Abstract
Membrane type-1 matrix metalloproteinase (MT1-MMP) is a key member of the matrix metalloproteinase (MMP) family. It participates in pericellular proteolysis of extracellular matrix (ECM) macromolecules and is essential for many biological and pathological processes, such as tumor development, angiogenesis and metastasis. A ligand that specifically binds to MT1-MMP may facilitate the labeling of this molecule, allow imaging at the cellular and organism levels, and provide a means for targeted drug delivery specific to MT1-MMP. A non-substrate MT1-MMP binding peptide was identified by screening a Ph. D.™ - 12 phage display peptide library and conjugated with near-infrared fluorescent (NIRF) dye Cy5.5 for tumor imaging. Peptide HWKHLHNTKTFL (denoted as MT1-AF7p) showed high MT1-MMP binding affinity. Computer modeling verified that MT1-AF7p binds to the MT-loop region of MT1-MMP and interacts with MT1-MMP through hydrogen bonding and hydrophobic interactions. MDA-MB-435 xenografts with high MT1-MMP expression had significantly higher tumor accumulation and better tumor contrast than the low MT1-MMP expressing A549 xenografts after intravenous injection of Cy5.5-MT1-AF7p. A novel non-substrate affinity peptide MT1-AF7p was found for MT1-MMP high affinity and specificity. Using NIRF imaging, we have demonstrated specific targeting of MT1-AF7p to MT1-MMP-expressing tumors. Thus, MT1-AF7p is an important tool for noninvasive monitoring of MT1-MMP expression in tumors, and it shows great potential as an imaging agent for MT1-MMP – positive tumors.
Keywords: matrix metalloproteinase, MT1-MMP (MMP-14), phage display peptide library, near-infrared fluorescence optical imaging
1. Introduction
The matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases that consist of 24 human MMPs, six of which are membrane bound [1]. The membrane type MMPs (MT-MMP) display the common structural domains of the MMP family, including a propeptide, a catalytic domain containing the Zn2+ - binding region, a hinge or linker region, and a hemopexin domain [2]. Four members of the MT-MMPs, MT1-, 2-, 3-, and 5-MMP (MMP-14, -15, -16, and -24), possess a type I transmembrane domain, while MT4-, and 6-MMP (MMP-17, -25) anchor to the membrane with a glycosylphosphatidylinositol (GPI) linkage. Membrane type-1 matrix metalloproteinase (MT1-MMP) has been shown to be a key member of the MMP family. It is also the most prominent member of the membrane type of MMP with much biological and pathological significance. MT1-MMP is intrinsically associated with the plasma membrane of normal and tumor cells and remodels the extracellular matrix (ECM) [3]. MT1-MMP displays a broad spectrum of activity against ECM components, such as type I and II collagens, fibronectin, vitronectin, laminin, fibrin and proteoglycan. MT1-MMP also activates pro-MMP-2 and pro-MMP-13 (pro-collagenase 3). This process requires the tissue inhibitor of MMP-2, which acts as an adaptor molecule mediating pro-MMP2 binding to MT1-MMP [4, 5]. In addition, MT1-MMP cleaves ECM proteins and controls the functionality of a number of cell adhesion and signaling receptors [6, 7]. It is directly involved in the cleavage of cell surface receptors including tissue transglutaminase, CD44, pro-αv integrin, syndecan-1, low-density lipoprotein (LDL) receptor-related protein and L-glycan [8, 9]. MT1-MMP knockout mice are dwarfs and most die from wasting by the time of early adulthood [10]. The mice also develop skeletal dysplasia, arthritis, severe osteopenia, and generalized soft tissue disorders. MT1-MMP-dependent proteolysis of both extracellular matrix components and cell surface adhesion receptors plays an essential role in physiological processes (wound healing; adipocyte differentiation; bone growth and remodeling) and pathological processes (arthritis; tumorigenesis; tumor growth; invasion; metastasis and angiogenesis) [9, 11].
The MT1-MMP is highly expressed in different cancers. For example, Takahashi et al. found that expression of MT1-MMP was elevated in 22 of 24 colon carcinomas that they examined [12]. It mediates pericellular proteolytic events that modulate cell attachment and motility [8], and its overexpression promotes migration, invasion and metastasis of cancer cells in vitro as well as in vivo [13, 14]. The enzyme has been shown to be essential for angiogenesis [15, 16]. Expression of MT1-MMP is crucial for cancer cell growth in a 3D collagen-based matrix [17], suggesting that MT1-MMP has important roles not only in cancer invasion but also in overall tumor progression [9].
Molecules capable of tracing MT1-MMP in vivo would be crucial to a detailed understanding of the nature of its expression, distribution, and its many biological and pathological functions. Furthermore, molecules that specifically target MT1-MMP would provide the potential for earlier detection and characterization of disease [18], targeted drug delivery [19, 20], and evaluation of treatment in diseases where MT1-MMP is overexpressed.
Phage display technology has been widely used to indentify cell surface receptor binding peptides by screening with immobilized, purified targets, intact cells or by in vivo selection [21–25]. A series of protease substrates of MT1-MMP have already been well documented [26, 27]. Ohkubo et al. searched for amino acid sequences cleaved by this protease using a hexamer substrate phage library and found that the consensus substrate sequences for MT1-MMP appeared to be P-X-G/P-L at the P3-P1’ site of the substrate [27]. Kridel et al. also identified a panel of optimal peptide substrates for MT1-MMP using substrate phage display. They divided the peptide substrates into different groups based on degree of selectivity for MT1-MMP. They showed that highly selective substrates lacked the characteristic Pro at the P3 position and instead contained an Arg at the P4 position, which is essential for efficient hydrolysis and selectivity for MT1-MMP [26].
Despite the success of identifying MT1-MMP substrates, no high affinity binding peptide for this protease has been reported. In order to find a peptide that can bind to MT1-MMP with high specificity, we used a recombinant form of MT1-MMP as a target to screen a random 12-mer phage-displayed library. We obtained a number of peptides with similar amino acid motifs as previously described, by using a substrate phage display [27]. To avoid repeat discovery of MT1-MMP substrates as affinity ligands, we abandoned the traditional approach of using recombinant protein as the screening target but used a peptide sequence that is specific to MT1-MMP as the target to screen the phage display peptide library. There are several advantages of using peptide as a phage display target: First, peptide libraries are readily available and the purity of the target peptide is higher than that of the other potential targets such as proteins or cells, therefore screening can be more effective. Second, the exact recognition site between the target and ligand is known, which facilitate the understanding of the ligand-target binding mechanism. Third, using a unique peptide from certain protein as target is more prone to get specific ligands to the protein.
In the present study, we screened for peptides that bind to a unique peptide sequence on the surface of MT1-MMP by in vitro panning of a phage display library and found a 12-mer peptide HWKHLHNTKTFL (MT1-AF7p) that binds MT1-MMP with high affinity and specificity. We further showed that the peptide can be used to image MT1-MMP expression in vivo. To the best of our knowledge, this is the first time that a non-substrate MT1-MMP affinity peptide has been found. We believe that it has good potential for use as a ligand in applications targeting this protease.
