Abstract
Secreted frizzled-related protein (sFRP)-1 is a Wnt antagonist that inhibits breast carcinoma cell motility, whereas the secreted glycoprotein thrombospondin-1 stimulates adhesion and motility of the same cells. We examined whether thrombospondin-1 and sFRP-1 interact directly or indirectly to modulate cell behavior. Thrombospondin-1 bound sFRP-1 with an apparent Kd = 48 nM and the related sFRP-2 with a Kd = 95 nM. Thrombospondin-1 did not bind to the more distantly related sFRP-3. The association of thrombospondin-1 and sFRP-1 is primarily mediated by the amino-terminal N-module of thrombospondin-1 and the netrin domain of sFRP-1. sFRP-1 inhibited α3β1 integrin-mediated adhesion of MDA-MB-231 breast carcinoma cells to a surface coated with thrombospondin-1 or recombinant N-module, but not adhesion of the cells on immobilized fibronectin or type I collagen. sFRP-1 also inhibited thrombospondin-1-mediated migration of MDA-MB-231 and MDA-MB-468 breast carcinoma cells. Although sFRP-2 binds similarly to thrombospondin-1, it did not inhibit thrombospondin-1-stimulated adhesion. Thus, sFRP-1 binds to thrombospondin-1 and antagonizes stimulatory effects of thrombospondin-1 on breast carcinoma cell adhesion and motility. These results demonstrate that sFRP-1 can modulate breast cancer cell responses by interacting with thrombospondin-1 in addition to its known effects on Wnt signaling.
Keywords: secreted frizzled-related proteins, netrin-binding proteins, thrombospondins, breast carcinoma
1. Introduction
Thrombospondin-1 (TSP1)* was initially described as a 450 kDa trimeric glycoprotein released from the α-granules of platelets [1]. In addition to modulating platelet activation [2, 3], TSP1 was the first identified endogenous angiogenesis inhibitor [4, 5]. Consistent with this activity, TSP1 expression was shown to inhibit tumor growth and metastasis [6]. Moreover, TSP1 expression is frequently lost during malignant transformation due to regulation of its expression by oncogenes, tumor suppressor genes, and hypermethylation [7–9]. However, TSP1 can become highly expressed by stromal fibroblasts and endothelial cells within tumors during tumor progression [10], which results in elevated levels of circulating TSP1 in some cancers [11] that may also inhibit tumor growth [12, 13]. In addition to modulating angiogenesis, tumor-associated and circulating TSP1 can also modulate tumor blood flow [14], anti-tumor innate immunity [15], and radiotherapy responses [16, 17].
The diverse and sometimes conflicting activities of TSP1 in the context of tumor progression may be rationalized in part by the complexity of TSP1 interactions with other proteins. Each subunit of TSP1 is composed of several domains with multiple ligand binding specificities [18]. TSP1 interacts with cell surface integrin and non-integrin receptors [19–22], heparan sulfate proteoglycans (HSPG)* [23], growth factors [24], and other bioactive molecules [25–27].
In the context of breast cancer, TSP1 expression appears to be a marker of aggressiveness and correlates with the microvessel density [28, 29] but is not related to p53 status or VEGF expression [30]. Endogenous TSP1 inhibits primary tumor growth and angiogenesis but promotes metastasis to the lung in the Pyt transgenic mouse breast cancer model [31]. TSP1 stimulates breast carcinoma cell adhesion and chemotaxis by engaging α3β1 integrin [32, 33]. TSP1 can also promote human breast cancer cell proliferation in vitro, but the mechanism remains to be elucidated [34, 35].
The secreted frizzled-related proteins (sFRPs)* comprise a family of 5 proteins that bear homology to frizzleds, the seven-pass transmembrane cell surface receptors for Wnts [36, 37]. sFRP-1, 2 and 5 are closely related and form one subgroup, and sFRP-3, and 4 form a second subgroup [37, 38]. Constitutive activation of Wnt signaling is common in neoplasia [39]; in particular, autocrine Wnt pathways contribute to the proliferative and metastatic properties of breast cancer cells [40, 41]. The sFRPs were first identified as Wnt antagonists, and accordingly they have been viewed as tumor suppressors [42–45]. Consistent with this assessment, four of the five human SFRP genes contain dense CpG islands that often are hypermethylated in many cancers, resulting in silencing of their expression [45]. Loss of sFRP-1 expression in breast cancer has been associated with decreased survival [46], and restoration of expression in colorectal and renal cell carcinoma lines attenuated the malignant phenotype [47, 48]. Similarly, ectopic sFRP-1 expression in MDA-MB-231 breast cancer cells suppressed tumor growth and metastasis [49]. In contrast to sFRP-1, sFRP-2 has positive effects on breast carcinoma cells and endothelial cells that imply a stimulation of tumor growth [37, 50– 52]
The sFRPs contain two distinct structural domains: a frizzled-related cysteine-rich domain (CRD)* and a netrin (NTR)* module [36, 37]. The CRD consists of 110–120 amino acid residues including ten invariant cysteines that form a conserved set of five disulfide bonds. The CRDs of frizzleds bind Wnt ligands [53], and initial studies suggested that sFRPs inhibit signaling by binding to Wnts via their CRDs [54, 55]. However, additional experiments demonstrated that sFRPs and frizzleds could associate with each other through CRD-CRD interactions, implying there are additional mechanisms of Wnt inhibition [55]. The NTR module is defined by a set of six characteristically spaced cysteines, stretches of hydrophobic and positively charged amino acids and, where three-dimensional structural data are available, two α-helices packed against a five-stranded β-barrel [56, 57]. NTR domains are found in the carboxyl (C) -terminus of netrins, laminin-related proteins in the extracellular matrix that control axon guidance. While the function of the NTR module in netrins is unknown, in the tissue inhibitors of metalloproteinases (TIMPs)* it mediates binding to their protease targets [56]. The NTR module also is present in type I procollagen C-proteinase enhancer proteins (pCOLCEs)*, complement proteins C3, C4 and C5 as well as other molecules [56].}. The NTR domain of sFRP-1 associates with Wnt proteins and modulates their activities [58, 59]. Besides its interaction with Wnts, the NTR domain is responsible for sFRP association with HSPG [58].
