by transfer of an excess electron from either Thy•− or Cyt(N3+H)•. One-electron reduction of Cyt(C5+H)• gives Cyt(C5+H)− via Reaction 6.3b. This anion, resulting from the two-consecutive one-electron reductions of Cyt, would be another precursor, which upon dissolution of the DNA film would give 5,6-diHCyt (Reaction 6.6c) and subsequently the detected product of 5,6-diHUra (Reaction 6.6b).
Cyt(C5+H)•, just like Cyt(C6+H, N3+H)•+, was relatively stable.48,49 For example, in crystalline d(CGCGAATTCGCG)2 (which is in B conformation), the total radical population decreases by about 60-76% upon warming from 4K to 240K . At 4K, Cyt(C6+H, N3+H)•+ and Cyt(C5+H)• account for ~30% of the total radical population. Upon warming to 240K, this fraction increases to ~60%. About half of the other ~40% is accounted for by Thy(C6+H)•. Therefore, Cyt(C6+H, N3+H)•+ and Cyt(C5+H)•, in addition to Thy(C6+H)•, are expected to persist until the films are dissolved. In the case of Cyt(C6+H, N3+H)•+, the dominate reaction in oxygenated solutions will be O2 addition (Reaction 6.5a) giving the peroxy radical Cyt(C5+O2)•. The peroxyl radicals abstract hydrogen to form the peroxide (Reactions 6.5b). We speculate that the reaction mechanism is like Reaction 3.5c in thymine, where the 5-peroxide of cytosine leads to 5-OHCyt, 5-OHUra, and other products.
Looking at the ratios in Table 1, we see that the yield of 5,6-diHUra (and perhaps 5-OHCyt) decreases when the hydration increases. The more striking result, however, is the order of magnitude increase in 5,6-diHUra and 5-OHCyt when comparing d(CTCTCGAGAG)2 with d(GCACGCGTGC)2, both at Γ = 2.5. Complementing this increase was a large decrease in the yield of 5,6-diHThy. We posit that Thy competes poorly for the excess electron when it is flanked by two Cyt compared to when it is flanked by two Gua. In addition, electrons diffusing out of the purine stack, GAGAG, first encounter Cyt, giving Cyt a positional advantage. Increasing hydration should inhibit transfer of the excess electron and reduce the preference for trapping by Cyt. This would be consistent with observations that radical yields increase with increasing hydration,50,51 a phenomenon that is likely related to the improved dielectric relaxation around the sites of electron loss and electron gain.
Balance between oxidation and reduction
Under the assumption that 8-oxoGua, 8-oxoAde, 5-OHMeUra, and fbr are products derived from an initial one-electron oxidation and that 5,6-diHThy, 5,6-diHUra, 5-OHCyt, and 5-OHUra are products derived from an initial one-electron reduction, the balance between these two families of products was calculated. Based on the results given in Table 3, there is reasonable balance. Interestingly, for d(CTCTCGAGAG)2 at Γ = 2.5 the family of products assigned to reduction is at least comparable to, and perhaps greater than, the family assigned to oxidation. This differs from prior observations on reduction/oxidation balance.17
Implication regarding charge transfer
When trying to understand the distribution of products derived from holes created by the direct effect of ionizing radiation, caution is required when extrapolating from experiments in which holes are injected by either photolytic techniques or chemical oxidants. Photolysis and gentle oxidants, like azide,52 are selective toward one-electron oxidation of Gua. The ensuing chemistry excludes the pyrimidines and is biased against Ade. But with ionizing radiation, holes are formed initially on all the bases. The fact that hole transfer from the other three bases to Gua is thermodynamically favorable does not mean it is kinetically favorable. The results presented here support a model in which the deprotonation rates for the radical cations of Thy, Cyt, and Ade compete favorably with the rates for hole transfer to Gua.
The rate of methyl deprotonation by the thymine radical cation competes with the rate of hole transfer from Thy to Gua; when Gua is not adjacent to Thy, deprotonation by Thy •+ is amplified. This observation is evidence that the rate of hole transfer to an adjacent base is comparable to the rate of methyl deprotonation by Thy •+.
The yield of 5-OHMeU is comparable to 8-oxoA. This is evidence that the hole transfer rate, from Thy to its base-paired Ade, is slower than the methyl deprotonation rate. Given #2 above, this indicates that the interstrand hole-transfer rate for Thy to Ade is slower than the intrastrand rate for hole transfer from Thy to Gua.
Even though Ade is sandwiched between two Gua, 8-oxoAde is formed. Therefore, the deprotonation rate of Ade is relatively fast compared to the Ade-to-Gua hole-transfer rate and this holds even when Gua is adjacent to Ade.