2. Materials and Methods
2.1. In vitro panning of MT1-MMP binding peptides
The panning procedure was done as described in Ph.D.-12™ phage display peptide library kit manual. Briefly, the synthetic MT1-MMP sequence of amino acid 160–174 (MT1-160p) was dissolved in 0.1 M NaHCO3 (pH 8.6) at a concentration of 100 µg/ml and coated onto a 35 mm polystyrene dish in a humidified incubator, stored at 4°C overnight. After being blocked by blocking buffer (0.1 M NaHCO3, pH 8.6, 5 mg/ml BSA, 0.02% NaN3) for 1 h at 37°C, 10 µl phage display peptide library (4×1010 phages) were diluted in 1 ml TBST (TBS containing 0.1% [v/v] Tween-20) and exposed for 1 h to the dish. After that, unbound phages were washed off with TBST ten times. The bound phages were collected by adding 2 ml of 0.2 M Glycine-HCl (pH 2.2) containing 1 mg/ml BSA to the plate for 10 min and neutralized with 150 µl of 1 M Tris-HCl (pH 9.1). One µl of collected phages was picked for phage tittering. For the amplification of selected phage clones to be used in the next round of panning, the remaining phages were mixed with 20 ml of ER2738 culture (at early log stage) and incubated at 37°C with vigorous shaking for 4.5 h. The culture was then centrifuged for 10 min at 10,000 rpm at 4°C. Then, the upper 80% of the supernatant was pipetted to a fresh tube and added to 1/6 volume of NaCl/PEG (2.5 M NaCl with 20% [w/v] PEG-8000). The phages were allowed to precipitate overnight at 4°C. After centrifugation for 10 min at 10,000 rpm, 4°C, the amplified phages were collected and dissolved in 200 µl of TBS buffer (50 mM Tris and 150 mM NaCl, pH 7.5) and the titer was determined on LB/IPTG/Xgal plates. This panning protocol was repeated two more times. In the fourth round of panning, the panning protocol was a little modified. Briefly, a 35 mm polystyrene dish was coated with 100 µg/ml MT1-160p. After being blocked by blocking buffer, the amplified phages in the third round were added to co-incubate with MT1-160p for 1 h at room temperature. Unbound phages were washed off by TBST for ten times. The bounded phages were washed off competitively by incubating with TBS containing MT1-160p at a concentration of 100 µg/ml for 1 h and were collected for amplification and titer determination. At the end of the fourth round of panning, the phage clones were analyzed by ELISA, and clones that displayed high binding ability to MT1-160p were amplified and, after in vitro replication, the appropriate DNA regions were sequenced, using −96 gIII sequencing primer, to determine the corresponding peptide sequences.
2.2. Phage capture ELISA
MT1-160p was diluted in bicarbonate/carbonate coating buffer (100 mM, pH 9.6) to a final concentration of 100 µg/ml. Aliquots (100 µl) of this solution were added to the 96-well plate and incubated at 4°C for overnight. After being washed with 250 µl phosphate buffered saline (PBS, 2.32 g/L Na2HPO4, 0.2 g/L KCl, 0.2 g/L K3PO4, 8.0 g/L NaCl, pH 7.4), each well was filled with 200 µl of PBS containing 1% BSA to block the nonspecific sites. Then 100 µl of selected phages were added into each well, and incubation was carried out at 37°C for 2 h to allow the phages to bind to MT1-160p. Then the plate was washed 3× by PBST (PBS containing 0.05% [v/v] Tween-20), and each well was filled with 80 µl of mouse anti-M13 phage antibody (1:1000 diluted by PBS) and incubated at 37°C for 1 h. After three washes with PBST, 100 µl of horseradish peroxidase-conjugated goat anti-mouse antibody (1:2500 diluted by PBS) was added to each well. As negative controls, three wells were coated by MT1-160p and blocked with 200 µl PBS containing 1% BSA without adding phages. Finally, 200 µl HRP substrate, 3, 3’, 5, 5’-tetramethylbenzidine (TMB) solution, was added to each well and kept in the dark at 37°C for 30 min. The reaction was stopped by adding 50 µl 2 M H2SO4 before the absorbance was measured by microtiter plate reader (Thermo Labsystems) at 450 nm.
2.3. Measurement of binding affinity
Apparent Kd value between MT1-MMP and MT1-AF7p (phage-displayed peptide binds to MT1-MMP) was obtained by immobilizing MT1-AF7p (2.5 µg, 1.6 nmol) on the microtiter plate and detected by gradient diluted recombinant human MT1-MMP catalytic domain from 2.5 µM to 11 nM. Rabbit anti-human MT1-MMP antibody was added and HRP-conjugated goat anti-rabbit antibody was used as the secondary antibody in the assay. Finally, 200 µl TMB solution was added to each well and kept in the dark at 37°C for 30 min. The reaction was stopped by 50 µl 2 M H2SO4, added before the absorbance was measured in microtiter plate reader (Thermo Labsystems) at 450 nm.
2.4. AFM measurement of the force between MT1-AF7p and MT1-160p
AFM sample preparation
MT1-AF7p was dissolved in PBS at a final concentration of 0.66 mg/ml and incubated at 37°C for 10 min. A 5 µl droplet of MT1-AF7p solution was deposited onto the surface of freshly cleaved mica for 1 min. Then, the surface was rinsed with 1 ml PBS in order to stabilize the MT1-160p in their 3D conformations for AFM imaging.
Functionalized AFM tips
Aldehyde-PEG-NHS was used as a linker between the tip and peptide. 3 mg of aldehyde-PEG-NHS was dissolved in 0.5 ml CHCl3 containing 30 µl triethylamine. The reaction lasted for 2 h. After that, tips were washed by CHCl3. One hundred µl MT1-160p at a concentration of 100 µg/ml was added onto the tip. Then, 2 µl 1 M NaCNBH3 (32 mg NaCNBH3 dissolved in 50 µl 100 mM NaOH, to which was added 450 µl H2O to make a final volume of 500 µl) was added into the reaction solution. The reaction lasted for 1 h. In the end, 5 µl ethanolamine was used to quench the non-reacted aldehyde group.
AFM imaging
AFM imaging was performed in AAC Mode with an AFM 5500 (Agilent Technologies, Santa Clara, CA). We used silicon nitride AFM probes (Veeco, Camarillo, CA) with nominal spring constants of 0.01 N/m. The scanning frequency was typically 1.5 Hz to 1.7 Hz per line. We obtained MT1-160p images at room temperature in aqueous solution. When measuring the force between the MT1-AF7p and MT1-160p, and between scrambled-AF7p and MT1-160p, the functionalized tips were used and scanned at a frequency lower than 1 Hz.
2.5. NMR study
The NMR study of the peptides was carried out on a Bruker 600 MHz magnet at Changchun Institute of Applied Chemistry. Each 1D 1H spectrum was collected with a spectral width of ~13 ppm using a WATERGATE pulse sequence at room temperature. The samples were dissolved in 10% D2O/90% H2O with a concentration of about 0.2 mM for each peptide. The spectra were recorded for the separated peptides MT1-AF7p, MT1-160p, MT1-AF7p-scr, MT1-AF7p/MT1-160p mixture (1:1) and MT1-AF7p-scr/MT1-160p mixture (1:1). The data were processed with an exponential line broadening 5 Hz window function.
