Abstract
7,8-dihydro-8-oxoguanine (8-oxoG) is a major oxidative lesion found in DNA. The 8-oxoguanine DNA glycosylases (Ogg) responsible for the removal of 8-oxoG are divided into three families: Ogg1, Ogg2 and AGOG. Since Ogg2 members are devoid of the recognition loop used by Ogg1 to discriminate between 8-oxoG and guanine it was unclear until recently how Ogg2 enzymes recognize the oxidized base. We present here the first crystallographic structure of an Ogg2 member, Methanocaldococcus janischii Ogg, in complex with a DNA duplex containing the 8-oxoG lesion. This structure highlights the critical role of the C-terminal lysine, strictly conserved in Ogg2, in the recognition of 8-oxoG. The structure also reveals that, similarly to Ogg1 glycosylases, Ogg2 undergoes a conformational change upon DNA binding. Furthermore, this work provides a structural rationale for the lack of opposite base specificity in this family of enzymes.
Keywords: 8-oxoguanine glycosylase, crystal structure, DNA repair, 8-oxoguanine, base excision repair
Introduction
One of the most abundant oxidative lesions in DNA is 7,8-dihydro-8-oxoguanine (8-oxoG). 1 8-oxoG results from the oxidation of guanine in DNA via oxidizing agents and ionizing radiation. 8-oxoG is highly mutagenic because of its ability to form a Hoogsteen pair with adenine (8-oxoG:A), which can lead to a G:C → T:A transversion mutations after replication. 2; 3 Cells have developed mechanisms to repair damaged bases in DNA. One of these mechanisms is known as the Base Excision Repair (BER) pathway. 4 8-oxoG is recognized and cleaved by two enzymes of the BER pathway, namely formamidopyrimidine-DNA glycosylase (Fpg) and 8-oxoguanine DNA glycosylase (Ogg). While Fpg is found mostly in bacteria, Ogg is the main 8-oxoG DNA glycosylase in eukaryotes and archaea.
The Ogg DNA glycosylases share the helix-hairpin-helix (HhH) signature motif of the HhH-GDP superfamily 5; 6 and are divided into three structurally distinct families: Ogg1, Ogg2 and archaeal GO glycosylase (AGOG). The Ogg1 family encompasses the largest number of members including the well-studied human OGG1 (hOGG1) 7; 8; 9; 10; 11; 12; 13 and the atypical bacterial Ogg1 from Clostridium acetobutylicum (CacOgg) 14; 15; 16. Even though the Ogg1 gene products can vary considerably in size these enzymes all share a similar tertiary fold composed of three domains. 14 Ogg2 was the last Ogg family to be structurally characterized and comprises primarily archaeal enzymes, such as Methanocaldococcus janischii Ogg (MjaOgg) and Sulfolobulus solfataricus Ogg (SsoOgg).17 Ogg2 glycosylases comprise two domains separated by the HhH motif and, in contrast to Ogg1, are less variable in size (± 207 amino acids). 18; 19; 20; 21 Ogg2 enzymes also display a less stringent opposite base specificity than Ogg1. 19; 21 Finally, members of the third family, AGOG, share a similar two-domain architecture with Ogg2. The HhH motif in AGOG, however, differs from that of Ogg1 or Ogg2. 22; 23
An obvious difference between 8-oxoG and guanine is the presence of an oxygen atom at the C8 atom of guanine to form a keto group. Surprisingly, Ogg1 enzymes did not exploit this feature to distinguish between guanine and 8-oxoG as there is no interaction between the enzyme and the C8-oxygen. However, as a consequence of C8 oxidation, an electron delocalizes from C8 to N7, which drives N7 to attract a proton. This second feature of 8-oxoG appears to be a major contributor to 8-oxoG recognition by Ogg1: The N7-H atom participates in a hydrogen bond with the main chain carbonyl of a conserved glycine (Gly42 in hOGG1 and Gly30 in CacOgg 16; 24) located in the αA-βB recognition loop of domain A. 14; 16; 24; 25 If G is bound instead of 8-oxoG, then the attractive interaction between the glycine carbonyl and the 8-oxoG N7-H is predicted to be replaced by a repulsive interaction between the same carbonyl and the N7 lone pair of G. 26 AGOG, on the other hand, seems to recognize the two features that distinguish 8-oxoG from guanine, interacting with both C8-oxygen and N7-H atom. 22 Moreover, the interactions between AGOG and 8-oxoG involve the side chains of two residues and not a main chain carbonyl as seen in Ogg1. Ogg2 members lack the αA-βB loop used by Ogg1 to specifically interact with 8-oxoG and until recently, it was unclear how Ogg2 would select for 8-oxoG. We previously reported the unliganded crystal structures of two members of the Ogg2 family, MjaOgg and SsoOgg, 17 and predicted that the conserved C-terminal lysine would play a crucial role in the distinction between 8-oxoG and G in this family of enzymes.
Here we describe the first structure of any Ogg2 enzyme bond to substrate DNA, a 15-mer DNA duplex containing the 8-oxoG lesion. The Methanocaldococcus janischii Ogg structure illustrates the crucial role of the conserved Ogg2 C-terminal lysine in 8-oxoG recognition. In addition, the structure revealed conformational changes upon binding DNA by MjaOgg of a magnitude similar to that reported for Ogg1. 16; 25 Furthermore, analysis of the interactions between the enzyme and the estranged base provides an explanation for the lack of opposite base specificity displayed by Ogg2 compared to hOGG1.
Results
Crystallization and structure determination of MjaOgg in complex with DNA containing 8-oxoG:C
A single crystal of MjaOggK129Q in complex with a 15-mer duplex DNA oligonucleotide containing 8-oxoG paired with C was used to collect a 2.7 Å data set at the Advanced Photon Source (See Table 1 for diffraction statistics). Crystals of the MjaOggK129Q/8-oxoG:C complex belong to the monoclinic space group P21. Diffraction data processing showed the crystal was twinned with a twin law of (h, -k, -h-l) and twin fraction of 0.288.