Thy is guarded against reduction when flanked by Cyt and/or Cyt preferentially captures electrons when they diffuse beyond the GAGAG purine track. This has two implications. i) The excess electron is relatively localized; it does not hop over Cyt. And, ii) the rate of intrastrand excess electron transfer is greater than the rate of interstrand transfer.
Figure 4.
Dose response data for Γ = 15 is shown as described in Figure 3 but with two differences: only one data set was fit and in one case (5-OHMeUra) k3 was constrained to ≤ 1000 MGy−1.
ACKNOWLEDGEMENTS
The authors thank Kermit R. Mercer for his excellent technical support, Richard J. Wagner for input on reaction mechanisms, and Paul J. Black for his careful editing of this manuscript. The project described was supported by Award Number R01CA032546 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
REFERENCES
- (1).Becker D, Adhikary A, Sevilla M. Mechanisms of Radiation-Induced DNA Damage: Direct Effects. In: Wishart JF, Rao BSM, editors. Recent Trends in Radiation Chemistry. World Scientific Publishing Co. Pte. Ltd.; Singapore: 2010. p. 509. [Google Scholar]
- (2).Von Sonntag C. Radiation-Induced DNA Damage: Indirect Effects. In: Wishart JF, Rao BSM, editors. Recent Trends in Radiation Chemistry. World Scientific Publishing Co. Pte. Ltd.; Singapore: 2010. p. 543. [Google Scholar]
- (3).Bernhard WA, Close DM. DNA damage dictates the biological consequence of ionizing radiation: the chemical pathways. In: Mozumder A, Hatano Y, editors. Charged particle and photon interactions with matter. Marcel Dekker; New York: 2003. p. 471. [Google Scholar]
- (4).Sevilla MD, Becker D, editors. ESR studies of radiation damage to DNA and related biomolecules. Vol. 19. The Royal Society of Chemistry; Cambridge, UK: 2004. p. 243. [Google Scholar]
- (5).Sevilla MD, Bernhard WA. Mechanisms of direct damage to DNA. In: Spotheim-Maurizot M, Mostafavi M, Douki T, Belloni J, editors. Radiation Chemistry: From Basics to Applications in Material and Life Sciences. EDP Sciences; Les Ulis, France: 2008. p. 191. [Google Scholar]
- (6).Bernhard WA, Barnes J, Mercer KR, Mroczka Radiation Research. 1994;140:199. [PubMed] [Google Scholar]
- (7).Sharma KK, Razskazovskiy Y, Purkayastha S, Bernhard WA. J. Phys. Chem. B. 2009;113:8183. doi: 10.1021/jp900803b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sharma KKK, Tyagi R, Purkayastha S, Bernhard WA. J. Phys. Chem. B. 2010;114:7672. doi: 10.1021/jp101717u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Purkayastha S, Milligan JR, Bernhard WA. Journal of Physical Chemistry B. 2005;109:16967. doi: 10.1021/jp0518409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Purkayastha S, Milligan JR, Bernhard WA. Radiation Research. 2007;168:357. doi: 10.1667/RR0964.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sharma KK, Purkayastha S, Bernhard WA. Radiation Research. 2007;167:501. doi: 10.1667/RR0847.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sharma KKK, Milligan JR, Bernhard WA. Radiation Research. 2008;170:156. doi: 10.1667/RR1277.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Sharma KKK, Bernhard WA. J. Phys. Chem. B. 2009;113:12839. doi: 10.1021/jp905750w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Swarts SG, Gilbert DC, Sharma KK, Razskazovskiy Y, Purkayastha S, Naumenko KA, Bernhard WA. Radiation Research. 2007;168:367. doi: 10.1667/RR1058.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Swarts SG, Sevilla MD, Becker D, Tokar CJ, Wheeler KT. Radiation Research. 1992;129:333. [PubMed] [Google Scholar]
- (16).Mercer KR, Bernhard WA. Journal of Magnetic Resonance. 1987;74:66. [Google Scholar]
- (17).Swarts SG, Becker D, Sevilla M, Wheeler KT. Radiation Research. 1996;145:304. [PubMed] [Google Scholar]
- (18).Debije MG, Bernhard WA. J. Phys. Chem. B. 2000;104:7845. [Google Scholar]
- (19).Steenken S. Chem. Rev. 1989;89:503. [Google Scholar]