2.6. Molecular modeling
Molecule simulations were performed on a Linux system using InsightII software package developed by Accelrys (InsightII, version 98.0. Accelrys Inc; San Diego, CA). The Extensible and Systematic Force Field (ESFF) was used for energy minimization and docking simulations. The three-dimensional structure of MT1-MMP (PDB code 1BQQ) was obtained from the Protein Data Bank (www.rcsb.org). The catalytic domain of MT1-MMP was selected based on knowledge that the enzymatic activity and substrate specificity of MMP catalytic domains were reported to be similar to full-length activated MMPs [26]. Before the docking procedure, water molecules and ligand (TIMP-2) were removed from the protein crystal structure using Swiss-Pdb Viewer. The Builder module of Insight II was used to modify the PDB files. The first step was that hydrogen atoms were added to the enzymes under the condition of pH 7.5, which was similar to the environment for testing the MT1-MMP binding of the MT1-AF7p. Similarly, the starting structure of MT1-AF7p was modeled by the Builder module. To release any internal strains in the structure of MT1-AF7p, we fixed the secondary structure and performed an energy minimization of 300 steps of conjugate gradient with a convergence value of 0.05 kcal mol−1 Å−1. In addition, in an explicit solvent model, TIP3P water was used with 5 water layers added to MT1-AF7p.
Automated molecular docking studies of the bioactive peptides (MT1-AF7p) at the MT1-MMP-binding site were performed with the Affinity module of the Insight II software package (Accelrys Inc.). After the energy minimization, the potentials and partial charges were calculated and assigned to the MT-MMP according to Insight II, however, the formal charges were assigned artificially with a value of +2.0 given to the catalytic zinc ion. The flexible sites of receptor (MT1-MMP) were selected using the amino acid sequence 160–174 of MT1-MMP (MT-160p), while the other atoms of the receptor were held rigid. After all of the above were done, automatic molecular dockings were performed without any constraints.
2.7. Fluorescent staining
For cell immunostaining, MDA-MB-435 cells were seeded into eight-well chamber at the concentration of 1×104 cells/well. The next day, cells were fixed by 90% cold ether for 20 min at −20°C. After being blocked by 10% BSA at 37°C, MDA-MB-435 cells were detected using 2 µg/ml rabbit anti-MMP14 primary antibody for 2 h at room temperature, and then visualized by Cy3 conjugated donkey anti-rabbit secondary antibody (1: 2000 dilution).
To label cells by Cy5.5-MT1-AF7p, 10 nM Cy5.5-MT1-AF7p was added to the cells after being fixed and blocked as described above. Cells were incubated with Cy5.5-MT1-AF7p at room temperature for 1 h. To confirm that MT1-AF7p bound specifically to MT1-MMP, fixed MDA-MB-435 cells were blocked with 10% BSA for 1 h and incubated with 5 µM MT1-AF7p before Cy5.5-AF7p (10 nM) was added. After washing steps, cells were mounted with 4′, 6-diamidino-2-phenylindole (DAPI)–containing mounting medium and observed with an epifluorescence microscope (Olympus, X81).
2.8. Cell culture and animal models
Human breast carcinoma cell line MDA-MB-435 was cultured in L-15 medium containing 10% (v/v) fetal bovine serum (Invitrogen) supplemented with penicillin (100 µg/ml) and streptomycin (100 µg/ml) at 37° C with 5% CO2. Human lung adenocarcinoma epithelial cell line A549 was cultured in DMEM (Invitrogen) containing 10% (v/v) fetal bovine serum supplemented with penicillin (100 µg/ml) and streptomycin (100 µg/ml) at 37° C with 5% CO2. MDA-MB-435 and A549 tumor models were established by subcutaneous injection of 5×106 cells into the right front flank of female athymic nude mice (Harlan Laboratories, Frederick, MD). The mice were used for optical studies when the tumor volume reached 100 – 300 mm3. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health (NIH) Guide for the Care and Use of Animals, and under protocols approved by the NIH Clinical Center Animal Care and Use Committee (CC/ACUC).
2.9. Tumor immunohistochemistry
MDA-MB-435 and A 549 tumors from the tumor-bearing nude mice were frozen in OCT embedding medium. Cryosections were cut into 4 µm and subjected to staining. Briefly, tumor slides were dried in the air and fixed with cold acetone for 20 min and dried again in the air for 30 min at room temperature. After blocking with 10% BSA for 30 min, the sections were incubated with rabbit anti-MT1-MMP antibody (10 µg/ml, Abcam, MA) for 60 min at room temperature in the dark, and then visualized with FITC-conjugated donkey anti-rabbit secondary antibody. Finally, the slices were mounted with DAPI-containing mounting medium under an epifluorescence microscope (Olympus, X81).
2.9. Whole-body small animal optical imaging
Optical image acquisition and analysis were done using a Maestro 2.10 in vivo imaging system (Cambridge Research & Instrumentation, Woburn, MA; excitation = 675 nm, emission = 695 nm). MDA-MB-435 and A549 tumor-bearing mice were injected via tail vein with 10 µM (1 nmol in 100 µl PBS) Cy5.5- AF7p (n = 3 per group). At different time points postinjection (2, 4, and 24 h), the mice underwent in vivo optical imaging. During the injection and image acquiring process, the mice were anesthetized with 2.5% isoflurane in oxygen at a flow of 1.5 L/min. After image acquisition, spectral unmixing yielded the pseudocolored images of the pure spectrum of Cy5.5. All the images were normalized to the same scales and analyzed using Maestro software. For quantitative comparison, regions of interest (ROIs) were drawn over tumors, and the total signal (counts/s) for each area was measured. Results were presented as mean for a group of 3–6 animals.
Ex vivo imaging of excised tumors and organs gave further confirmation of the targeting specificity of MT1-AF7p. Female athymic nude mice bearing MDA-MB-435 and A549 tumors were injected with 1 nmol Cy5.5-AF7p to evaluate the distribution of MMP-14 affinity peptide in tumor tissues and the major organs. At 4 h postinjection, the tumor-bearing mice were sacrificed and their major organs, tissues and tumors were harvested, and placed on black paper for ex vivo imaging. The results were presented as the average scaled signal from the organs and tumors. Values were expressed as mean ± SD for a group of three animals.
2.10. Tumor histochemistry
To investigate the localization of Cy5.5-MT1-AF7p in tumor, MDA-MB-435 and A549 tumor mice injected with 1 nmol of Cy5.5-MT1-AF7p were sacrificed after 2 hours blood circulation. The tumors were collected and made into frozen tissue blocks. These tumor specimens were subsequently sectioned with a thickness of 10 µm. The tumor slices were observed directly after mounting with DAPI-containing mounting medium under an epifluorescence microscope (Olympus, X81). Fluorescence pictures were taken using Cy5.5 filter settings (excitation = 675 nm, emission = 695 nm).
2.11. Statistical analysis
Results were expressed as mean ± SD. Two-tailed paired and unpaired Student’s t tests were used to test differences within groups and between groups, respectively. P values < 0.05 were considered statistically significant.
