Abstract
Somatic loss of wild-type alleles can produce disease traits such as neoplasia. Conversely, somatic loss of disease-causing mutations can revert phenotypes, however these events are infrequently observed. We demonstrate that ichthyosis with confetti, a severe, sporadic skin disease, is associated with thousands of revertant clones of normal skin that arise from loss of heterozygosity on chromosome 17q via mitotic recombination. This enabled mapping and identification of disease-causing mutations in keratin 10 (KRT10); all result in frameshifts into the same alternative reading frame, producing an arginine-rich C-terminal peptide that redirects keratin 10 from the cytokeratin filament network to the nucleolus. The general rarity of spontaneous reversion and the specific absence of reversion of other dominant mutations in KRT10 implicate the frameshift peptide in the appearance of revertants. These results may have ramifications for reversion of other mutations.
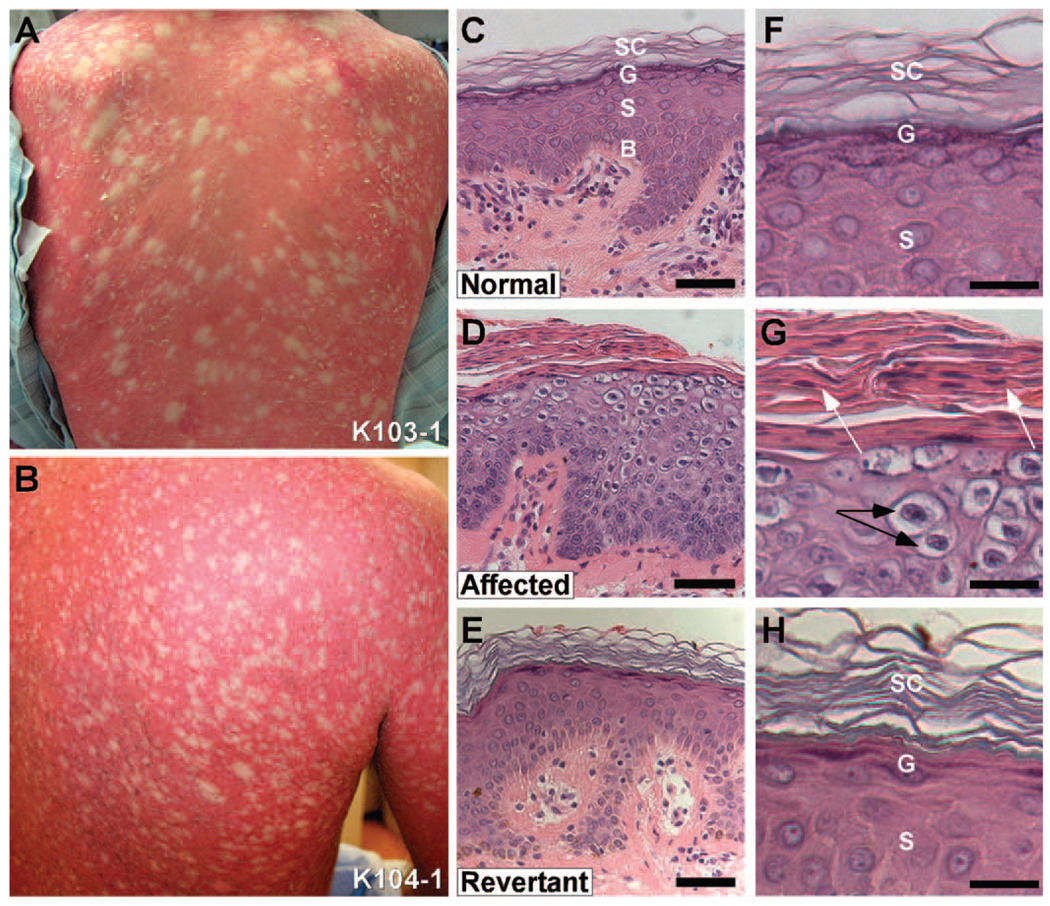
Ichthyosis with confetti (IWC; also known as ichtyose en confettis, congenital reticular ichthyosiform erythroderma, and ichthyosis variegata) is a very rare, sporadic severe skin disease of unknown cause (1–3). Affected subjects are born with erythroderma (red skin) owing to defective skin barrier function, prominent scale, and palmoplantar keratoderma (thickening of skin on palms and soles). Poor skin integrity leads to bacterial infections and, frequently, early death. Early in life, hundreds to thousands of pale confetti-like spots appear across the body surface and increase in number and size with time (Fig. 1A,B). Histology of ichthyotic skin shows epidermal thickening and disordered differentiation above the basal layer, with perinuclear vacuolization, lack of a granular layer, and hyperkeratosis (thickening) with retained nuclei in the stratum corneum (Fig. 1C,D).