Table 1.
| MjaOggK129Q/8- oxoG:C |
|
|---|---|
| Data collection | |
| Wavelength (Å) | 1.0332 |
| Resolution (Å) a | 19.84-2.7 (2.8-2.7) |
| Space group | P21 |
| Unit-cell parameters a,b,c (Å) | 54.80, 150.03, 90.27 |
| β(°) | 107.53 |
| Total reflection | 137275 (12247) |
| Unique reflection | 35975 (3160) |
| Redundancy | 3.8 (3.9) |
| Completeness (%) | 94.2 (81.2) |
| I/σ(I) | 13.2 (2.2) |
| Rmerge (%) | 10.7 (70.7) |
| Refinement | |
| Rcryst (%) | 18.1 (30.4) |
| Rfree (%)b | 22.4 (34.4) |
| Rmsd from ideal bond length (Å)/angles (°) | 0.006/1.19 |
| Average B factors (Å2) | 63.3 |
| Twinning | |
| Twin law | (h,-k,-h-l) |
| Twin fraction | 0.288 |
| Ramachandran plot (%) | |
| Most favored regions | 89.0 |
| Allowed regions | 10.5 |
| Disallowed regions | 0.5 |
High-resolution shell is shown in parentheses.
Rfree was calculated with 5% of the reflections not used in refinement
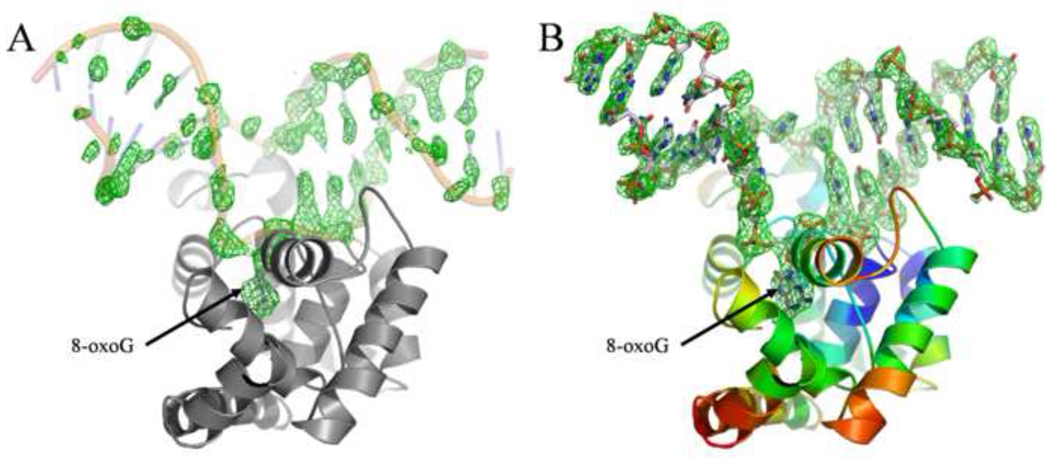
A Matthews coefficient calculation 27 indicated the possibility of four molecules by asymmetric unit. Phenix.automr 28 found the correct solution for the four molecules with LLG=1294 and R=0.354 using the apo-MjaOgg structure 17 as search model. The resulting electron density map was well defined and density was observed for the four protein/DNA complexes (see figure 1). The final model was refined to a Rwork/Rfree of 0.180/0.224 and rmsd of 0.006Ǻ and 1.191 for bonds length and angles, respectively. The four copies of the MjaOgg complexes are nearly equivalent (overall rmsd =0.4 Å). We describe below the interactions observed in molecule A.
Fig. 1.
A) Fo-Fc electron density map at 2.5σ level calculated after molecular replacement (i.e. before any model refinement). The DNA backbone from the refined model is superimposed. B) Fo-Fc electron density map at 3σ level calculated around the DNA molecule. MjaOgg is colored according to Cα B-factors (blue to red, from high to low). DNA is colored using standard CPK colors. Figure 1, 2, 4, 5 and 6 were prepared using PyMOL. 40
Overall structure description
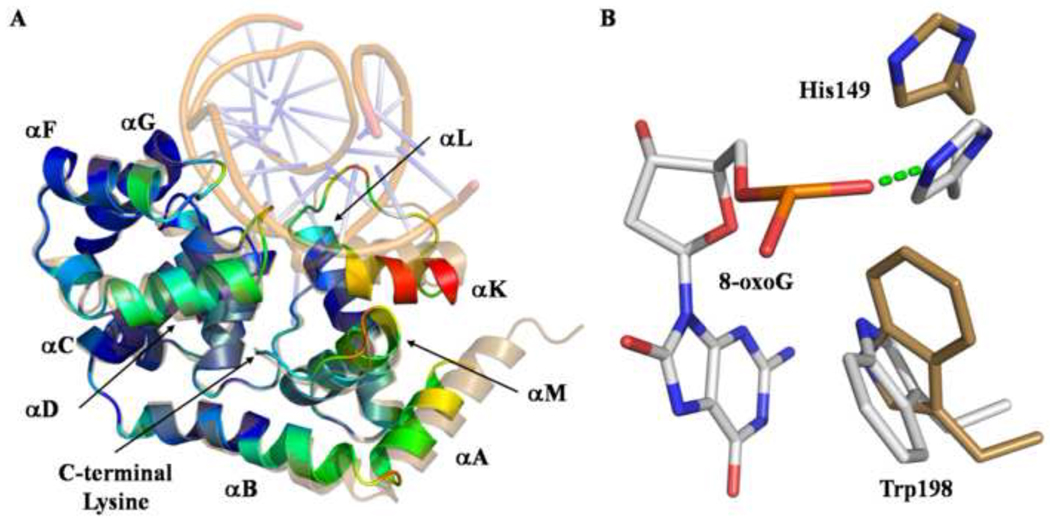
MjaOgg is composed of two domains separated by the central HhH motif considered to be the fingerprint of this family of DNA repair glycosylases. 29 The N-terminal domain forms an orthogonal helix bundle and encompasses helices αB to αJ, whereas the C-terminal domain comprises helices αA and αK to αM. 17 The DNA binds in the groove between the two domains and the HhH motif (see below). The 8-oxoG base is flipped out the DNA helix and lies in the substrate binding site. The space vacated by 8-oxoG is filled by Asn49 of the small αC–αD loop. The DNA molecule is bent by an angle of about 26° (calculated with CURVES 5.1 30) centered at the lesion site. A sodium ion is found at the junction of the hairpin of the HhH motif and a phosphate group from the DNA backbone. The identity of the metal ion was deduced based on geometry (octahedral), bound distances (average of 2.5 Å) and the previously determined structure of CacOgg. 16