3. Results
3.1. Identification of a unique peptide sequence from MT1-MMP
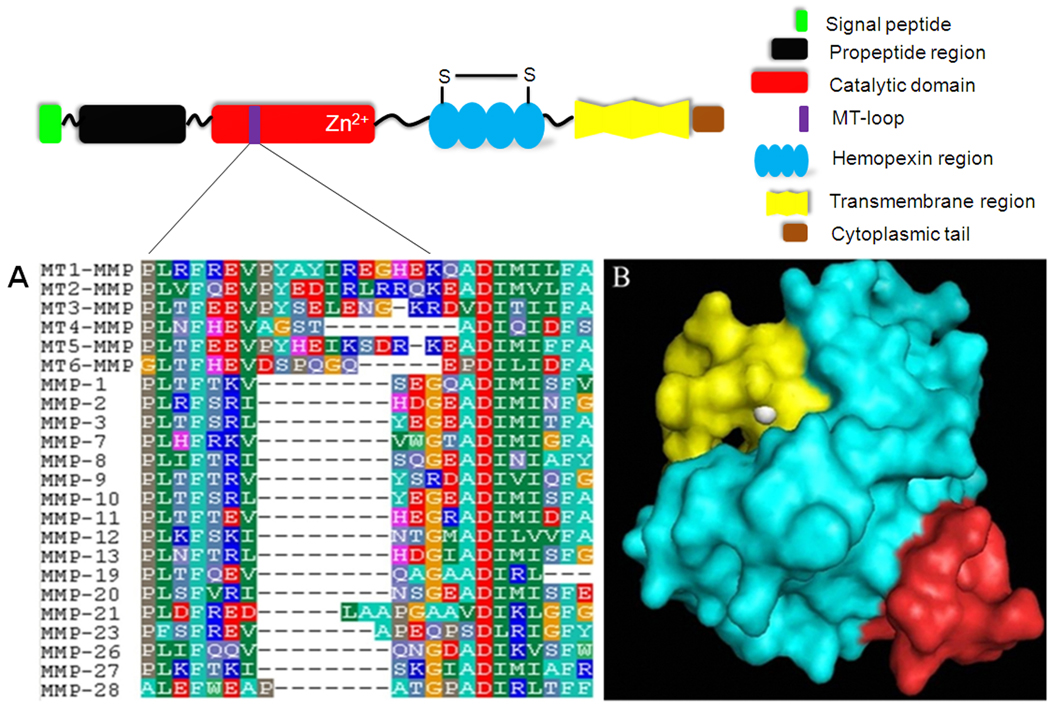
All human MMPs sequences were aligned and compared using BioEdit software. A unique sequence, 160REVPYAYIREGHEKQ174, located on MT1-MMP was identified. This segment mostly belongs to the MT-loop of MT1-MMP, an insertion of eight amino acids with residues 163PYAYIREG170 in between strands βII and III in the catalytic domain, which is a characteristic structure of membrane type MMPs with a transmembrane domain (Fig. 1A) [28]. In addition, this sequence is located on the surface of MT1-MMP, and thus can be easily accessed by an affinity ligand (Fig. 1B). We designated the 160REVPYAYIREGHEKQ174 peptide sequence as MT1-160p and used it to screen the phage display peptide library for a MT1-MMP binding peptide.
Fig. 1.
(A) Partial sequences alignment for MMPs. Numbering is according to the sequence of MT1-MMP. The peptide sequence 160REVPYAYIREGHEKQ174 is unique to MT1-MMP. Amino acids are represented as single-letter codes. The gap (represented by “−”) is auto-generated by the BioEdit software. (B) The 3D structure of MT1-MMP is shown as an electrostatic potential surface with the target peptide sequence 160REVPYAYIREGHEKQ174 colored in red. The catalytic center, in yellow, is located on the surface of the MT1-MMP. Zinc is shown in white.
3.2. Phage display identification of MT1-MMP binding peptide
Phage clones that can bind to MT1-160p were isolated by four rounds of panning. In each round, the unbound phages were cleared by washing with stepwise increases in tris-buffered saline and Tween-20 (TBST), and then the bound phages were amplified in ER2738 for the following round of panning. ELISA was used to measure the affinity between phages and the MT1-160p. The phages with stronger binding abilities were significantly enriched after each round of panning. Phages with high affinity for the target were analyzed for their apparent Kd with MT1-160p. A peptide displayed on the surface of phage with a sequence of HWKHLHNTKTFL, which had the lowest apparent Kd value (0.075 nM) for MT1-160p was designated as MT1-AF7p.
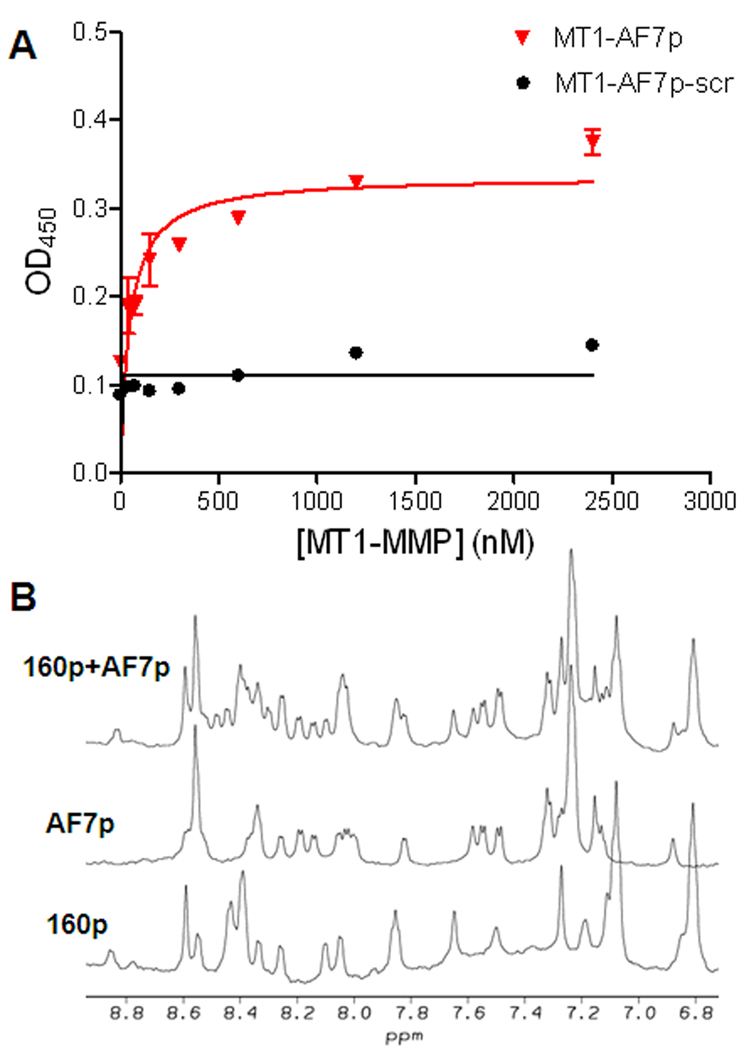
The MT1-MMP binding affinity of MT1-AF7p was determined by ELISA. We coated a 96-well plate with MT1-AF7p and incubated with serial diluted concentrations of MT1-MMP. The bound MT1-MMP was detected by an MT1-MMP polyclonal antibody and HRP-conjugated goat anti-rabbit secondary antibody. A scrambled peptide MT1-AF7p-scr was coated as a control. As shown in Fig. 2A, the apparent Kd value for MT1-AF7p was 47.4 nM, while MT1-AF7p-scr did not show any binding with MT1-MMP.
Fig. 2.
(A) The MT1-MMP binding affinity of MT1-AF7p and MT1-AF7p-scr. An apparent Kd value was measured by ELISA and analyzed by GraphPad Prism 4.0 software. The apparent Kd between MT1-AF7p and MT1-MMP is 47.41 nM. MT1-AF7p-scr did not show any binding with MT1-MMP. (B) The 1D NMR spectra of MT1-AF7p, MT1-160p, and their mixtures. The peak changes were indicated by arrows in the spectrum. A novel peak appeared at about 8.5 ppm in the MT1-160p/MT1-AF7p mixture, while a peak at about 8.0 ppm for MT1-AF7p and a peak at about 7.2 ppm for MT1-160p were missing in the spectrum of the mixture.