Fig. 1.
Frequent revertants in Ichthyosis with confetti. (A, B) The backs of an 18 year-old female subject (103-1) and 42 year-old male (104-1) show background redness and scaling with hundreds of white, normal-appearing "confetti" spots. (C) Histology of normal human skin showing basal layer (labeled ‘B’), stratum spinosum (‘S’), granular layer (‘G’) and stratum corneum (‘SC’). (D) Affected skin shows loss of differentiation of all layers above the basal layer and hypercellularity with increased epidermal thickness. There is no granular layer, marked peri-nuclear vacuolization in the suprabasal epidermis, and retention of cell nuclei in the stratum corneum. (E) Revertant skin shows normalization of epidermal thickness and architecture, with normal granular layer, normal spinous layer, and stratum corneum. (Scale bars in C–E = 50 µm) (F) High power of spinous layer in normal epidermis with intercellular spines visible, overlying granular layer with purple keratohyalin granules and basket weave stratum corneum. (G) High power view of affected skin shows peri-nuclear vacuolization (black arrows), lack of keratohyalin granules, and retained nuclei (white arrows) in the stratum corneum. (H) High power view of revertant skin shows normal spinous layer with intercellular spines, granular layer with purple keratohyalin granules in keratinocytes, and basket weave stratum corneum. (Scale bars in F–H = 25 µm)
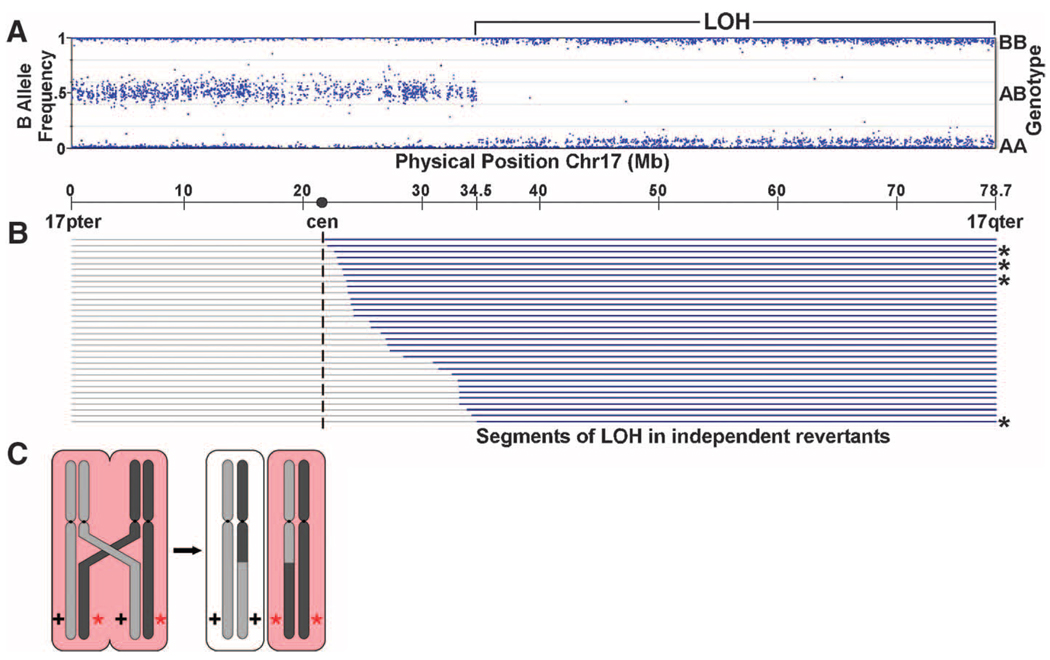
We studied 7 kindreds with characteristic IWC (Fig. S1). In 5 kindreds, there was a single affected offspring of unaffected, unrelated parents, and in two an affected parent had affected offspring. Biopsy of confetti spots in different kindreds revealed that these have normal histology (Fig. 1E,H), consistent with each representing a revertant from clonal expansion of a normal stem cell. This observation suggested that IWC might be caused by dominant mutations that are lost in revertant spots, and the high frequency of reversion suggested deletion, gene conversion, or recombination as possible mechanisms. To test this, we compared genotypes of DNA from blood and cultured keratinocytes from biopsies of diseased and revertant skin of subject 106-1 typed on Illumina arrays (4). In contrast to blood and disease keratinocytes, revertant DNA showed a single large segment of copy-neutral LOH on chromosome 17q extending from 34.5 Mb to the