8-oxoG binding site
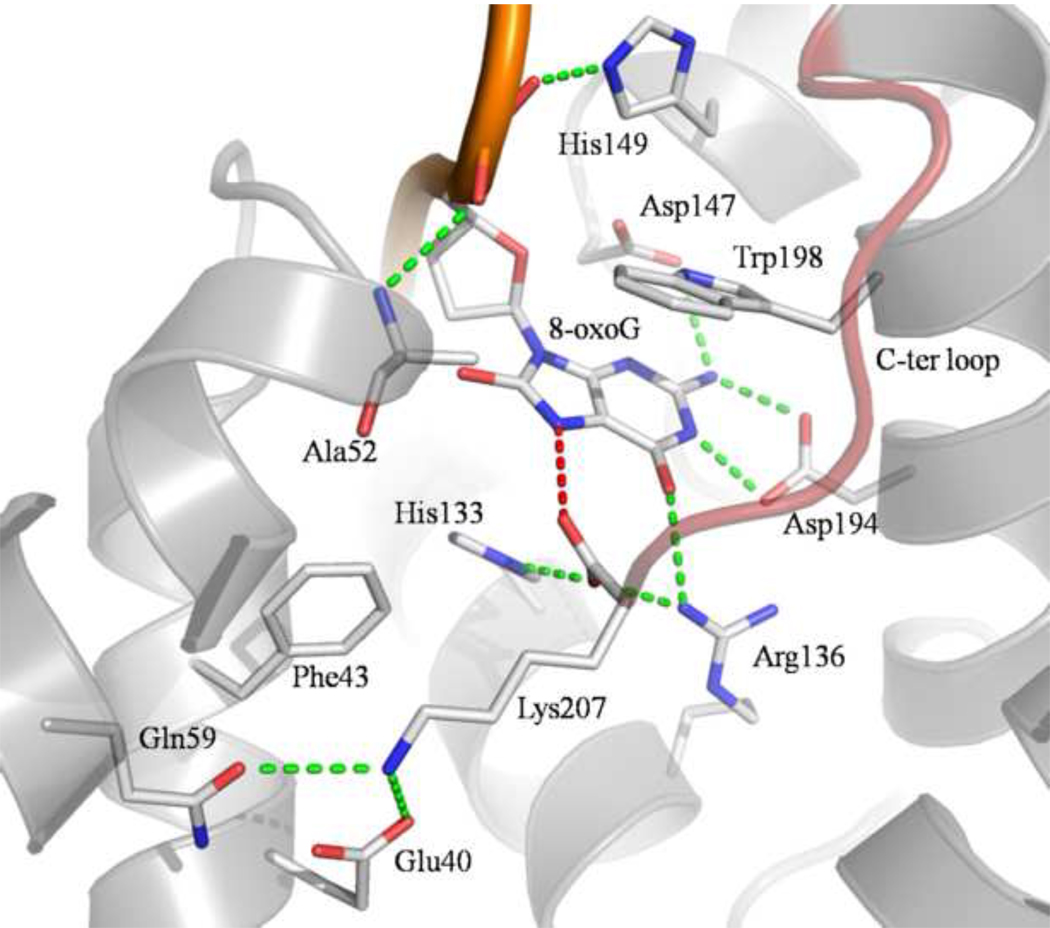
The 8-oxoG binding site is a deep polar pocket located at the interface between the two domains of MjaOgg. Once in this pocket, 8-oxoG engages in several H-bonds with residues from both domains (see figure 2). Briefly, the six-membered ring of the 8-oxoG base interacts with the side chains of Arg136, Asp147, and Asp194. In addition, the main chain N of Ala52 and side chain of His149 interact with the phosphate group of 8-oxoG. Finally, the N7-H atom of 8-oxoG is H-bonded to the C-terminal carboxyl group of Lys207. This hydrogen bond mimics the interaction between the main chain carbonyl of a conserved glycine (Gly42 in hOGG1) of the αA-βB loop and the damaged base in hOGG1. This H-bond is critical for the ability of Ogg1, and now Ogg2 enzymes, to distinguish between 8-oxoG and guanine. It is noteworthy that, similarly to Ogg1 enzymes 16; 24, Ogg2 does not interact with the C8 oxygen atom of 8-oxoG. Furthermore, in addition to the hydrophilic interactions described above, 8-oxoG is sandwiched between His133 and Trp198. Trp198 is conserved among Ogg2 family members and plays a similar stacking role as the conserved Phe319 in hOGG1.
Fig. 2.
Close-up view of MjaOgg residues interacting with 8-oxoG and Lys207. Only residues involved in H-bond and stacking interactions with 8-oxoG or Lys207 are depicted. The H-bond between the C-terminal lysine (Lys207) carboxyl and the 8-oxoG N7-H atom is colored in red. This H-bond is a critical molecular determinant for 8-oxoG recognition. 17 The C-terminal loop is colored in red.
Interactions of MjaOgg with DNA and the estranged base
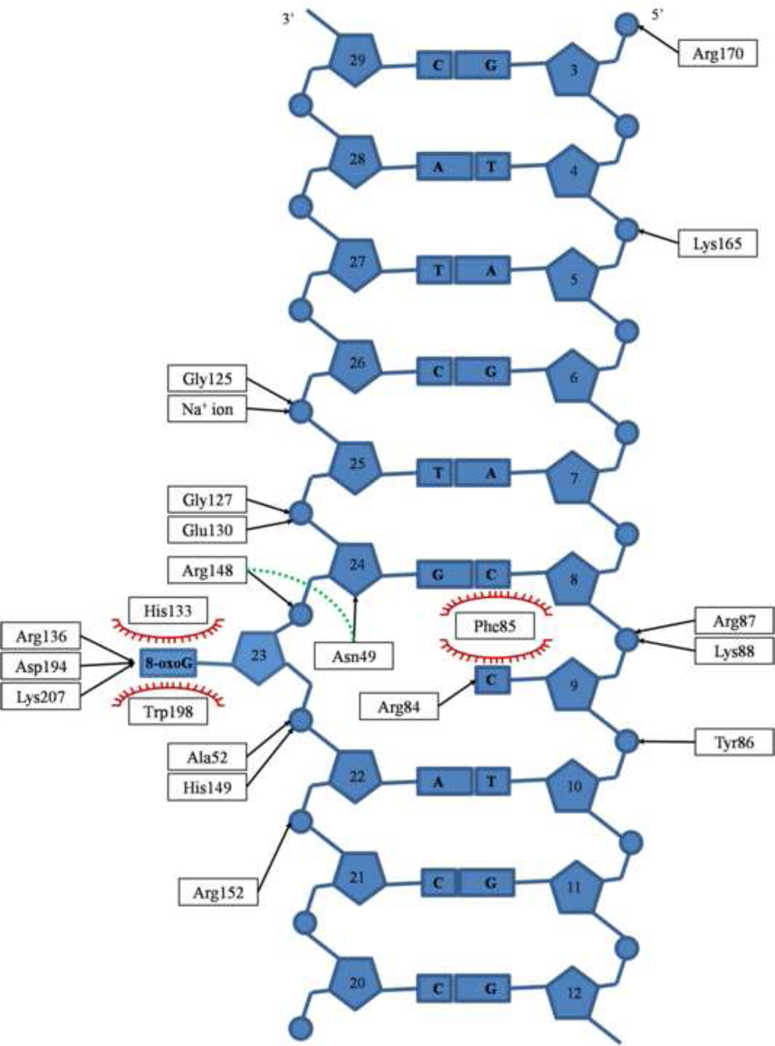
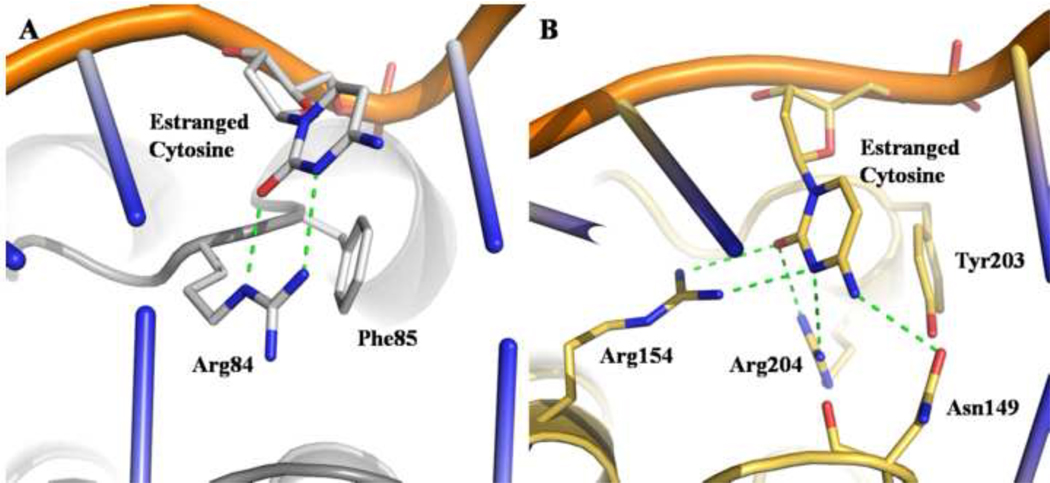
The DNA duplex participates in H-bonds with several protein residues. Most of these interactions involve the 8-oxoG lesion. The remaining hydrophilic contacts are made with the DNA backbone and the estranged base. The protein-DNA interactions are summarized in figure 3. Briefly, a total of 17 amino acids contact 8 nucleotides. The majority of hydrophilic contacts between DNA and MjaOgg are to the DNA strand containing the 8-oxoG lesion and primarily to the DNA phosphate groups. Only two bases, 8-oxoG and the estranged C, are H-bonded to the protein. In addition, the amino group of residue Asn49 interacts with the deoxyribose of the 8-oxoG 3’-neighboring base. Similarly to Ogg1 enzymes, an aromatic residue (Phe85) is inserted between the estranged base and its 5’- neighbor, thereby creating the bend in the DNA. Other than the stacking interaction with Phe85, the estranged base shows very little contact with the protein, interacting with a single residue, Arg84, via two H-bonds. (Figure 4)