The comparison of 1D NMR spectra of MT1-AF7p, MT1-160p, and MT1-AF7p/MT1-160p mixture was shown in Fig. 2B. The majority of the resonances from the backbone amide protons in all three spectra are located in a narrow region of 7.5–8.6 ppm, indicating that the peptides are in random coil states either with or without the existence of the other peptide. However, several chemical shift changes were observed in the spectrum of the mixture (the spectrum labeled with 160p+AF7p) compared to those of the individual peptides (the spectrum labeled with AF7p and 160p, respectively). For example as arrow indicated in the spectrum, the resonance at 8.45 ppm in the spectrum of the mixture was not observed in either spectrum of the individual peptides. At the same time, the resonance at about 8.0 ppm in the spectrum of AF7p and the resonance at about 7.2 ppm in the spectrum of 160p disappeared in the spectrum of the mixture. The results strongly suggest that two peptides interact with each other in solution, otherwise the NMR spectrum of the mixture should be a simple add-up of the spectra of the two individual peptides. On the other hand, the mixture of scrambled peptide MT1-AF7p-scr and MT1-160p had minimal NMR spectrum changes from the overlay of these two peptide spectra. Small chemical shift movement at about 8.3 ppm is probably due to variation in viscosity or pH variation, or nonspecific sticking, rather than effective interactions (Fig. S1). The interaction between MT1-AF7p and MT1-160p was further studied by atomic force microscopy (AFM) (Fig. S2). The two lines in Fig. S2 show the direction of measuring motion: approach (red) and retraction (blue). The force between 160p and AF7p was calculated to be 304 pN. By the same method, the force between MT1-AF7p-scr and MT1-160p was negligible, again confirming the strong interactions between MT1-AF7p and MT1-160p.
3.3. Molecular modeling of interaction between MT1-AF7p and MT1-MMP
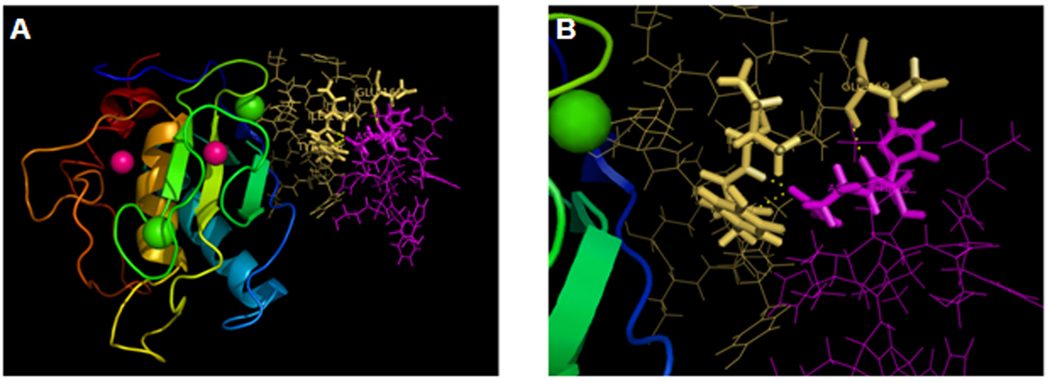
A molecular modeling experiment using Insight II software package revealed that when MT1-AF7p binds to MT1-MMP at lowest energy, 29 amino acids belonging to MT1-MMP interact with MT1-AF7p within a distance of 10 Å. Among these 29 amino acids, 13 have the lowest interaction energy (Etotal < −1 kcal/mol) (Table 1). A closer look at these amino acids shows that most of them are within MT1-160p, which strongly supports our results obtained by phage display (Fig. 3). Within a 5 Å radius, 13 amino acids of MT1-MMP had the lowest range of interaction energies, specifically the following amino acid residues in MT1-AF7p: His4, His6, Asn7, Thr8, Lys9, and Leu12. Among them, His4, His6, Asn7, and Leu12 of MT1-AF7p showed the lowest interaction energy, which indicates that they bind to MT1-MMP more tightly than the others. In line with this result, docking experiment also revealed that MT1-AF7p forms three hydrogen bonds with MT1-MMP: Asn7 of MT1-AFp with Try166 of MT1-MMP, Asn7 of MT1-AFp with Ile167 of MT1-MMP, and His6 of MT1-AFp with Glu169 of MT1-MMP.
Table 1.
Molecular docking shows most reaction amino acids between MT1-MMP and MT1-AF7p are located within the MT1-160p sequence.
| Amino Acid | Evdw (kcal/mol) | EEle (kcal/mol) | ETotal (kcal/mol) |
|---|---|---|---|
| GLN129 | −0.81938 | −0.852713 | −1.67209 |
| PRO163 | −0.203475 | −1.22109 | −1.42457 |
| TYR164 | −2.94284 | −0.0409189 | −2.99076 |
| ALA165 | −3.12668 | −4.24512 | −7.3718 |
| TYR166 | −6.67669 | −8.68291 | −15.3596 |
| ILE167 | −2.12323 | −10.4497 | −12.573 |
| ARG168 | 0.935912 | −9.12259 | −8.18668 |
| GLU169 | −5.83048 | −11.6427 | −17.4732 |
| GLY170 | −0.808551 | −4.90689 | −5.71544 |
| HIS171 | −0.323898 | −3.42 | −3.7439 |
| GLU172 | −0.33641 | −4.03144 | −4.36789 |
| GLN174 | −0.160664 | −4.29591 | −4.45656 |
| ALA175 | −0.10965 | −2.10506 | −2.21471 |
Fig. 3.
Molecular modeling of interaction between MT1-AF7p and MT1-MMP. (A) MT1-MMP and MT1-AF7p. The zinc atoms in the MT1-MMP are shown in pink and the calcium atoms are shown in green. The MT-loop of MT1-MMP is shown as yellow sticks and MT1-AF7p is colored in pink. (B) A closer view of the interface of the interaction between MT1-AF7p and MT1-MMP.
3.4. In vivo imaging of MT1-MMP using Cy5.5-MT1-AF7p
We used in vivo optical imaging studies to investigate the MT1-MMP targeting efficiency of MT1-AF7p in live mice. Optical imaging is becoming an increasingly important tool for visualizing molecular events in vivo. Near-infrared fluorescent (NIRF) dyes allow the imaging of deep tissues in rodents because of the relatively high penetration, and low tissue absorption and scattering of near-infrared wavelengths [24, 29]. Here, Cy5.5 was conjugated onto the N-terminate of MT1-AF7p.