telomere at 78.7 Mb (Fig. 2A, Fig. S2). Three additional revertant spots from this subject also showed copy-neutral LOH extending from proximal 17q to the telomere, each with different inferred start-sites for LOH (Fig. 2B), thereby excluding simple genetic mosaicism. These findings are consistent with mitotic recombination as the mechanism of LOH (Fig. 2C). In each revertant the same parental haplotype was lost, consistent with loss of a dominant mutation. We then analyzed 28 revertant spots from 5 additional patients. Again, all revertants showed copy-neutral LOH on 17q extending to the telomere (Fig. 2B). Sites of inferred recombination are distinct and are confined to the interval from 21.7 Mb (near the centromere) to 34.5 Mb. These observations suggest that IWC is genetically homogeneous and localize the disease locus to a 99.9% confidence interval, calculated from the position of the most distal recombinant and the number of independent recombinants, to the 34.5 to 37.7 Mb interval on 17q. This interval is notable for a gene cluster encoding 28 type-1 keratins and 24 keratin-associated proteins (5).
Fig. 2.
Revertant spots show loss of heterozygosity on 17q. (A) Genotypes on chromosome 17 from revertant keratinocytes of IWC subject 106-1 are shown. From 17pter to 34.5M base pairs, genotypes show the expected heterozygosity with genotypes identical to blood and disease keratinocyte DNA (Fig. S2), while from 34.5M base pairs to 17qter, genotypes are homozygous with no change in diploid copy number (Fig. S2). (B) Results of genotyping keratinocytes of 32 revertant spots from 7 unrelated IEC subjects (106-1 revertants denoted with *). Gray lines = genomic segments with heterozygous genotypes matching blood DNA; blue lines = segments showing copy-neutral LOH. These results are consistent with IWC being caused by a dominant allele distal to 34.5M bp that is lost by mitotic recombination, as depicted in panel (C).
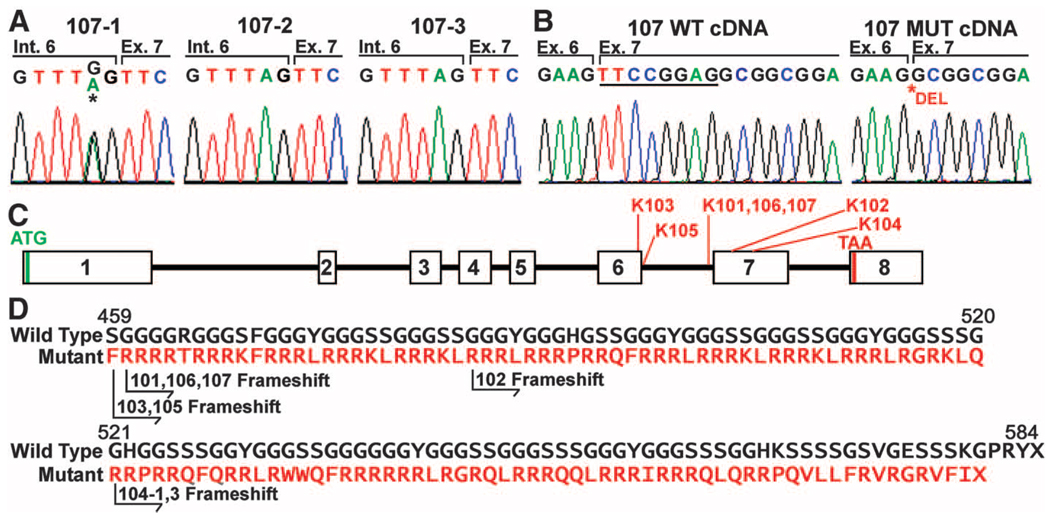
Assuming that affected offspring of unaffected parents harbor de novo mutations in the IWC gene, we conducted Illumina sequencing of overlapping PCR amplicons spanning the entire critical interval in a parent-offspring trio. At mean 95× per base coverage, ~95% of all the bases in the 99.9% confidence interval were read at least 10 times in each subject, enabling high quality genotype calls. The affected subject had a