Fig. 3.
Schematic representation of MjaOgg-DNA interactions. Phe85 is wedged between the estranged cytosine and its 5’- neighbor, whereas 8-oxoG is sandwiched between His133 and Trp198 (Stacking interactions are depicted with red spoked arcs). Only Arg84 interacts with the estranged cytosine. The sodium ion interaction with DNA is shown and the H-bond between Asn49 and Arg148 over the DNA backbone is depicted with a green dash line.
Fig. 4.
MjaOgg and hOGG1 interactions with the estranged cytosine. A) MjaOgg residues involved in H-bond and stacking interactions are depicted. Only Arg84 is involved in binding to the estranged base whereas Phe85 stacks against the cytosine. B) In hOGG1 three residues (Asn149, Arg154, and Arg204) participate in H-bond interactions with the estranged cytosine.
Structural reorganization upon DNA binding
Similarly to Ogg1 enzymes, which display a significant reorganization of the C-terminal domain upon binding to DNA or an 8-oxoguanosine nucleoside 14; 16; 25, the C-terminal domain of Ogg2 undergoes a conformational change upon DNA binding (see figure 5a). Most of this movement is localized in loop αC–αD, loop αF–αG, helix αK and the long loop between αL and αK. This is not surprising since several residues of these secondary structure elements are involved in H-bonding interactions with the DNA molecule. To a lesser extent, helices αA, αB, αD and αL shift towards the 8-oxoG binding site. Several side chains undergo some reorganization to interact with the DNA. Among these, His149 and Trp198 move drastically in order to bind and/or stabilize 8-oxoG in the active site (figure 5b). In contrast the position of the C-terminal Lys207 is not altered by DNA binding.
Fig. 5.
A) Superposition of apo-MjaOgg (RCSB PDBid:3FHF 17) and MjaOgg/DNA complex. Apo-MjaOgg is shown in sand color while the MjaOgg/DNA complex is colored according to the RMSD between the two models (blue for low RMSD and red for high RMSD). DNA is shown in transparent mode for clarity. The C-terminal Lys207 superimposes perfectly in both the apo- and liganded forms. B) Close-up view of residues that undergo a large shift upon binding 8-oxoG (apo-MjaOgg amino acids are shown in beige and the MjaOgg/DNA complex residues are depicted in standard CPK colors).