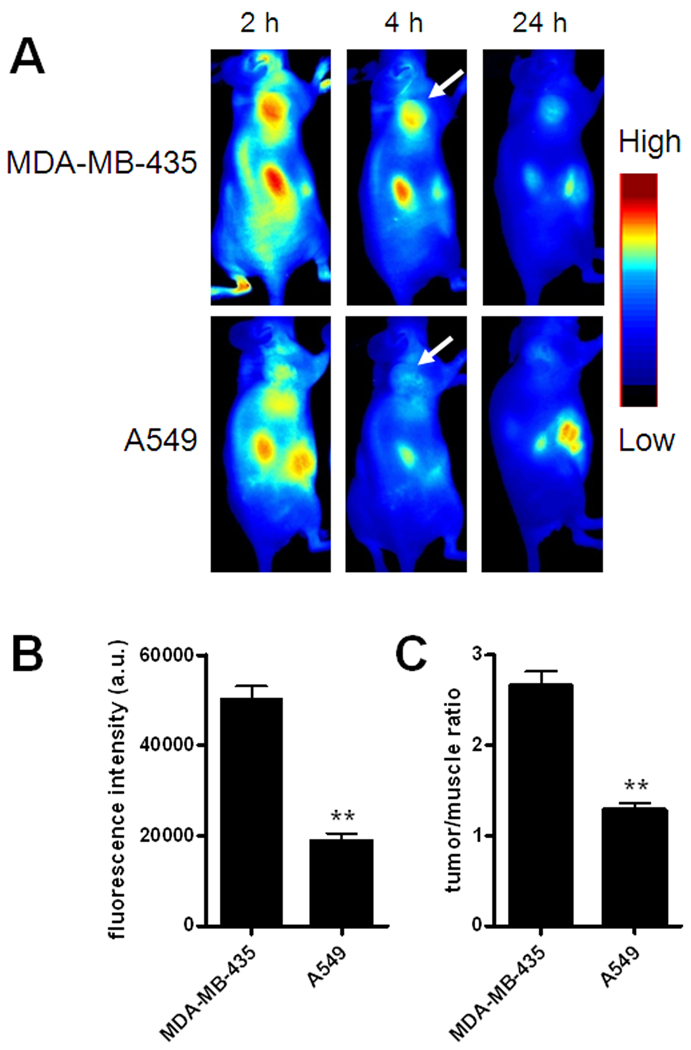
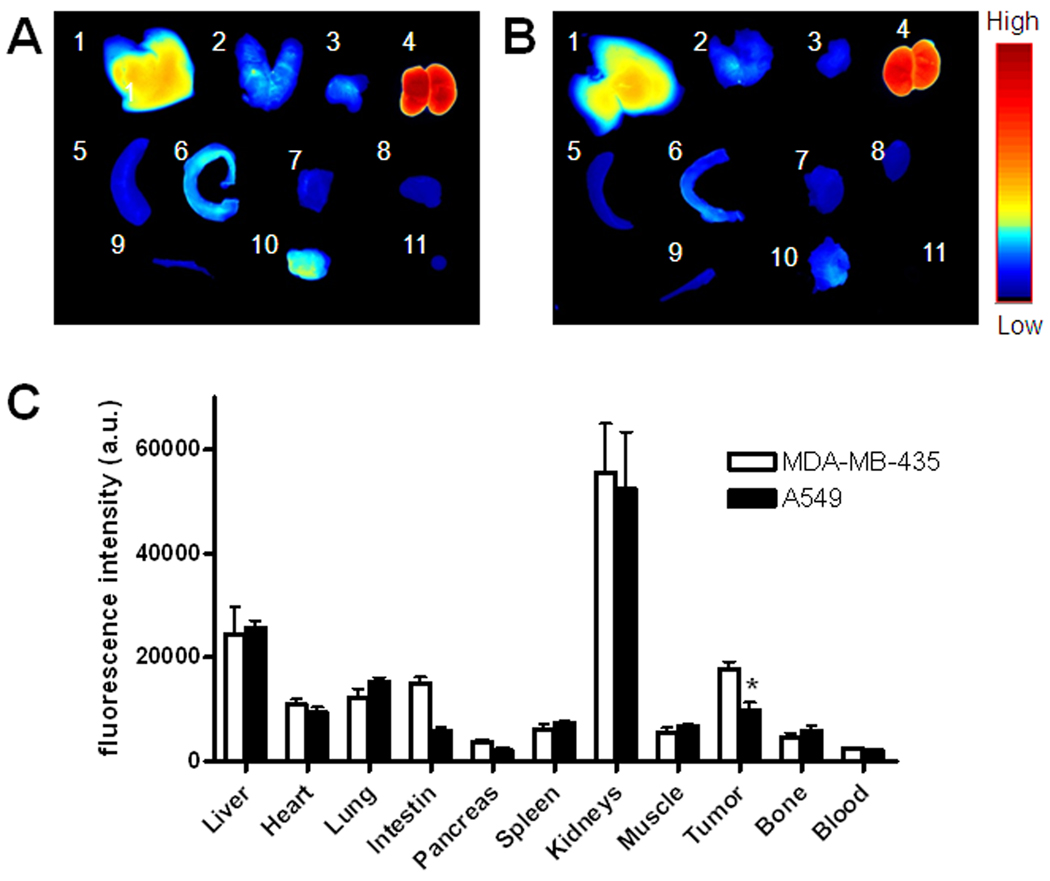
To evaluate the specificity of Cy5.5-AF7p to MT1-MMP in vivo, MDA-MB-435 breast cancer model with high MT1-MMP expression and A549 lung cancer model with low MT1-MMP expression (Fig. S3–4) were chosen. As shown in Fig. 4, MDA-MB-435 xenografts had significantly higher tumor accumulation and better tumor contrast than the A549 xenografts (p < 0.05, n = 3/group) at 4 h postinjection of 1 nmol of Cy5.5-MT1-AF7p. To investigate the biodistribution of Cy5.5-AF7p in the MDA-MB-435 and A549 tumors and normal tissues, we performed ex vivo optical imaging of tissues obtained at this 4 h time point (Fig. 5). Fluorescence intensities were calculated from whole organs, as well as from tissues. The ex vivo quantification corroborated the observations from in vivo optical imaging region-of-interest (ROI) analysis. It was also of note that Cy5.5-MT1-AF7p had a prominent renal clearance. Histological analysis of tumor cryosections also showed MT1-AF7p-specific targeting of MT1-MMP in tumor slices (Fig. S5). Taken together, Cy5.5-MT1-AF7p showed MT1-MMP-specific targeting potential in tumors. By optical imaging techniques, we have demonstrated the targeting capability of MT1-AF7P. However, more precise quantitative data on tumor uptake and pharmacokinetics will require radiolabeled MT1-AF7p in conjunction with such nuclear medicine modalities as positron emission tomography (PET) and single-photon emission computed tomography (SPECT), but this will be the subject of a subsequent study.
Fig. 4.
Near-infrared optical imaging of MT1-MMP by Cy5.5-MT1-AF7p in different tumor models. (A) Representative sagittal views of MDA-MB-435 and A549 tumor-bearing mice at different time points after intravenous injection of Cy5.5-MT1-AF7p. High MT1-MMP expressing MDA-MB-435 tumors show persistently high uptake of Cy5.5-MT1-AF7p, whereas low MT1-MMP expressing A549 tumors have some initial tumor uptake due to passive targeting followed by rapid clearance. White arrows indicate the tumors. (B) Region of interest (ROI) analysis of MDA-MB-435 and A549 tumors at 4 h postinjection (p.i.) of 1 nmol of Cy5.5-MT1-AF7p. **, p < 0.01. (C) Tumor/muscle ratios at 4 h p.i. **, p < 0.01.
Fig. 5.
(A–B) Representative images of dissected organs and tissues of tumor–bearing nude mice sacrificed at 4 hours after intravenous injection of Cy5.5-AF7p into MDA-MB-435 (A) and A549 (B) tumor-bearing mice at a dose of 1 nmol per mouse. 1, liver; 2, lung; 3, pancreas; 4, kidneys; 5, spleen; 6, small intestine; 7, muscle; 8, heart; 9, bone; 10, tumor; 11, blood. (C) Biodistribution of MT1-AF7p in MDA-MB-435 and A549 tumor-bearing nude mice at 4 h p.i. (mean ± SD, n = 3/group).
4. Discussion
Phage display has been used to identify optimal substrates for MT1-MMP and other MMPs including MMP-2, MMP-9, MMP-11, and MMP-26 [30–32]. The substrates and peptide inhibitors are being used to develop molecular probes to image and inhibit tissue remodeling associated with tumor cell invasion [33]. However, limited research has been done to find a non-substrate peptide that specifically binds to individual MMPs, other than to MMP-9 [34]. Our findings here add a non-substrate affinity peptide to MT1-MMP, and further study of whether MT1-AF7p can modulate the cellular functions of it would be of great interest.