single de novo mutation (Table S1), which was confirmed by Sanger sequencing (Fig. 3A). The mutation abolishes the canonical splice acceptor site of intron 6 of keratin 10 (KRT10) (TAG to TGG mutation). Moreover, the mutation was absent in revertant spots (Fig. S3). Sequencing of KRT10 transcripts from mutant keratinocyte cDNA revealed a wild-type and a mutant isoform showing splicing at an AG site at bases 7–8 in the normal exon 7, leading to an 8 base deletion (Fig. 3B,C). This results in a frameshift at normal codon 458 leading to 119 aberrant amino acids followed by termination at codon 577. The frameshift peptide has an extremely skewed amino acid composition, with 67 arginine residues (Fig. 3D).
Fig. 3.
Mutations in KRT10 (keratin 10) cause IWC. (A) Sanger sequencing confirms de novo mutation in KRT10 from Illumina sequencing that is absent in the parents (107-2 and 107-3) but present in the affected offspring (107-1) that abolishes the splice acceptor site of intron 6. (B) Abnormal splicing of KRT10. cDNA from diseased keratinocytes of 107-1 shows 2 splice forms, one wild-type (WT) and one using an AG splice acceptor that deletes 8 bases from WT cDNA (underlined). (C) The genomic structure of KRT10 is shown. The locations of mutations found in IWC kindreds are indicated. (D) IWC frameshifts all produce an arginine-rich C-terminal peptide. The normal sequence of the C-terminal 226 amino acids of keratin 10 is shown; below, in red, the sequence of the frameshift peptides found in IWC are shown, with the position of the frameshift in each kindred indicated.
Sequencing of KRT10 from genomic DNA and disease keratinocyte cDNA in the 6 other IWC kindreds identified de novo mutations in all four simplex kindreds and transmitted mutations in the two multiplex kindreds (Table S2, Figs. 3C,D, Fig. S4). Most interestingly, all mutations resulted in cDNAs encoding frameshifts that enter the same alternative C-terminal reading frame (Figs. 3C,D S5). Mutations included two additional intron 6 splice acceptor mutations, an intron 6 splice donor site mutation that results in skipping of exon 6, two frameshift mutations in exon 7, and an exon 6 mutation that creates a premature splice donor site. All of these mutations are absent among control chromosomes, and each is lost in revertant spots (Fig. S3). These findings provide unequivocal evidence that mutations in KRT10 cause IWC.