Screening of a peptide phage display library resulted in the identification of a peptide binding motif for sFRP-1 with micromolar affinity [60]. The core of this motif, DGR, is present in the type 3 calcium-binding repeats of TSP1 [18], suggesting that these proteins might interact. Here we report that sFRP-1 binds to TSP1 with high affinity, although not via the DGR motif. This binding is shared by sFRP-2 but not sFRP-3. The interaction primarily involves the NTR module of sFRP-1 interacting with the N-module of TSP1. sFRP-1 specifically disrupts integrin-mediated cell adhesion of MDA-MB-231 breast carcinoma cells to surfaces coated with TSP1 or its N-module, and blocks TSP1-mediated migration of breast carcinoma cells. These activities suggest that physical and functional interactions of TSP1 with sFRP-1 have pathophysiological relevance for breast cancer progression.
2. Materials and methods
2.1. Reagents
Human TSP1 and fibronectin were purified from platelets and plasma, respectively, obtained from the National Institutes of Health Department of Transfusion Medicine [61, 62]. Monomeric and trimeric recombinant regions of TSP1 (Fig. 3) expressed in insect cells and prepared as described [63, 64] were provided by Dr. Deane Mosher, University of Wisconsin. Monomeric recombinant N-module containing residues 1 to 250 of mature TSP1 was prepared as previously described [65]. A glutathione-S-transferase (GST)* fusion protein expressing the von Willebrand factor type C (vWC)* domain of TSP1 (provided by Dr. Jack Lawler, Harvard University, Boston, MA) was prepared as described [66]. Recombinant human sFRP-1 and sFRP-2 were prepared as described [58, 67]. Recombinant human sFRP-3 was purchased from R&D Systems (Minneapolis, MN). Type I collagen was purchased from Inamed (Fremont, CA).
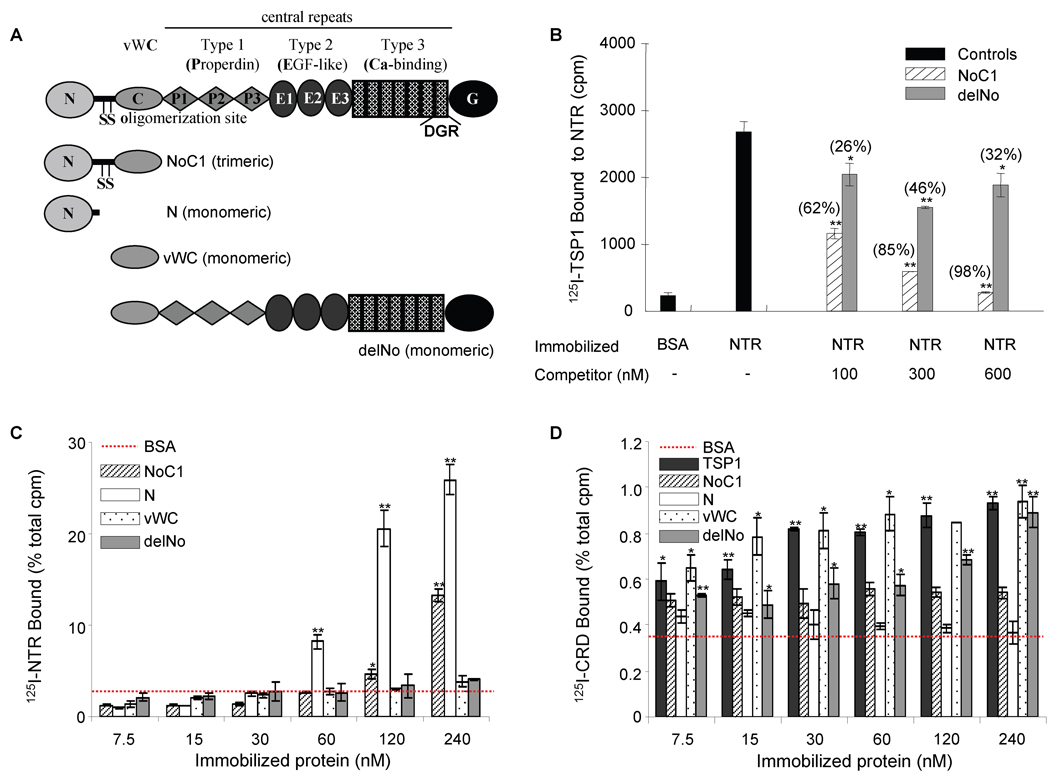
Figure 3. Localization of the sFRP-1 binding site in TSP1.