In recent years, it has been demonstrated that MT1-MMP activity is regulated by endocytosis [35, 36]. Mechanism studies have provided extensive evidence for endocytosis of MT1-MMP through both clathrin-dependent and caveolae-mediated pathways [7, 9, 37], suggesting that MT1-MMP is an ideal target for drug targeting. The drug can be delivered to the cell surface that overexpress MT1-MMP, then internalized through this molecule and function within the cell., This strategy can be used to selectively target and kill tumor cells that overexpress MT1-MMP or newly formed neovasculature. Besides using MT1-AF7p to mediate imaging of MT1-MMP, our next goal is to study this peptide for its effect in MT1-MMP targeted drug delivery.
One common structural feature within the group of type I transmembrane MT-MMPs (MT1, -2, -3, and -5-MMP), which is lacking in the other MMP members, is an insertion of eight amino acids in the catalytic domain termed an ‘MT-loop’ [38]. The MT-loop connects strands βII and βIII in the catalytic domain. Crystal structure of the complex formed by the MT1-MMP with the tissue inhibitor of MMP-2 reveals that this ‘MT-loop’ chain forms a bulge on the surface of the molecule that is quite flexible due to lack of the direct support by the molecular surface [38]. Murphy and co-workers have extensively characterized the ‘MT-loop’ for its role in proMMP-2 activations by MT1-MMP [28]. According to mutation/deletion/kinetic experiments in a TIMP2-independent cell-free system and in a TIMP2-dependent cellular activation mode of proMMP-2, MT-loop mutants were kinetically impaired in their ability to activate proMMP-2 when compared with wild-type MT1-MMP, indicating that the MT-loop is required for efficient MMP-2 activation [28]. Furthermore, it is reported that MT1-MMP is involved in energy metabolism and pericellular proteolysis in migrating cancer cells by interacting with adenine nucleotide translocator (ANT) via a non-proteolytic process [39]. Whether MT1-AF7p will interfere with these processes remain needs to be tested.
New peptide-based molecular probes to facilitate cancer detection are rapidly evolving [40–42], examples include RGD analogs [43] and MMP substrate based activatable probes [44]. In this paper we report for the first time a non-substrate affinity peptide ligand for MT1-MMP. The identified MT1-AF7p peptide possesses high affinity and specificity for MT1-MMP. Using NIRF imaging, we have shown specific targeting of MT1-AF7p to MT1-MMP-expressing tumor xenografts in nude mice in vivo, which is consistent with ex vivo analyses of their tumors and organs. Our approach is a practical means to search for affinity peptides to other type I transmembrane MT-MMPs that can be used for labeling and targeting these molecules. Our findings have laid the foundation for further use of this MT1-MMP affinity peptide in peptide mediated drug delivery that targets MT1-MMP.
Supplementary Material
Acknowledgement
We thank Dr. Henry S. Eden for proof-reading the manuscript. This work was supported in part by the Intramural Research Program (IRP), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) and the National Science Foundation of China (Grant No. 31070669 and 81028009) and National High-Tech R&D Program (863 Program, No. 2009AA03Z309).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 2.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9(8):893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 4.Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem. 1995;270(10):5331–5338. doi: 10.1074/jbc.270.10.5331. [DOI] [PubMed] [Google Scholar]
- 5.Hernandez-Barrantes S, Shimura Y, Soloway PD, Sang QA, Fridman R. Differential roles of TIMP-4 and TIMP-2 in pro-MMP-2 activation by MT1-MMP. Biochem Biophys Res Commun. 2001;281(1):126–130. doi: 10.1006/bbrc.2001.4323. [DOI] [PubMed] [Google Scholar]
- 6.Murphy G, Knauper V, Cowell S, Hembry R, Stanton H, Butler G, Freije J, Pendas AM, Lopez-Otin C. Evaluation of some newer matrix metalloproteinases. Ann N Y Acad Sci. 1999;878:25–39. doi: 10.1111/j.1749-6632.1999.tb07672.x. [DOI] [PubMed] [Google Scholar]
- 7.Munshi HG, Stack MS. Reciprocal interactions between adhesion receptor signaling and MMP regulation. Cancer Metastasis Rev. 2006;25(1):45–56. doi: 10.1007/s10555-006-7888-7. [DOI] [PubMed] [Google Scholar]
- 8.Seiki M. Membrane-type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett. 2003;194(1):1–11. doi: 10.1016/s0304-3835(02)00699-7. [DOI] [PubMed] [Google Scholar]
- 9.Itoh Y, Seiki M. MT1-MMP: a potent modifier of pericellular microenvironment. J Cell Physiol. 2006;206(1):1–8. doi: 10.1002/jcp.20431. [DOI] [PubMed] [Google Scholar]
- 10.Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, Ward JM, Birkedal-Hansen H. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99(1):81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 11.Barbolina MV, Stack MS. Membrane type 1-matrix metalloproteinase: substrate diversity in pericellular proteolysis. Semin Cell Dev Biol. 2008;19(1):24–33. doi: 10.1016/j.semcdb.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi M, Tsunoda T, Seiki M, Nakamura Y, Furukawa Y. Identification of membrane-type matrix metalloproteinase-1 as a target of the beta-catenin/Tcf4 complex in human colorectal cancers. Oncogene. 2002;21(38):5861–5867. doi: 10.1038/sj.onc.1205755. [DOI] [PubMed] [Google Scholar]
- 13.Ueda J, Kajita M, Suenaga N, Fujii K, Seiki M. Sequence-specific silencing of MT1-MMP expression suppresses tumor cell migration and invasion: importance of MT1-MMP as a therapeutic target for invasive tumors. Oncogene. 2003;22(54):8716–8722. doi: 10.1038/sj.onc.1206962. [DOI] [PubMed] [Google Scholar]
- 14.Szabova L, Chrysovergis K, Yamada SS, Holmbeck K. MT1-MMP is required for efficient tumor dissemination in experimental metastatic disease. Oncogene. 2008;27(23):3274–3281. doi: 10.1038/sj.onc.1210982. [DOI] [PubMed] [Google Scholar]
- 15.Sounni NE, Devy L, Hajitou A, Frankenne F, Munaut C, Gilles C, Deroanne C, Thompson EW, Foidart JM, Noel A. MT1-MMP expression promotes tumor growth and angiogenesis through an up-regulation of vascular endothelial growth factor expression. FASEB J. 2002;16(6):555–564. doi: 10.1096/fj.01-0790com. [DOI] [PubMed] [Google Scholar]
- 16.Hiraoka N, Allen E, Apel IJ, Gyetko MR, Weiss SJ. Matrix metalloproteinases regulate neovascularization by acting as pericellular fibrinolysins. Cell. 1998;95(3):365–377. doi: 10.1016/s0092-8674(00)81768-7. [DOI] [PubMed] [Google Scholar]