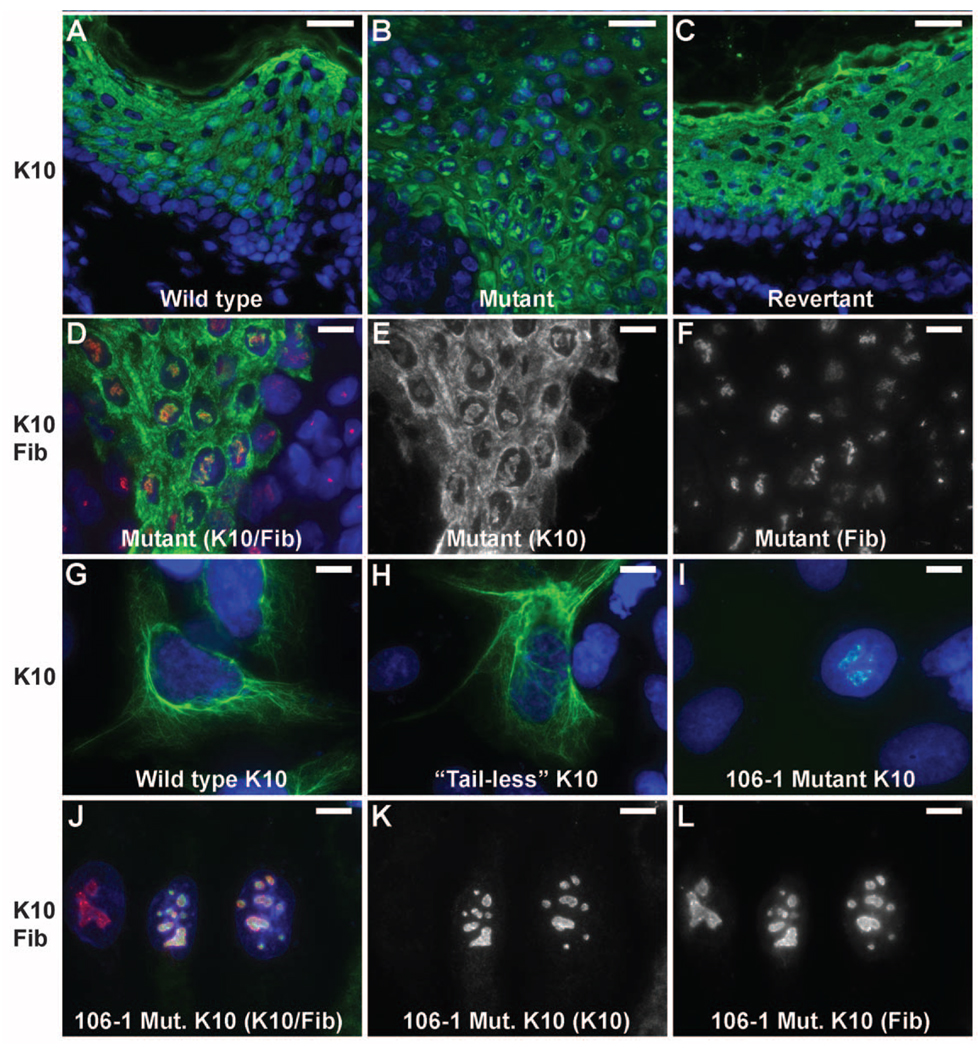
K10 is highly expressed in the suprabasal layers of the epidermis and forms heterodimers with keratin 1 (6, 7), which then assemble to form 10 nm intermediate filaments (8). In diseased skin, keratin 10 levels are reduced (Fig. S6A, B), and electron microscopy reveals a marked reduction in the total number of cytokeratin filaments and poor investment of desmosomes with filaments (Fig. S7). In addition, however, K10 is also mislocalized in diseased skin, with prominent nuclear localization in discrete foci that prove to be nucleoli by co-staining with fibrillarin (Fig. 4A–F); this nuclear localization is not seen in normal or revertant skin. Keratin 1 shows similar mislocalization in diseased skin (Fig. S6C–E). Similarly, while wild-type and C-terminal truncated K10 localize to the cytoplasmic filament network when expressed in PLC cells, K10 harboring disease-causing frameshifts localizes virtually exclusively to the nucleolus (Fig. 4G–L).
Fig. 4.
Mutant keratin 10 is redirected to nucleoli in vivo and in vitro. (A–C) Images of normal, mutant and revertant skin stained with DAPI and antibodies to keratin 10 reveal discrete foci of nuclear keratin 10 in mutant skin which are absent in normal and revertant skin. (Scale bars = 50 µm). (D–F) Co-staining with the nucleolar marker fibrillarin shows that keratin 10 is in the nucleolus. (Scale bars = 10 µm) (G–I) Constructs bearing wild-type keratin 10 (G), keratin 10 truncated at the beginning of the tail domain (amino acid 459) (H) and keratin 10 with the kindred 106 frameshift mutation beginning at codon 460 (I) were expressed in PLC cells and stained with DAPI and monoclonal antibody to keratin 10. Wild-type and ‘tailless’ K10 integrate into the cytoplasmic filament network while the IWC mutant K10 localizes to nucleoli as shown by costaining with fibrillarin (J–L). (Scale bars in G–L = 10 µm).