20 µg/ml of recombinant NTR domain were absorbed onto microtiter plate wells (B), and 125I-TSP1 binding was measured at 37°C in the presence of competing concentrations of the indicated unlabelled recombinant regions of TSP1 (as shown in A). Nonspecific sites were blocked with 3% (w/v) BSA in DPBS containing Ca2+ and Mg2+. Data were normalized and presented in brackets as percent of inhibition. The results are representative of three independent experiments, each performed in triplicate. 125I-NTR domain (C) and 125I-CRD (D) binding to the indicated concentrations of TSP1 or TSP1 recombinant regions coated onto microtiter wells was determined as above. Data were normalized and are presented as percent of total cpm. The results are representative of four (125I-NTR) and two (125I-CRD) independent experiments, each performed in triplicate.
2.2. Recombinant expression and purification of CRD and NTR domain
cDNAs encoding the CRD and NTR domain along with a small amount of flanking sequence were generated by PCR using full-length human sFRP-1 cDNA as template and the indicated primers for the CRD (5’-CCGCTCGAGAAAAGACGCTTCTACACCAAGCCACCT -3’, 5’-GCTCTAGATCATCACGTCATGGCGATGCAGACGTCCCCCT -3’) and NTR domain (5’-CGGAATTCGTGTGTCCTCCCTGTGACAACGAG-3’, 5’-GCTCTAGATCATCACTTAAACACGGACTGAAAGGTGGGGC -3’). After the fidelity of PCR products was verified by sequence analysis, the cDNAs were digested with XhoI/XbaI (CRD) or EcoRI/XbaI (NTR) and subcloned into the pICZaA expression vector (Invitrogen). Competent Pichia pastoris cells were transformed with these constructs using Pichia EasyComp Kit (Invitrogen). Transformed clones were grown in 5 ml BMGY medium overnight at 30°C, 275 rpm. Cultures were transferred into 500 ml BMGY medium in a 2 liter flask with deep baffles and incubated overnight at 30°C, 250 rpm. Cells were pelleted by centrifugation and resuspended at 80 g/liter in BMMY medium supplemented with 1% methanol/day for 3–5 days to induce expression of the recombinant protein (cultures maintained at 30°C, 250 rpm).
All purification steps were performed at 4°C. CRD culture medium was concentrated 10-fold by ultrafiltration using a YM3 membrane (Millipore), dialyzed against solution A (20mM Tris-HCl, pH 7.4), filtered through a 0.44 µm membrane and applied to a 5 ml HiTrap Q FF column (GE Healthcare) equilibrated with solution A. After washing the column with 50–100 ml of solution A, protein was eluted with a linear gradient of increasing NaCl (solution B: 20mM Tris-HCl, pH 7.4, 1M NaCl). The CRD either bound weakly (eluting with 3–10% solution B) or remained in the flow through. The early eluting fractions containing the CRD and flow through were combined, dialyzed in solution A and loaded on a freshly equilibrated HiTrap Q FF column. The CRD was retained on the resin and eluted with 3–10% solution B. NTR culture medium was filtered through a 0.44 µm membrane and loaded directly on a HiTrap Heparin HP column (GE Healthcare) equilibrated with solution A' (25mM phosphate buffer, pH7.4). After washing the resin with 10–20 column volumes of solution A', NTR protein was eluted with a NaCl step gradient in fractions containing ~ 1 M NaCl (solution B': 25mM phosphate buffer, pH7.4, 2M NaCl). Purified protein was visualized by SDS-PAGE, followed by staining with Coomassie blue. The mass and amino-terminal sequence of the CRD and NTR domain were confirmed by MALDI-TOF and Edman degradation, respectively.
2.3. Solid Phase Binding Assays
50 µl of the indicated concentrations of full-length recombinant sFRP-1 or sFRP-1 fragments, CRD and NTR domain, sFRP-2, and sFRP-3 were adsorbed onto Immulon® HB (ThermoLabsystems, Franklin, MA) microtiter strips by incubation in Dulbecco’s PBS (DPBS) without Ca2+ and Mg2+, for 16 h at 4°C. Nonspecific sites were blocked with 3% (w/v) BSA (Sigma-Aldrich) in DPBS with 0.9 mM CaCl2 and 0.5 mM MgCl2, at room temperature for 1 h. The wells were emptied and 50 µl/well of 125I-TSP1 (0.5 µg/ml, 2.5–8 µCi/µg) were added alone or in the presence of the indicated unlabeled ligands as competitor in DPBS with Ca2+ and Mg2+, containing 0.5% (w/v) BSA, and 0.1 mM phenylmethylsulfonyl fluoride, and incubated at room temperature, or 37°C for 3 h. The wells were washed with the same cold buffer, and the bound radioactivity was quantified using a gamma counter (PerkinElmer Life Sciences). For some experiments recombinant TSP1 fragments were immobilized as above, and binding of 125I-NTR domain (293 ng/ml, 3.4–4.4 µCi/µg)and 125I-CRD (250 ng/ml, 5.3 µCi/µg) was determined. For calculation of equilibrium binding constants, binding was determined in triplicate at each concentration to wells coated with the indicated ligands and corrected for nonspecific binding determined at each ligand concentration using wells only coated with BSA.