Discussion
Structural comparison of the MjaOgg/DNA complex with the apo-MjaOgg model
It has been shown previously that some structural elements, mostly from the C-terminal domain, of Ogg1 enzymes undergo a conformational change upon binding to DNA or even upon binding to a simple nucleoside. 14; 25 We now show that Ogg2 glycosylases behave similarly, exhibiting a conformational change of the C-terminal domain and of loop αC–αD from the N-terminal domain upon binding DNA. Globally these conformational changes are triggered by the formation of several hydrophilic contacts between the deep groove of MjaOgg and the DNA duplex. The DNA molecule interacts mostly with helices αK and αM and with loops αC–αD, αF–αG, αL-αK and the C-terminal loop (figure 5). However, helices αA, αB and αD undergo some reorganization despite the fact that they are not directly involved in binding DNA. These helices seem to move to accommodate and maintain their interactions with the structural elements that interact with DNA. A H-bond is formed over the backbone of the DNA lesion between Asn49 and Arg148. The MjaOgg-Asn49 residue is conserved in both Ogg2 and Ogg1 families (hOGG1-Asn149 and CacOgg-Asn127), but instead of forming a “bridge” over the DNA backbone, the hOGG1 counterpart is involved in estranged base recognition (see section 4.3). It is noteworthy that the C-terminal Lys207 superimposes very well in both the apo- and MjaOgg/DNA complex forms: The side chain of Lys207 is involved in several hydrophilic interactions stabilizing its conformation and essentially locking it in position. More importantly, the carboxy terminal group of Lys207 occupies the same position in both forms (apo and DNA-bound). This is reminiscent of what was described for hOGG1 where Gly42, the residue that interacts with the N7-H of 8-oxoG via its main chain carbonyl group, adopts the same backbone conformation in apo- and DNA bound structures. Verdine and collaborators have proposed that this “hardwiring” of the position of Gly42 is essential for the distinction between G and 8-oxoG, which arises from the repulsion between the Gly-42 amide and the N7 of G and the attractive interaction between the same amide and the protonated N7 of 8-oxoG. 31
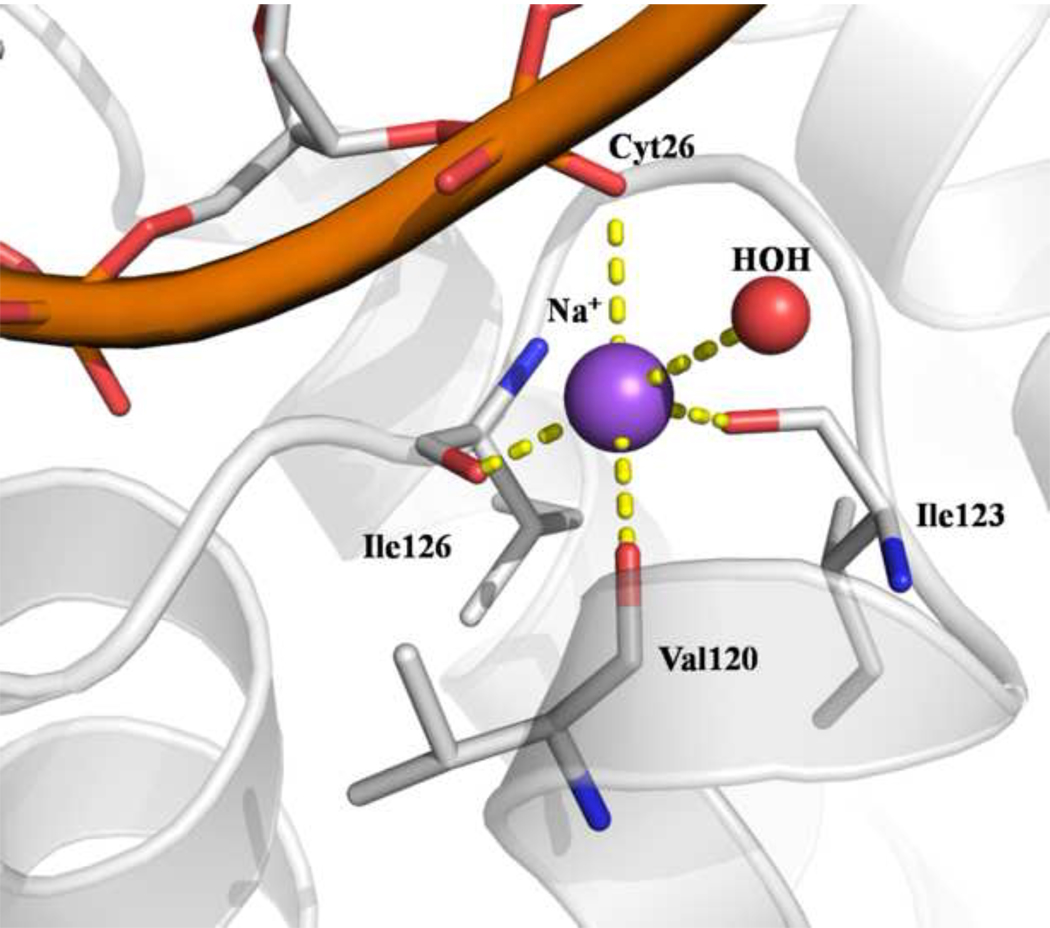
One major feature of enzymes bearing the HhH motif is their ability to bind one metal cation between the hairpin and DNA. 32 Characterization of the HhH motif suggested that DNA binding is dependent on the presence of a metal ion. We indeed observe a sodium ion between the DNA phosphate and carbonyl groups of the hairpin from the HhH motif of MjaOgg. The sodium ion interacts with the backbone carbonyl groups of Val120, Ile123 and Ile126, in addition to a phosphate oxygen of Cytosine 26 and a water molecule (see figure 6). These interactions occur only in the presence of DNA since no ion was observed in the apo-MjaOgg model. Similar metal ion interactions between the HhH and DNA were reported for CacOgg/DNA complexes 16 but, surprisingly, not for the hOGG1/DNA complex (PDBID 1EBM) 24. Examination of the electron density map of the 1EBM model reveals density near the HhH motif, which was assigned in the model as a water molecule (A426) but could be attributed to a cation, based on geometry and distance. Indeed more recent crystal structures of hOGG1 DNA complexes include a metal ion (Ca2+) at the junction between the hairpin and the DNA (PDB ID 2NOB, 2NOE; 31).
Fig. 6.
Sodium ion interactions at the junction between DNA and the hairpin from the HhH motif of MjaOgg.