The results demonstrate that IWC is caused by dominant mutations in keratin 10 that all produce an arginine-rich C-terminal peptide that confers mislocalization of the protein to the nucleolus. This mislocalization provides a mechanism for disruption of the keratin filament network, which in turn contributes to loss of barrier function. The observed abnormalities in differentiation seem unlikely to be attributable simply to loss of the keratin network and suggest that the mutant K10 may have broader effects to disrupt cellular physiology. K10 has previously been proposed to play a role in cell cycle regulation (9). It will be interesting to assess whether the IWC mutations disrupt general cellular functions such as ribosome biogenesis, protein synthesis, the cell cycle, DNA repair and replication.
The nucleolar localization of mutant keratin 10 is likely attributable to RNA binding owing to the extremely arginine-rich frameshift peptide and the high concentration of ribosomal RNA in the ribosome assembly factory; virtually all RNA binding proteins have arginine rich motifs that interact with the phosphate backbone of RNA, and the specific arginine-rich sequences capable of binding to RNA are diverse (10, 11). Similarly, arginine-rich motifs also contribute to nuclear localization (12). While we have not seen localization of the frameshift peptide to other RNA- or DNA- containing structures, we cannot exclude such possibilities.
IWC is perhaps most remarkable for its exceptionally high frequency of spontaneous reversion, with more than a thousand revertants in many subjects. The mechanism - mitotic recombination - represents the complement of a mechanism for producing somatic homozygosity for tumor suppressor mutations (13–15). The revertants in IWC are clonal, detectable in the first year of life, and widely distributed in both sun-exposed and unexposed skin. These recombination events simultaneously create homozygous mutant cells, but we see no phenotypic evidence of these, suggesting they are cell-lethal or contribute little to the epidermal surface. Somatic reversion has previously been reported for several other disorders (16). These include recessive diseases such as Bloom syndrome and Fanconi anemia and X-linked Wiscott-Aldrich syndrome in which 10–20% of patients show some revertant blood cells (17–19). These revertant blood cell clones arise by various mechanisms including intragenic recombination, gene conversion, second-site complementation and direct reversion. In Bloom syndrome these revertant clones contributed to refined mapping of the Bloom locus, analogous to the mapping approach used herein (20). Similarly, mutations underlying a substantial number of other severe dominant skin diseases have been identified, including keratitis-ichthyosis-deafness syndrome, progressive symmetric erythrokeratoderma, ichthyosis bullosa of Siemens, and dominant dystrophic epidermolysis bullosa; to our knowledge, only a single revertant clone, produced by second-site complementation, has been reported for these diseases (21). Similarly, a fraction of patients with recessive skin disorders have been reported to have revertant patches of skin comprising 1–6 reported patches in a total of 7 patients. In cases where the mechanism was established, all arose by second site mutation or gene conversion (22–27). Moreover, no revertant clones have been reported, or seen in our clinics, in patients with dominant negative or recessive mutations in keratin 10 that cause a distinct disease, epidermolytic ichthyosis (also known as epidermolytic hyperkeratosis) (Fig. S8) (28). These exceptions underscore the infrequency of spontaneous reversion and the generally low frequency of mitotic recombination as a mechanism of reversion. In particular, the absence of reversion of other dominant missense mutations in KRT10 implicate the IWC frameshift mutations in the appearance of revertant clones.
The high frequency of somatic reversion of IWC requires either that revertant stem cells clones are under strong positive selection, or that the rate of production of revertant clones is markedly elevated, or both. The persistence of revertant clones requires that reversion must be present in epidermal stem cells. Epidermal stem cell units have been estimated to populate a fixed area of approximately 0.25 – 0.5 mm2 in human skin (29, 30). The revertant clones we observe in adults with IWC increase in size with time and reach up to 4 cm, consistent with positive selection. Nonetheless, rare revertants in other skin diseases have achieved very large size (22–27), arguing that positive selection is not likely the sole rate-limiting step in production of detectable revertants, and suggesting an increased rate of mitotic recombination in IWC. Similarly, the fact that none of the previously described revertants in other skin diseases but all of the IWC revertants have occurred via mitotic recombination lends support to an effect on the rate of mitotic recombination.