2.4. Cell Culture
Cell cultures were maintained at 37°C, 5% CO2, in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) (Biosource), 2 mM L-glutamine, 100 units/ml penicillin, and 100 µg/ml streptomycin (Gibco). Three human tumor cell cultures were used. MDA-MB-231 human breast carcinoma cells (American Type Culture Collection), MDA-MB-468 human breast carcinoma cells (kindly provided by Dr. Lisa A. Ridnour, National Cancer Institute, NIH, Bethesda, MD), and A2058 human melanoma cells [68].
2.5. Cell Adhesion Assays
TSP1, the trimeric recombinant region of TSP1 (NoC1), type I collagen, and fibronectin, diluted in DBPS with Ca2+ and Mg2+, were coated onto 96-well flat-bottom plates (NUNC MaxiSorp) overnight at 4°C. The wells were emptied, filled with DPBS containing 1% BSA (w/v), and incubated at room temperature for 30 min to block nonspecific adherence to plastic. The indicated concentrations of full-length recombinant human sFRP-1 or sFRP-1 fragments, CRD and NTR domain, and sFRP-2 diluted in RPMI medium containing 0.1%BSA, and 0.1 mM MnCl2 when TSP1 is used, were added in 50 µl, and the plate was incubated at 37°C for 1 h. MDA-MB-231 cells were washed in serum-free medium and resuspended at 1×106 cells/ml in RPMI/0.1% BSA. 50 µl of cell suspension were added to each well containing 50 µl of full-length recombinant human sFRP-1, sFRP-1 fragments or sFRP-2, and the plate was incubated at 37°C for 60 min. Non-adherent cells were removed by washing. The number of adherent cells was quantified using the previously described colorimetric hexosaminidase assay [69]. All samples were run in triplicate. Same experimental conditions were used to dynamically record cell attachment and spreading on TSP1 using RT-CES system (ACEA Biosciences). Measurements were automatically collected by the analyzer every 3 min for up to 2 h. Alternatively, TSP1, NoC1, type I collagen, and fibronectin were adsorbed (triplicates of 8-µl drops) onto polystyrene dishes (Falcon 1008) by incubating overnight at 4 °C. The drops were removed, and the dishes were blocked with 1% BSA/DPBS for 30 min. MDA-MB-231 cells were washed in serum-free medium and resuspended at 2×106 cells/ml in RPMI/0.1% BSA. 100 µl of cell suspension were added to each dish containing 10 µg/ml of sFRP-1 or sFRP-2 in RPMI/0.1% BSA. For activation cells were treated with 0.1 mM MnCl2. After incubation for 1 h at 37 °C in 5% CO2, the dishes were washed 2 times with DPBS and fixed for 30 min with 1% glutaraldehyde/DPBS. After staining with Diff-Quik, cells were imaged using an Olympus IX70 microscope, and pictures were taken using a Spot Insight cooled digital camera (Diagnostic Instruments, Sterling Heights, MI). Filopodial length was quantified using Image J software (200 µm ≈ 1,600 pixels). n=9 cells/group from three 600x fields (3 to 7 filopodia per cell) were averaged.
2.6. Wound Repair Cell Motility Assay
A standard scratch wound repair assay was used to assess the effects of sFRP-1 and TSP1 on migration of a human breast carcinoma cell line [70]. Briefly, MDA-MB-231 cells were seeded on 24-well plates and grown to confluence. The monolayers were pre-incubated with full-length recombinant human sFRP-1 and TSP1 in complete RPMI medium for 45 min. Monolayers were scratched in a line using a standard 100 µl pipette tip. Marked sections of the wounds were imaged over time using an Olympus IX70 microscope, and pictures of four wound edges per condition were taken at 1h using a Spot Insight cooled digital camera (Diagnostic Instruments, Sterling Heights, MI). Migration of cells into the wound was quantified as the percentage of recovered area using Image J software.
2.7. Cell Migration Assay
Cell migration of MDA-MB-231, MDA-MB-468 and A2058 cells was monitored in real time using an impedance-based system (xCELLigence, Roche Applied Science). A 165 µl volume of medium containing 10% FBS (chemoattractant) and 35 µl of medium containing 1% BSA were added to the lower and upper chamber of the CIM-Plate 16, respectively, and incubated at 37°C, 5% CO2 for 1h. A 100 µl cell suspension of 4×105 cells/ml was then added to the upper chamber, and measurements were automatically collected by the analyzer every 5 min for up to 4 h.
2.8. Data Analysis
Scafit version 3.10 of the Ligand program [71] was used for the quantitative analysis of competitive binding. All data are shown as mean ± SD except where indicated. Significance was determined with one-tailed distribution Student's t test analysis. The difference was considered significant (*) when P ≤ 0.05 and (**) when P ≤ 0.001.