Molecular determinants for 8-oxoG recognition by Ogg2
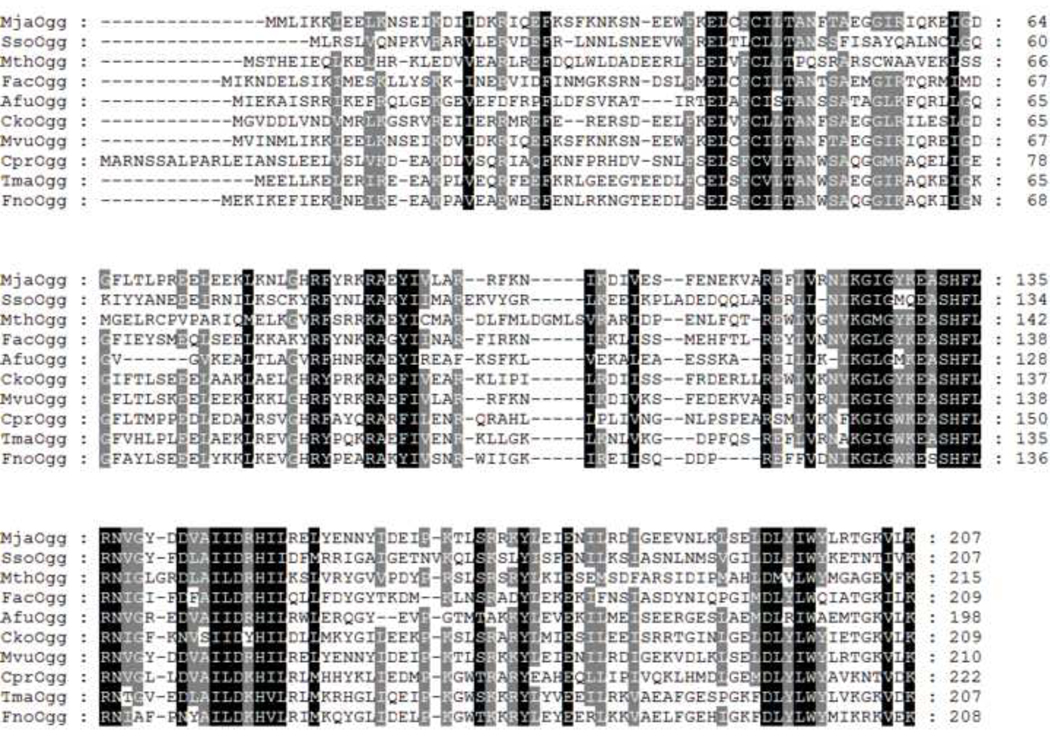
Most DNA glycosylases bind their DNA substrate and flip the damaged base into a deep cavity lined with residues that participate in lesion recognition and catalysis 33. In Ogg2 8-oxoG is flipped out from the DNA helix and bound in a cavity located at the junction between the two domains. The 8-oxoG lesion differs from guanine by the presence of a keto group on the C8 atom. However, Ogg2 enzymes did not exploit this difference to recognize the damaged base but rather use the proton addition on the N7 atom to distinguish between the two bases. It has been established that Ogg1 mainly discriminates between guanine and 8-oxoG via a H-bond between a conserved glycine, from the N-terminal domain, and the N7-H atom of 8-oxoG. 14; 24; 26 Since Ogg2 members are devoid of the N-terminal domain of Ogg1, the mechanism for guanine/8-oxoG distinction was unclear until recently. We previously surmised that the C-terminal lysine might play a crucial role in binding 8-oxoG by comparing apo-MjaOgg with hOGG1 and CacOgg bound to DNA. 17 We found that the C-terminal loop of Ogg2 superimposes very well onto the Ogg1 loop (αA-βB) involved in 8-oxoG recognition. More specifically, we noticed that Lys207 of Ogg2 overlaps with the conserved glycine in Ogg1 responsible for binding the N7-H atom of 8-oxoG. The C-terminal lysine is absolutely conserved in the Ogg2 family (figure 7), supporting its critical involvement in 8-oxoG binding. A variant of MjaOgg missing the last three residues was shown to have no detectable glycosylase activity on 8-oxoG-containing DNA 17 By crystallizing MjaOgg in complex with a DNA oligonucleotide bearing the 8-oxoG lesion, we now confirm the critical role of the lysine residue in G/8-oxoG distinction by means of a H-bond between the carboxyl group of Lys207 and the N7-H atom of 8-oxoG.
Fig. 7.
Protein sequence alignment of ten Ogg2 glycosylases from different species, showing conserved areas (Black shaded areas indicate strictly conserved residues whereas gray shade signifies partial conservation). The different species are as follows: MjaOgg, Methanocaldococcus janischii; SsoOgg, Sulfolobulus solfataricus; MthOgg, Methanosaeta thermophila; FacOgg, Ferroplasma acidarmanus; AfuOgg, Archaeoglobus fulgidus; CkoOgg, Candidatus Korarchaeum; MvuOgg, Methanocaldococcus vulcanius; CprOgg, Coprothermobacter proteolyticus; TmaOgg, Thermotoga maritima; and FnoOgg, Fervidobacterium nodosum.
As mentioned above, the C-terminal lysine is strictly conserved in Ogg2 enzymes. The conservation of a lysine in the ultimate position may be puzzling at first glance because only the carboxyl group of this residue is involved in a H-bond with 8-oxoG; any residue at this position would presumably be capable to partake in the same interaction through its carboxyl group. The evolutionary pressure to keep a lysine must reside elsewhere. As seen on figure 2, Lys207 is involved in multiple interactions with several polar residues in addition to 8-oxoG. One of the two Lys207 carboxyl oxygens is stabilized via two H-bonds with two conserved residues (His133 and Arg136). These interactions help orient the other oxygen atom for interaction with N7-H of 8-oxoG. The NH3+ moiety is bound by Gln59 and by the negatively charged and strictly conserved Glu40. The aliphatic part of Lys207 stacks against the conserved Phe43. The stabilization of both NH3+ and COO− moieties of Lys207 may help the proper folding of the C-terminal loop in a position suitable for the binding of 8-oxoG. The C-terminal lysine of Ogg2 may therefore serve a dual role: The NH3+ group helps stabilize the C-terminal loop in the proper conformation for substrate binding while the Lys207 carboxyl group interacts specifically with 8-oxoG.
Although a direct interaction between a protein oxygen and the N7-H of 8-oxoG has been observed in Ogg1 and Ogg2 enzymes, another hypothesis has been put forth to explain the guanine/8-oxoG distinction by Ogg glycosylases: The dipole change on the C8 and N7 atoms of 8-oxoG compared to guanine has been proposed to be involved in the specific recognition of the lesion by hOGG1. 26 The hOGG1 structural characterization supports this idea because the 8-oxoG binding site appears to complement the dipole observed on 8-oxoG. However, one of the residues involved in the hOGG1 binding site dipole (Cys253-S−) is not conserved in Ogg2 and is replaced by an histidine (MjaOgg-His133), implying that the dipole effect may be less critical in Ogg2.