Both mechanisms would be most readily explained by effects of the mutant peptide in epidermal stem cells: toxic effects could give revertants a survival or replicative advantage; effects on DNA replication, repair or cell cycle could also promote mitotic recombination. While keratin 10 is classically regarded as an early differentiation marker, there is evidence that a small proportion of basal cells, which contain rare epidermal stem cells, express KRT10. Moreover, purified putative stem cells of the interfollicular epidermis and follicular bulge show substantial KRT10 expression in proliferating cells, supporting this possibility (30–32).
Genetic therapies for dominant diseases have focused on correction of mutations (33) or inhibition of mutant protein synthesis by antisense or interfering RNAs (34). The present results demonstrate high frequency spontaneous reversion of dominant mutations in vivo by mitotic recombination. While the potential of adverse effects from producing homozygosity at undesired loci across the genome must be carefully considered in conjunction with the potential benefit of reversion, these results nonetheless suggest the possibility that reversion of other mutations by similar mechanisms might be induced and/or selected for therapeutic benefit.
Supplementary Material
References and Notes
- 1.Camenzind M, Harms M, Chavaz O, Saurat JH. Ichythose en confettis. Ann. Dermatol. Venereol. 1984;111:675. [PubMed] [Google Scholar]
- 2.Brusasco A, et al. A case of congenital reticular ichthyosiform erythroderma – ichthyosis ‘en confetis’. Dermatology. 1994;188:40. doi: 10.1159/000247084. [DOI] [PubMed] [Google Scholar]
- 3.Marghescu S, Anton-Lamprecht I, Rudolph PO, Kaste R. Congenital reticular ichthyosiform erythroderma. Hautarzt. 1984;35:522. [PubMed] [Google Scholar]
- 4.Information on materials and methods is available on Science Online.
- 5.UCSC genome browser, build hg18 of the human genome, http://genome.ucsc.edu/cgi-bin/hgGateway, of the human genome.
- 6.Steinert PM. The two-chain coiled-coil molecule of native epidermal keratin intermediate filaments is a type I-type II heterodimer. J. Biol. Chem. 1990;265:8766. [PubMed] [Google Scholar]
- 7.Zhou X-M, Idler WW, Steven AC, Roop DR, Steinert PM. The complete sequence of the human intermediate filament chain keratin 10. Subdomainal divisions and model for folding of end domain sequences. J. Biol. Chem. 1988;263:15584. [PubMed] [Google Scholar]
- 8.Steinert PM. Analysis of the mechanism of assembly of mouse keratin 1/keratin 10 intermediate filaments in vitro suggests that intermediate filaments are built from multiple oligomeric units rather than a unique tetrameric building block. J. Struct. Biol. 1991;107:175. doi: 10.1016/1047-8477(91)90020-w. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, et al. An unexpected role for keratin 10 end domains in susceptibility to skin cancer. J. Cell Sci. 2006;119:5067. doi: 10.1242/jcs.03298. [DOI] [PubMed] [Google Scholar]
- 10.Draper DE. Themes in RNA-protein recognition. J. Mol. Biol. 1999;293:255. doi: 10.1006/jmbi.1999.2991. [DOI] [PubMed] [Google Scholar]
- 11.Burd CG, Dreyfuss G. Dreyfuss, Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 12.Sugaya M, Nishino N, Katoh A, Harada K. Amino acid requirement for the high affinity binding of a selected arginine-rich peptide with the HIWC Rev-response element RNA. J. Pept. Sci. 2008;14:924. doi: 10.1002/psc.1027. [DOI] [PubMed] [Google Scholar]
- 13.Knudson G. Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U.S.A. 1971;68:820. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavenee WK, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 15.O’Keefe C, McDevitt MA, Maciejewski JP. Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood. 2010;115:2731. doi: 10.1182/blood-2009-10-201848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J. Med. Genet. 2003;40:721. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellis NA, et al. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am. J. Hum. Genet. 1995;57:1019. [PMC free article] [PubMed] [Google Scholar]
- 18.Kalb R, Neveling K, Nanda I, Schindler D, Hoehn H. Fanconi anemia: causes and consequences of genetic instability. Genome Dyn. 2006;1:218. doi: 10.1159/000092510. [DOI] [PubMed] [Google Scholar]