3. Results
3.1. TSP1 binds to sFRP-1 and sFRP-2 but not sFRP-3
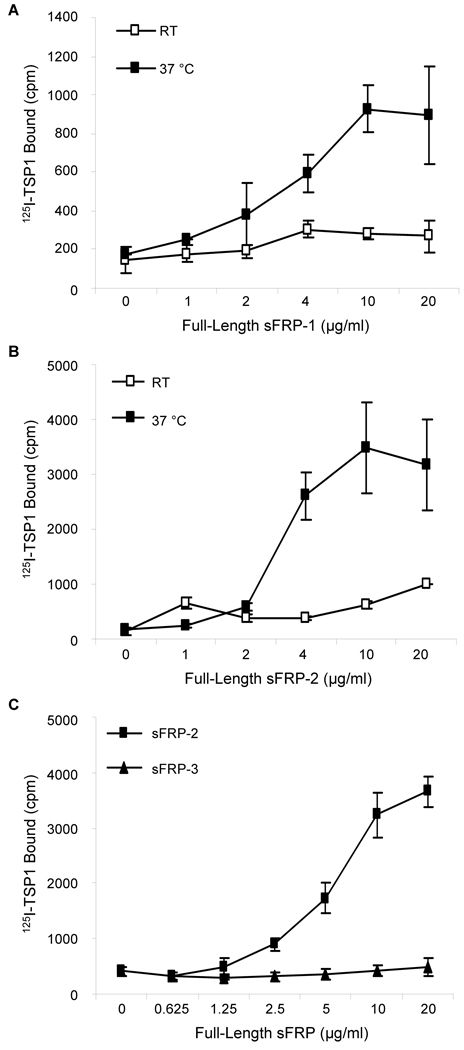
To investigate the interaction between TSP1 and sFRPs, we labeled human platelet TSP1 with 125I and studied its binding to recombinant human sFRPs immobilized on polystyrene. In the presence of divalent cations, TSP1 binding to immobilized full-length sFRP-1 was saturable and dose-dependent at 37°C, but much weaker at room temperature (Fig. 1A). 125I-TSP1, similarly bound in a dose- and temperature-dependent manner to the related family member human sFRP-2, with higher binding also obtained at 37°C (Fig. 1B). In contrast, recombinant human sFRP-3 showed no significant interaction with 125I-TSP1 at 37°C (Fig. 1C).
Figure 1. Temperature dependence of 125I-TSP1 binding to recombinant sFRP-1 and sFRP-2.
Solid phase binding assays were assessed to quantify 125I-TSP1 (0.5 µg/ml, 50 µl/well) binding to full-length recombinant human sFRP-1 (A), sFRP-2 (B) and sFRP-3 (C) coated onto microtiter plate wells. Nonspecific binding sites were blocked with 3% (w/v) BSA in DPBS containing Ca2+ and Mg2+, at room temperature for 1 h. Binding was performed at room temperature or 37°C in a humidified atmosphere at pH 7.4 for 3 h. The results are representative of two independent experiments performed in triplicate.
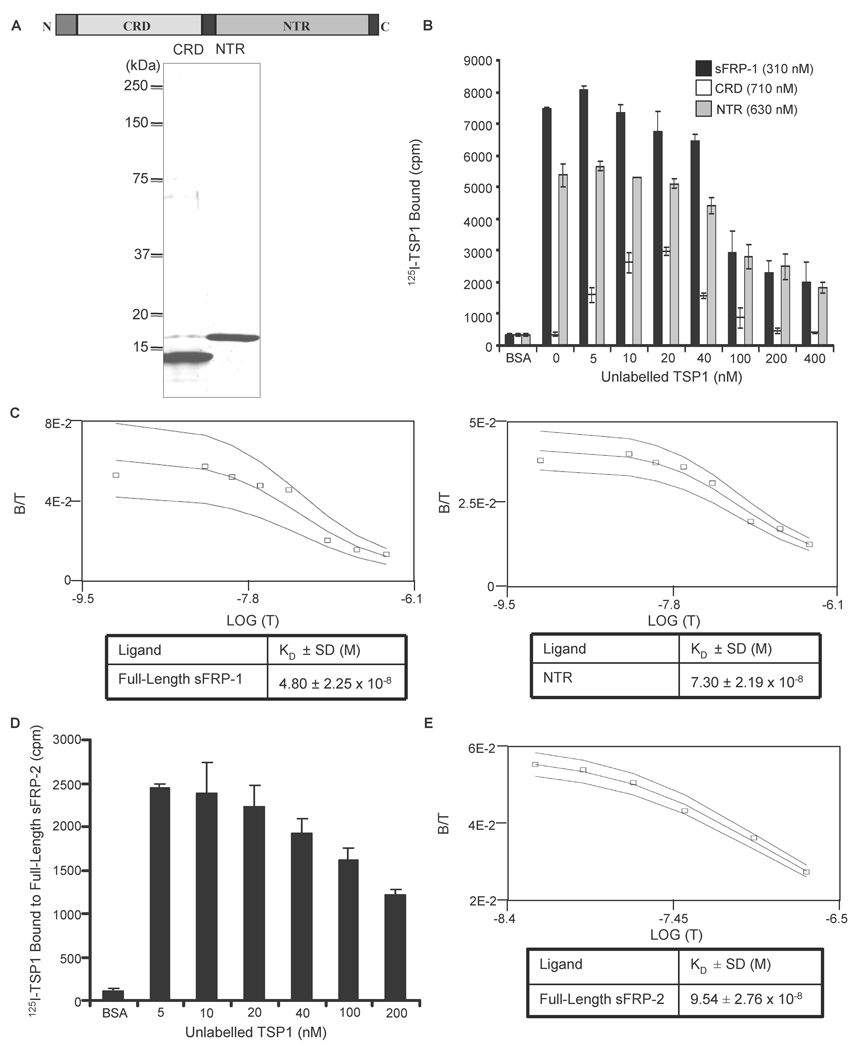
Binding constants for sFRP-1 and sFRP-2 were determined by homologous displacement experiments using unlabelled TSP1 (Fig. 2). Based on analysis of binding data in the presence of divalent cations at 37°C using the Ligand program [71], TSP1 bound with apparent Kd values of 48 nM and 95 nM to immobilized full-length sFRP-1 (Fig. 2B&C left) and sFRP-2 (Fig. 2D&E), respectively.