- (20).Steenken S, Telo JP, Novais HM, Candeias LP. J. Am. Chem. Soc. 1992;114:4701. [Google Scholar]
- (21).Steenken S, Jovanovic SV. J. Am. Chem. Soc. 1997;119:617. [Google Scholar]
- (22).Steenken S. Free Radical Research Communications. 1992;16:349. doi: 10.3109/10715769209049187. [DOI] [PubMed] [Google Scholar]
- (23).Hole EO, Nelson WH, Close DM, Sagstuen E. J. Chem. Phys. 1987;86:5218. [Google Scholar]
- (24).Rakvin B, Herak JN, Voit K, Hüttermann J. Radiat. Environ. Biophys. 1987;26:1. doi: 10.1007/BF01211360. [DOI] [PubMed] [Google Scholar]
- (25).Yan M, Becker D, Summerfield S, Renke P, Sevilla MD. J. Phys. Chem. 1992;96:1983. [Google Scholar]
- (26).Wang W, Razskazovskii Y, Sevilla MD. Int. J. Radiat. Biol. 1997;71:387. doi: 10.1080/095530097144003. [DOI] [PubMed] [Google Scholar]
- (27).Shukla LI, Adhikary A, Pazdro R, Becker D, Sevilla MD. Nucleic Acids Research. 2004;32:6565. doi: 10.1093/nar/gkh989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Close DM, Nelson WH. Radiation Research. 1989;117:367. [PubMed] [Google Scholar]
- (29).Kar L, Bernhard WA. Radiation Research. 1983;93:232. [Google Scholar]
- (30).Steenken S. Biol. Chem. 1997;378:1293. [PubMed] [Google Scholar]
- (31).Spalletta RA, Bernhard WA. Radiation Research. 1992;130:7. [PubMed] [Google Scholar]
- (32).Spalletta RA, Bernhard WA. Radiation Research. 1993;133:143. [PubMed] [Google Scholar]
- (33).Sevilla MD, Engelhardt ML. Faraday Discuss. Chem. Soc. 1977;63:255. [Google Scholar]
- (34).Weiland B, Hüttermann J. Int.J.Radiat.Biol. 1998;74:341. doi: 10.1080/095530098141483. [DOI] [PubMed] [Google Scholar]
- (35).Schmidt J. J. Chem. Phys. 1975;62:370. [Google Scholar]
- (36).Hole EO, Nelson WH, Sagstuen E, Close DM. Radiation Research. 1998;149:109. [PubMed] [Google Scholar]
- (37).Purkayastha S, Milligan JR, Bernhard WA. Journal of Physical Chemistry B. 2006;110:26286. doi: 10.1021/jp065489i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Wang W, Yan M, Becker D, Sevilla MD. Radiation Research. 1994;137:2. [PubMed] [Google Scholar]
- (39).Weiland B, Hüttermann J, Van Tol J. Acta Chem. Scand. 1997;51:585. [Google Scholar]
- (40).Bernhard WA. Journal of Physical Chemistry. 1989;93:2187. [Google Scholar]
- (41).Ormerod MG. Int. J. Radiat. Biol. 1965;9:291. doi: 10.1080/09553006514550341. [DOI] [PubMed] [Google Scholar]
- (42).Wang W, Sevilla MD. Radiation Research. 1994;138:9. [PubMed] [Google Scholar]
- (43).Salovey R, Shulman RG, Walsh WM., Jr. J. Chem. Phys. 1963;39:839. doi: 10.1063/1.1734334. [DOI] [PubMed] [Google Scholar]
- (44).Bernhard WA. Solid-state radiation chemistry of DNA: the bases. In: Lett JT, Adler H, editors. Advances in Radiation Biology. Vol. 9. Academic Press; New York: 1981. p. 199. [Google Scholar]
- (45).Bernhard W, Snipes W. J. Chem. Phys. 1967;46:2848. doi: 10.1063/1.1841130. [DOI] [PubMed] [Google Scholar]
- (46).Park Y, Li Z, Cloutier P, Sanche L, Wagner JR. Radiation Research. 2011;175:240. doi: 10.1667/rr2381.1. [DOI] [PubMed] [Google Scholar]
- (47).Sagstuen E, Hole EO, Nelson WH, Close DM. J. Phys. Chem. 1992;96:8269. [Google Scholar]
- (48).Debije MG, Bernhard WA. Journal of Physical Chemistry A. 2002;106:4608. [Google Scholar]
- (49).Debije MG, Close DM, Bernhard WA. Radiation Research. 2002;157:235. doi: 10.1667/0033-7587(2002)157[0235:rdidid]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Becker D, Sevilla MD, Wang W, LaVere T. Radiation Research. 1997;148:508. [Google Scholar]
- (51).Milano MT, Bernhard WA. Radiation Research. 1999;151:39. [PMC free article] [PubMed] [Google Scholar]
- (52).Milligan JR, Aguilera JA, Nguyen TTD, Ward JF, Kow YW, He B, Cunningham RP. Radiation Research. 1999;151:334. [PubMed] [Google Scholar]
- (53).Roginskaya M, Bernhard WA, Marion RT, Razskazovskiy Y. Radiation Research. 2005;163:85. doi: 10.1667/rr3288. [DOI] [PMC free article] [PubMed] [Google Scholar]