- 19.Davis BR, Candotti F. Revertant somatic mosaicism in the Wiskott–Aldrich Syndrome. Immunol. Res. 2009;44:127. doi: 10.1007/s12026-008-8091-4. [DOI] [PubMed] [Google Scholar]
- 20.Ellis NA, et al. Bloom syndrome gene product is homogous to RecQ helicases. Cell. 1995;83:655. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 21.Smith FJD, Morley SM, McLean WHI. Novel mechanism of revertant mosaicism in Dowling-Meara epidermolysis bullosa simplex. J. Invest. Dermatol. 2004;122:73. doi: 10.1046/j.0022-202X.2003.22129.x. [DOI] [PubMed] [Google Scholar]
- 22.Pasmooij AM, Pas HH, Bolling MC, Jonkman MF. Revertant mosaicism in junctional epidermolysis bullosa due to multiple correcting second-site mutations in LAMB3. J. Clin. Invest. 2007;117:1240. doi: 10.1172/JCI30465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasmooij M, Pas HH, Deviaene FC, Nijenhuis M, Jonkman MF. Multiple correcting COL17A1 mutations in patients with revertant mosaicism of epidermolysis bullosa. Am. J. Hum. Genet. 2005;77:727. doi: 10.1086/497344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonkman MF, et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88:543. doi: 10.1016/s0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- 25.Darling TN, Yee C, Bauer JW, Hintner H, Yancey KB. Revertant mosaicism, partial correction of a germ-line mutation in COL17A1 by a frame-restoring mutation. J. Clin. Invest. 1999;103:1371. doi: 10.1172/JCI4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Almaani N, et al. Revertant mosaicism in recessive dystrophic epidermolysis bullosa. J. Invest. Dermatol. 2010;130:1937. doi: 10.1038/jid.2010.64. [DOI] [PubMed] [Google Scholar]
- 27.Schuilenga-Hut PH, et al. Partial revertant mosaicism of keratin 14 in a patient with recessive epidermolysis bullosa simplex. J. Invest. Dermatol. 2002;118:626. doi: 10.1046/j.1523-1747.2002.01715.x. [DOI] [PubMed] [Google Scholar]
- 28.Rothnagel JA, et al. Mutations in the rod domains of keratins 1 and 10 in epidermolytic hyperkeratosis. Science. 1992;257:1128. doi: 10.1126/science.257.5073.1128. [DOI] [PubMed] [Google Scholar]
- 29.Ghazizadeh S, Taichman LB. Organization of stem cells and their progeny in human epidermis. J. Invest. Dermatol. 2005;124:367. doi: 10.1111/j.0022-202X.2004.23599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen UB, Lowell S, Watt FM. The spatial relationship between stem cells and their progeny in the basal layer of human epidermis, a new view based on whole-mount labeling and lineage analysis. Development. 1999;126:2409. doi: 10.1242/dev.126.11.2409. [DOI] [PubMed] [Google Scholar]
- 31.Ohyama M, et al. Characterization and isolation of stem cell-enriched human hair follicle bulge cells. J. Clin. Invest. 2006;116:249. doi: 10.1172/JCI26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koçer SS, Djurić PM, Bugallo MF, Simon SR, Matic M. Transcriptional profiling of putative human epithelial stem cells. BMC Genomics. 2008;9:359. doi: 10.1186/1471-2164-9-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 34.Shen J, et al. Suppression of ocular neovascularization with siRNA targeting VEGF receptor 1. Gene Ther. 2006;13:225. doi: 10.1038/sj.gt.3302641. [DOI] [PubMed] [Google Scholar]
- 35.Acknowledgements: We thank the patients studied and their families for their invaluable contributions to this study. We thank Lynn Boyden and Susan Baserga for helpful discussions, Nancy DeSilva, Shrikant Mane and the Yale Center for Genome Analysis for assistance in development of critical methodologies, and Mark Ceneri for the identification of a previously unreported IWC kindred. Funding: The NIAMS/Yale Skin Diseases Research Center provided cell culture support for this work. K.A.C. is supported by a K08 award from NIAMS/NIH and a fellowship from the Foundation for Ichthyosis and Related Skin Types. Supported in part by a National Center for Research Resources High End Instrumentation Grant, the Yale Clinical and Translational Science Award, and the Yale Center for Human Genetics and Genomics. R.P.L. is an Investigator of the Howard Hughes Medical Institute. Competing interests: The authors declare that they have no competing interests.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.