Figure 2. Equilibrium binding of 125I-TSP1 to sFRP-1 and sFRP-2.
Schematic diagram of sFRP-1 modular structure and Coomassie blue-stained SDS-polyacrylamide gel containing purified CRD and NTR domain of sFRP-1 (A). 125I-TSP1 binding to full-length sFRP-1 or the sFRP-1 domains (CRD and NTR domain) was performed in microtiter plate wells coated with 50 µl/well of 10 µg/ml of each protein. Nonspecific binding was blocked with 3% (w/v) BSA in DPBS containing Ca2+ and Mg2+, at room temperature for 1 h. Binding was measured at 37°C in the presence of competing concentrations of unlabelled TSP1 (B). The Ligand program was used for quantitative analysis of the binding data. Results for sFRP-1 and NTR domain (C left and right, respectively) are presented as displacement plots, and are representative of three (sFRP-1) and two (NTR domain) independent experiments performed in triplicate. Microtiter plate wells were coated using 50 µl/well of 5 µg/ml of full-length sFRP-2. Nonspecific binding was blocked with 3% (w/v) BSA in DPBS containing Ca2+ and Mg2+, at room temperature for 1 h. 125I-TSP1 binding was measured at 37°C in the presence of the indicated concentrations of unlabelled TSP1 (D). The Ligand program was used for quantitative analysis of competitive binding. Results are presented as displacement plots (E), and are representative of two independent experiments performed in triplicate.
3.2. TSP1 binds preferentially to the NTR module of sFRP-1
To determine the role of the individual sFRP-1 domains in binding to TSP1, we generated recombinant CRD and NTR module using a Pichia pastoris expression system. We chose this system because a prior study had demonstrated efficient production and proper folding of the Frizzled-type CRD from Ror1 [72]. The recombinant CRD product spanned the entire CRD motif and included a small amount of additional sFRP-1 sequence at either end (R49 to T169, numbering as reported in [44]). The recombinant NTR domain extended from V184 to the C-terminus of sFRP-1 (K313), and contained a residual segment of sequence (EAEAEF) from the yeast alpha mating factor at the N-terminus. Milligram quantities of purified proteins were isolated from a few hundred ml of culture fluid using ion exchange (CRD) or heparin affinity (NTR) chromatography, and protein purity was assessed by SDS-polyacrylamide gel electrophoresis (Fig. 2A). When immobilized on polystyrene, the NTR domain strongly bound to TSP1 with an apparent Kd of 73 nM (Fig. 2B&C right). 125I-TSP1 showed no significant binding to the immobilized CRD derivative (Fig. 2B). However, addition of unlabeled TSP1 surprisingly stimulated binding at low concentrations and then inhibited this binding at higher concentrations. The molecular basis for this unusual biphasic response is unknown, but such binding curves are characteristic of a self-associating ligand [73].
3.3. The N-module of TSP1 binds to the NTR module of sFRP-1
TSP1 is a disulfide-bonded trimer of subunits composed of an N-module followed by the oligomerization domain, a vWC domain, three thrombospondin type 1 repeats (properdin), and a signature domain comprising three epidermal growth factor (EGF)-like type 2 repeats, the type 3 calcium-binding repeats and a lectin-like C-terminal globe (Fig. 3A) [74]. Two approaches were used to define the sFRP-1 binding domain within TSP1. Binding of 125I-TSP1 to immobilized NTR domain was significantly (p<0.001) inhibited in a dose-dependent manner by the trimeric N-terminal construct NoC1 (Fig. 3B). Recombinant delNo, lacking only the N-module and oligomerization site, inhibited moderately but showed no dose-dependence.
The binding site in TSP1 was further mapped using a reverse assay where we quantified 125I-NTR domain binding to recombinant regions of TSP1 immobilized on polystyrene. Only the two constructs containing the N-module of TSP1 bound 125I-NTR domain in a dose-dependent manner (Fig. 3C). In contrast 125I-CRD bound minimally to immobilized TSP1, and no significant binding to the N-module was detected (Fig. 3D). The highest binding of 125I-CRD was to recombinant vWC and to the delNo, which also contains this domain. Note, however, that 125I-CRD binding was an order of magnitude less than that observed with 125I-NTR. Therefore, binding of the CRD domain to TSP1 is relatively weak and may be mediated by the vWC module, but the N-module of TSP1 mediates high affinity binding to the NTR domain of sFRP-1.