Lack of estranged base specificity in Ogg2 glycosylases
Two previous studies have shown that Ogg2 glycosylases display very little preference for the base opposite the lesion. 19; 21 The majority of Ogg2 enzymes are found in archaea (mostly anaerobic) and in a few anaerobic bacteria. The exposure of these organisms to oxygen is likely to be sporadic and thus the formation of 8-oxoG a rare event. There was therefore little evolutionary pressure to maintain an estranged base specificity opposite 8-oxoG similar to that of Ogg1. The availability of the MjaOgg structure in complex with DNA provides a unique opportunity to further investigate the lack of estranged base specificity of Ogg2 enzymes. As shown in figure 4A, the estranged cytosine interacts with only two residues, Arg84 and Phe85. Phe85 appears to have a structural equivalent in all three Ogg families, being either a phenylalanine or a tyrosine. The aromatic residue stacks against the 8-oxoG base and therefore cannot distinguish between the four possible DNA bases. Similarly to CacOgg (a peculiar bacterial Ogg1 enzyme with little preference for the base opposite the damage during base excision), MjaOgg binds the estranged base with only one strictly conserved arginine (Arg84) (in MjaOgg the arginine NHε and NH1 interact with the cytosine O2 and N3 rather than NH1 and NH2 in CacOgg). In contrast to CacOgg and MjaOgg, hOGG1 interacts with the estranged base via two arginine residues: Arg 204, which corresponds to MjaOgg Arg84, and Arg154. Arg154 has been shown to play a crucial role in opposite base selection. 24 Although the naturally occurring hOGG1 R154H variant has a glycosylase activity similar to wild-type its opposite base specificity is reduced: it can excise 8-oxoG opposite A nearly as efficiently as opposite C. In CacOgg the residue corresponding to hOGG1 Arg154 is a methionine (Met132). We showed recently that substituting arginine for the methionine increases specificity for 8-oxoG:C relative to 8-oxoG:A 16 These findings indicate that the specificity for C in hOGG1 arises primarily from Arg154, a residue that is missing in Ogg2 enzymes. In Ogg1, another conserved residue (hOGG1-Asn149; CacOgg-Asn127) is involved in the binding of the estranged base. This residue has a structural equivalent in Ogg2 enzymes (MjaOgg-Asn49), which instead of binding the estranged base participates in a H-bond with Arg148, thereby holding the DNA in the enzyme groove. The paucity of interactions between MjaOgg with C is such that the estranged base binding site should accommodate any other natural DNA base at the estranged position. Biochemical studies have shown that AGOG also shows no preference for the estranged base. 23 Analysis of the lack of specificity for the estranged base in AGOG will have to await a structure of AGOG with substrate DNA.
Concluding remarks
We presented here the first crystal structure of an Ogg2 enzyme in complex with a DNA duplex containing the 8-oxoG lesion opposite cytosine. We confirmed our earlier hypothesis regarding the crucial role of the Ogg2 C-terminal lysine in 8-oxoG recognition via a protein carbonyl - 8-oxoG N7-H hydrogen bond interaction reminiscent of that described in hOGG1. We also showed that Ogg2 undergoes a conformational change upon DNA binding similar to that described for Ogg1 glycosylases. We observed that metal ion coordination within the HhH motif accompanies the binding of DNA. Finally, the MjaOgg structure in complex with DNA provides an explanation for the lack of estranged base specificity of Ogg2 enzymes.
Material and methods
Protein expression and purification
The open reading frame of MjaOgg was cloned using Nde1-Xho1 restriction sites of a pET-22b vector (Novagen). A penta-histidine-tag sequence was inserted before the first ATG codon of MjaOgg while the stop codon was conserved. A glycosylase deficient variant (K129Q) was generated by site directed mutagenesis (QuikChange, Stratagene). After overnight expression at room temperature with 0.1mM IPTG, the bacterial pellet was sonicated in lysis buffer (50 mM Na-Phosphate pH 8.0, 300 mM NaCl, 5 mM imidazole). The centrifuged lysate was loaded on a Ni-NTA column (GE Healthcare) and eluted using an imidazole gradient (0 → 300 mM). Pooled fractions of MjaOgg were dialyzed in 50 mM Tris-HCl pH 7.6, 150 mM NaCl and 1 mM β-mercaptoethanol then loaded on an SP column (GE Healthcare) and eluted with a NaCl gradient (0.1 → 1.5 M). The purified protein was concentrated to ~ 4 mg/ml prior to crystallization.
Crystallization of MjaOgg in complex with a 15mer 8-oxoG:C DNA molecule
Crystals of MjaOggK129Q in complex with DNA were obtained by hanging-drop vapor diffusion at 12°C. DNA oligonucleotides (15-mer) were ordered from Midland Certified Reagent Co. (Midland, TX) and purified on an acrylamide gel. The sequences were as follows: 5’-ACG-TCC-AXG-TCT-ACC-3’ and 5’-TGG-TAG-ACC-TGG-ACG-3’ where X is 8-oxoG. Protein and duplex DNA were mixed in a 1:1 molar ratio. Crystals grew in 2 µL drops containing a 1:1 ratio of protein/DNA mix and well solution (16% (w/v) PEG-8000, 0.1 M Tris-HCl pH 6.5 and 0.1 M NaCl). Typical crystals grew to dimensions suitable for X-ray diffraction experiments (200 × 70 × 70 µm3) in about two weeks.
X-ray analysis of diffraction experiments
A complete data set was collected at 100 K and at a wavelength of 1.0332 Å at beamline 23-ID-B at the Argonne Advanced Photon Source. Diffraction data were indexed using XDS 34 and scaled with XSCALE. The data could be indexed in either monoclinic P2 or centered orthorhombic C222 space groups with unit-cell parameters a=54.80 Ǻ, b=150.03 Ǻ, c=90.27 Ǻ β=107.53° and a=54.8 Ǻ, b=171.78 and c=150.03 Ǻ, respectively. Examination of systematically absent reflections revealed a two-fold screw axis, leaving P21 and C2221 as possible space groups. However, scaling statistics were slightly worse in C2221 (Rmerge=0.116) compared to P21 (Rmerge=0.107). The space group was judged to be P21 and a Matthews coefficient calculation 27 suggested four molecules per asymmetric unit for an estimated solvent content of 52%. Further analysis using phenix.xtriage 28 revealed the presence of a pseudo-merohedral twinning with twin law = (h,-k,-h-l) and a twin fraction of 0.29 and confirmed the monoclinic space group P21. Twinning occurs when two crystals of different relative orientations inter-grow. MjaOgg crystals display a rare pseudo-merohedral twinning meeting the condition: c cosβ = −a/2, i.e. the two unit cells of the twin domain share the same a and b axes, but in opposite directions. Nevertheless, twinned data have been used successfully for structure determination by molecular replacement. 35; 36
Structure determination of MjaOgg/DNA complex
The structure of the MjaOgg/DNA complex was obtained by molecular replacement using the coordinates of apo-MjaOgg (RCSB PDBid 3FHF 17). The structure was solved with phenix.automr 28 by using both the twin law and twin fraction. A very clear Fo-Fc electron density map corresponding to DNA (including 8-oxoG and opposite base) was observed immediately after molecular replacement for each of the four molecules by asymmetric unit (see figure 1). The initial model was submitted to rigid body refinement and one cycle of simulated annealing at 3000 K followed by energy minimization and B-factor refinement cycle with CNS twinning inputs. 37 Afterwards, the model was refined with CNS 37 and phenix.refine 28 by simple energy minimization followed by isotropic B-factors refinement (restrained and individual) and corrected by manual rebuilding using COOT. 38 The quality of the model was verified with PROCHECK and COOT. 38; 39 The penultimate residue, Leu206, has (phi, psi) angles in a disallowed region of the Ramachandran plot. This residue immediately precedes the C-terminal lysine, which is locked in position by several H-bond interactions with both DNA and protein residues and might therefore induce a strained conformation for Leu206.