- 17.Hotary KB, Allen ED, Brooks PC, Datta NS, Long MW, Weiss SJ. Membrane type I matrix metalloproteinase usurps tumor growth control imposed by the three-dimensional extracellular matrix. Cell. 2003;114(1):33–45. doi: 10.1016/s0092-8674(03)00513-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhao T, Harada H, Teramura Y, Tanaka S, Itasaka S, Morinibu A, Shinomiya K, Zhu Y, Hanaoka H, Iwata H, Saji H, Hiraoka M. A novel strategy to tag matrix metalloproteinases-positive cells for in vivo imaging of invasive and metastatic activity of tumor cells. J Control Release. 2010;144(1):109–114. doi: 10.1016/j.jconrel.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Manjappa AS, Chaudhari KR, Venkataraju MP, Dantuluri P, Nanda B, Sidda C, Sawant KK, Ramachandra Murthy RS. Antibody Derivatization and Conjugation Strategies: Application in Preparation of Stealth Immunoliposome to Target Chemotherapeutics to Tumor. J Control Release. 2010 Nov 20; doi: 10.1016/j.jconrel.2010.11.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Lowery A, Onishko H, Hallahan DE, Han Z. Tumor-targeted delivery of liposome-encapsulated doxorubicin by use of a peptide that selectively binds to irradiated tumors. J Control Release. 2010 Nov 12; doi: 10.1016/j.jconrel.2010.11.006. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. 2010;110(5):3196–3211. doi: 10.1021/cr900317f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X, Niu G, Yan Y, Yang M, Chen K, Ma Y, Chan N, Shen B, Chen X. Phage display-derived peptides for osteosarcoma imaging. Clin Cancer Res. 2010;16(16):4268–4277. doi: 10.1158/1078-0432.CCR-10-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Q, Liu S, Niu G, Chen K, Yan Y, Liu Z, Chen X. Phage display peptide probes for imaging early response to bevacizumab treatment. Amino Acids. 2010 Mar 16; doi: 10.1007/s00726-010-0548-9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, Xie J, Chen X. Peptide-based probes for targeted molecular imaging. Biochemistry. 2010;49(7):1364–1376. doi: 10.1021/bi901135x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Purushotham S, Lee JY, Na MH, Park H, Oh SJ, Park RW, Park JY, Lee E, Cho BC, Song MN, Baek MC, Kwak W, Yoo J, Hoffman AS, Oh YK, Kim IS, Lee BH. In vivo imaging of tumor apoptosis using histone H1-targeting peptide. J Control Release. 2010;148(3):283–291. doi: 10.1016/j.jconrel.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Kridel SJ, Sawai H, Ratnikov BI, Chen EI, Li W, Godzik A, Strongin AY, Smith JW. A unique substrate binding mode discriminates membrane type-1 matrix metalloproteinase from other matrix metalloproteinases. J Biol Chem. 2002;277(26):23788–23793. doi: 10.1074/jbc.M111574200. [DOI] [PubMed] [Google Scholar]
- 27.Ohkubo S, Miyadera K, Sugimoto Y, Matsuo K, Wierzba K, Yamada Y. Identification of substrate sequences for membrane type-1 matrix metalloproteinase using bacteriophage peptide display library. Biochem Biophys Res Commun. 1999;266(2):308–313. doi: 10.1006/bbrc.1999.1816. [DOI] [PubMed] [Google Scholar]
- 28.English WR, Holtz B, Vogt G, Knauper V, Murphy G. Characterization of the role of the "MT-loop": an eight-amino acid insertion specific to progelatinase A (MMP2) activating membrane-type matrix metalloproteinases. J Biol Chem. 2001;276(45):42018–42026. doi: 10.1074/jbc.M107783200. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Liu S, Niu G, Wang F, Chen X. Optical imaging of integrin alphavbeta3 expression with near-infrared fluorescent RGD dimer with tetra(ethylene glycol) linkers. Mol Imaging. 2010;9(1):21–29. [PMC free article] [PubMed] [Google Scholar]
- 30.Kridel SJ, Chen E, Kotra LP, Howard EW, Mobashery S, Smith JW. Substrate hydrolysis by matrix metalloproteinase-9. J Biol Chem. 2001;276(23):20572–20578. doi: 10.1074/jbc.M100900200. [DOI] [PubMed] [Google Scholar]
- 31.Pan W, Arnone M, Kendall M, Grafstrom RH, Seitz SP, Wasserman ZR, Albright CF. Identification of peptide substrates for human MMP-11 (stromelysin-3) using phage display. J Biol Chem. 2003;278(30):27820–27827. doi: 10.1074/jbc.M304436200. [DOI] [PubMed] [Google Scholar]
- 32.Park HI, Turk BE, Gerkema FE, Cantley LC, Sang QX. Peptide substrate specificities and protein cleavage sites of human endometase/matrilysin-2/matrix metalloproteinase-26. J Biol Chem. 2002;277(38):35168–35175. doi: 10.1074/jbc.M205071200. [DOI] [PubMed] [Google Scholar]
- 33.Kuhnast B, Bodenstein C, Haubner R, Wester HJ, Senekowitsch-Schmidtke R, Schwaiger M, Weber WA. Targeting of gelatinase activity with a radiolabeled cyclic HWGF peptide. Nucl Med Biol. 2004;31(3):337–344. doi: 10.1016/j.nucmedbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 34.Bjorklund M, Heikkila P, Koivunen E. Peptide inhibition of catalytic and noncatalytic activities of matrix metalloproteinase-9 blocks tumor cell migration and invasion. J Biol Chem. 2004;279(28):29589–29597. doi: 10.1074/jbc.M401601200. [DOI] [PubMed] [Google Scholar]
- 35.Jiang A, Lehti K, Wang X, Weiss SJ, Keski-Oja J, Pei D. Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc Natl Acad Sci U S A. 2001;98(24):13693–13698. doi: 10.1073/pnas.241293698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Annabi B, Lachambre M, Bousquet-Gagnon N, Page M, Gingras D, Beliveau R. Localization of membrane-type 1 matrix metalloproteinase in caveolae membrane domains. Biochem J. 2001;353(Pt 3):547–553. doi: 10.1042/0264-6021:3530547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J Cell Physiol. 2004;200(1):2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez-Catalan C, Bode W, Huber R, Turk D, Calvete JJ, Lichte A, Tschesche H, Maskos K. Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 1998;17(17):5238–5248. doi: 10.1093/emboj/17.17.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radichev IA, Remacle AG, Sounni NE, Shiryaev SA, Rozanov DV, Zhu W, Golubkova NV, Postnova TI, Golubkov VS, Strongin AY. Biochemical evidence of the interactions of membrane type-1 matrix metalloproteinase (MT1-MMP) with adenine nucleotide translocator (ANT): potential implications linking proteolysis with energy metabolism in cancer cells. Biochem J. 2009;420(1):37–47. doi: 10.1042/BJ20090082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu L, Niu G, Fang X, Chen X. Preclinical molecular imaging of tumor angiogenesis. Q J Nucl Med Mol Imaging. 2010;54(3):291–308. [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S, Xie J, Chen X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem Rev. 2010;110(5):3087–3111. doi: 10.1021/cr900361p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li ZJ, Wu WK, Ng SS, Yu L, Li HT, Wong CC, Wu YC, Zhang L, Ren SX, Sun XG, Chan KM, Cho CH. A novel peptide specifically targeting the vasculature of orthotopic colorectal cancer for imaging detection and drug delivery. J Control Release. 2010;148(3):292–302. doi: 10.1016/j.jconrel.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 43.Zhang X, Xiong Z, Wu Y, Cai W, Tseng JR, Gambhir SS, Chen X. Quantitative PET imaging of tumor integrin alphavbeta3 expression with 18F-FRGD2. J Nucl Med. 2006;47(1):113–121. [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Ryu JH, Park K, Lee A, Lee SY, Youn IC, Ahn CH, Yoon SM, Myung SJ, Moon DH, Chen X, Choi K, Kwon IC, Kim K. Polymeric nanoparticle-based activatable near-infrared nanosensor for protease determination in vivo. Nano Lett. 2009;9(12):4412–4416. doi: 10.1021/nl902709m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.