3.4. sFRP-1 inhibits TSP1-mediated breast cancer cell adhesion and migration
Based on the strong binding between TSP1 and sFRP-1 and -2, we examined the potential functional consequences of this interaction on known biological activities of the proteins using the malignant breast cancer model MDA-MB-231, which express very low TSP1 mRNA and protein [75, 76] and no sFRP-1 mRNA [77].
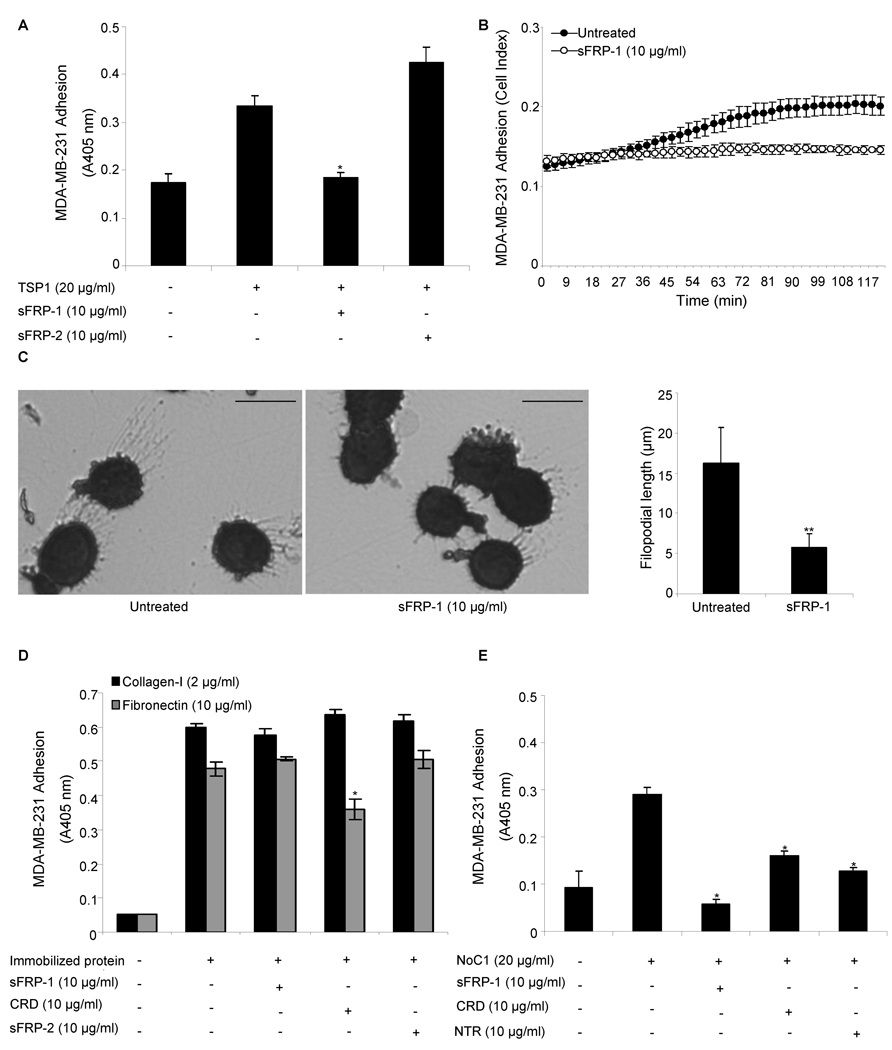
Adhesion of breast carcinoma cells to immobilized TSP1 or its N-terminal domain is mediated by α3β1 integrin [33, 78]. Therefore, we asked whether intact sFRP-1 and sFRP-2 were able to inhibit α3β1-mediated MDA-MB-231 cell adhesion on immobilized TSP1 or its N-terminal domain. First, we performed adhesion assays on substrates coated with intact TSP1 (20 µg/ml) in the presence of soluble sFRP-1 or sFRP-2 (10 µg/ml) and found that MDA-MB-231 cell adhesion on TSP1 was significantly inhibited (p<0.05) by intact sFRP-1 (93 % inhibition) (Fig. 4A–B) but not by sFRP-2 (Fig. 4A).
Figure 4. sFRP modulation of breast cancer cell adhesion on TSP1.
MDA-MB-231 breast cancer cell adhesion on immobilized TSP1 (20 µg/ml) (A–C), type-I collagen (2 µg/ml) or fibronectin (10 µg/ml) (D), NoC1 (20 µg/ml) (E) was assessed in the absence or presence of 10 µg/ml of full-length recombinant sFRP-1, CRD and NTR domain of sFRP-1, or sFRP-2. After 60 min incubation, cell adhesion was quantified by colorimetric detection of cell-associated hexosaminidase activity (A, D–E), using the RT-CES system (ACEA Biosciences) (B) or stained with Diff-Quik and imaged (C; bar = 15 µm). Representative images are shown, and filopodia length (mean ± SD) was quantified using Image J software (right panel) as described in Materials and Methods. The results are representative of three independent experiments, each performed in triplicate.