Acknowledgements
We thank Dr. Pierre Aller and Karl Zahn for collecting the diffraction data set at the APS synchrotron. GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357. This research was supported by National Institutes of Health grants R01CA33657 and P01CA098993 awarded by the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Protein Data Bank accession codes
Atomic coordinates and structure factor amplitudes have been deposited with the Protein Data Bank and are available under the following accession code: 3KNT
References
- 1.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 2.Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature. 1987;327:77–79. doi: 10.1038/327077a0. [DOI] [PubMed] [Google Scholar]
- 3.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 4.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash HM, Bruner SD, Scharer OD, Kawate T, Addona TA, Spooner E, Lane WS, Verdine GL. Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr Biol. 1996;6:968–980. doi: 10.1016/s0960-9822(02)00641-3. [DOI] [PubMed] [Google Scholar]
- 6.Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 8.Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 9.Bjørås M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J. 1997;16:6314–6322. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagashima M, Sasaki A, Morishita K, Takenoshita S, Nagamachi Y, Kasai H, Yokota J. Presence of human cellular protein(s) that specifically binds and cleaves 8-hydroxyguanine containing DNA. Mutat Res. 1997;383:49–59. doi: 10.1016/s0921-8777(96)00045-6. [DOI] [PubMed] [Google Scholar]
- 11.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faucher F, Robey-Bond SM, Wallace SS, Doublié S. Structural characterization of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase in its apo form and in complex with 8-oxodeoxyguanosine. J Mol Biol. 2009;387:669–679. doi: 10.1016/j.jmb.2009.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robey-Bond SM, Barrantes-Reynolds R, Bond JP, Wallace SS, Bandaru V. Clostridium acetobutylicum 8-oxoguanine DNA glycosylase (Ogg) differs from eukaryotic Oggs with respect to opposite base discrimination. Biochemistry. 2008;47:7626–7636. doi: 10.1021/bi800162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faucher F, Wallace SS, Doublié S. Structural basis for the lack of opposite base specificity of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase. DNA Repair (Amst) 2009;8:1283–1289. doi: 10.1016/j.dnarep.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faucher F, Duclos S, Bandaru V, Wallace SS, Doublié S. Crystal structures of two archaeal 8-oxoguanine DNA glycosylases provide structural insight into guanine/8-oxoguanine distinction. Structure. 2009;17:703–712. doi: 10.1016/j.str.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A, Scott JL, Geoghagen NS, Venter JC. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 19.Gogos A, Clarke ND. Characterization of an 8-oxoguanine DNA glycosylase from Methanococcus jannaschii. J Biol Chem. 1999;274:30447–30450. doi: 10.1074/jbc.274.43.30447. [DOI] [PubMed] [Google Scholar]
- 20.Faucher F, Duclos S, Bandaru V, Wallace SS, Doublie S. Crystal structures of two archaeal 8-oxoguanine DNA glycosylases provide structural insight into guanine/8-oxoguanine distinction. Structure. 2009;17:703–712. doi: 10.1016/j.str.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im EK, Hong CH, Back JH, Han YS, Chung JH. Functional identification of an 8-oxoguanine specific endonuclease from Thermotoga maritima. J Biochem Mol Biol. 2005;38:676–682. doi: 10.5483/bmbrep.2005.38.6.676. [DOI] [PubMed] [Google Scholar]
- 22.Lingaraju GM, Sartori AA, Kostrewa D, Prota AE, Jiricny J, Winkler FK. A DNA glycosylase from Pyrobaculum aerophilum with an 8-oxoguanine binding mode and a noncanonical helix-hairpin-helix structure. Structure. 2005;13:87–98. doi: 10.1016/j.str.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Sartori AA, Lingaraju GM, Hunziker P, Winkler FK, Jiricny J. Pa-AGOG, the founding member of a new family of archaeal 8-oxoguanine DNA-glycosylases. Nucleic Acids Res. 2004;32:6531–6539. doi: 10.1093/nar/gkh995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 25.Bjørås M, Seeberg E, Luna L, Pearl LH, Barrett TE. Reciprocal "flipping" underlies substrate recognition and catalytic activation by the human 8-oxoguanine DNA glycosylase. J Mol Biol. 2002;317:171–177. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- 26.Banerjee A, Yang W, Karplus M, Verdine GL. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 2005;434:612–618. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 27.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 29.Denver DR, Swenson SL, Lynch M. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol. 2003;20:1603–1611. doi: 10.1093/molbev/msg177. [DOI] [PubMed] [Google Scholar]
- 30.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 31.Radom CT, Banerjee A, Verdine GL. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem. 2007;282:9182–9194. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier H, Sawaya MR. Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis. Biochemistry. 1996;35:12778–12787. doi: 10.1021/bi960790i. [DOI] [PubMed] [Google Scholar]
- 33.Huffman JL, Sundheim O, Tainer JA. DNA base damage recognition and removal: new twists and grooves. Mutat Res. 2005;577:55–76. doi: 10.1016/j.mrfmmm.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 35.Hamburger AE, West AP, Jr, Bjorkman PJ. Crystal structure of a polymeric immunoglobulin binding fragment of the human polymeric immunoglobulin receptor. Structure. 2004;12:1925–1935. doi: 10.1016/j.str.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Rudolph MG, Wingren C, Crowley MP, Chien YH, Wilson IA. Combined pseudo-merohedral twinning, non-crystallographic symmetry and pseudo-translation in a monoclinic crystal form of the gammadelta T-cell ligand T10. Acta Crystallogr D Biol Crystallogr. 2004;60:656–664. doi: 10.1107/S0907444904002239. [DOI] [PubMed] [Google Scholar]
- 37.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 40.DeLano WL. http://www.pymol.org. San Carlos, CA, USA: 2008. "The PyMOL Molecular Graphics System.". [Google Scholar]