α3β1-mediated adhesion on TSP1 is associated with formation of filopodia [33, 78]. Consistent with this previous report, most breast carcinoma cells attaching on TSP1 showed prominent filopodia (Fig. 4C left panel). Addition of soluble sFRP-1, but not sFRP-2 (data not shown), significantly reduced (p<0.001) filopodia formation (Fig. 4C). sFRP-1 and sFRP-2 had no effect on the morphology of MDA-MB-231 cells attaching on type I collagen (data not shown). Furthermore, the inhibition of MDA-MB-231 cell adhesion mediated by immobilized TSP1 is specific because the same concentration of sFRP-1 did not alter MDA-MB-231 cell adhesion on immobilized type I collagen, mediated by α2β1 integrin, or fibronectin, mediated by α5β1 integrin (Fig. 4D), and did not alter MDA-MB-468 cell adhesion on immobilized TSP1 (data not shown).
Because TSP1 has α3β1 integrin binding sites in the N-module and its type 1 repeats [79], we next examined substrates coated with the trimeric N-terminal construct NoC1 (20 µg/ml) and found that MDA-MB-231 cell adhesion on NoC1 was significantly inhibited (p<0.05) to basal levels by intact sFRP-1 (10 µg/ml) (Fig. 4E). If this inhibition resulted exclusively from sFRP-1 binding to the N-module of TSP1 and blocking α3β1 integrin binding, the NTR domain but not the CRD should replicate this activity. However, at 10 µg/ml both NTR and CRD partially inhibited MDA-MB-231 cell adhesion on immobilized NoC1 (p<0.05; Fig. 4E). Notably, the same concentration of CRD also partially inhibited adhesion on immobilized fibronectin but not on type I collagen (Fig. 4D). Therefore, the CRD module of sFRP-1 may indirectly inhibit adhesion mediated by α3β1 and α5β1 through its binding to Wnt or frizzleds. At an equimolar concentration to that where sFRP-1 showed specific inhibition of adhesion on TSP1, CRD (5 µg/ml) did not significantly inhibit adhesion on NoC1 (data not shown). Therefore, inhibition by sFRP-1 of adhesion on NoC1 and TSP1 may be partially dependent on direct sFRP-1 binding to the N-module of TSP1.
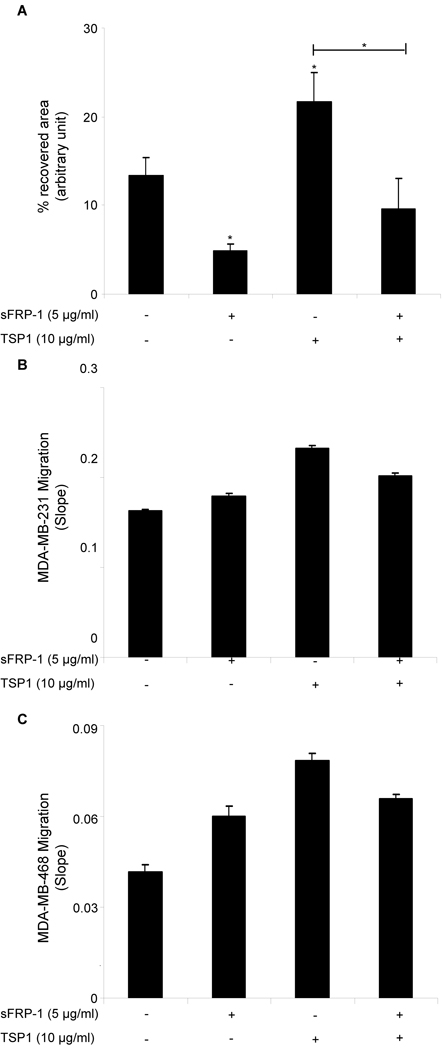
The Wnt signaling pathway mediates cell proliferation, migration, differentiation, adhesion, and survival, and its deregulation has been associated with breast and many other cancers [80]. Ectopic expression of sFRP-1 lowers the migratory potential of MDA-MB-231 breast cancer cells in a wound healing assay [49]. We confirmed, using a scratch wound assay, that full-length recombinant human sFRP-1 at 5 µg/ml significantly inhibits MDA-MB-231 cell motility (p<0.05; Fig. 5A). In response to 10 µg/ml of TSP1 cells migrated significantly more rapidly into the wound area compared with untreated breast cancer cells, but 45 min pre-incubation with 5 µg/ml of sFRP-1 was sufficient to block the stimulatory effect of TSP1 on MDA-MB-231 cell motility (p<0.05; Fig. 5A). Furthermore, we confirmed, using an impedance-based cell migration assay, that addition of 10 µg/ml of TSP1 in the upper chamber increased migration of MDA-MB-231 and MDA-MB-468 breast carcinoma cells towards 10% FBS (Fig. 5B–C). Addition of 5 µg/ml of full-length recombinant human sFRP-1 was sufficient to block the stimulatory effect of TSP1 on MDA-MB-231 and MDA-MB-468 cell migration (Fig. 5B–C). Note, however, that recombinant human sFRP-1 alone did not inhibit MDA-MB-231 and MDA-MB-468 cell migration (Fig. 5B–C). Similarly, addition of 5 µg/ml of full-length recombinant human sFRP-1 was sufficient to block the stimulatory effect of TSP1 on A2058 human melanoma cell migration (data not shown).
Figure 5. sFRP-1 inhibits TSP1-mediated breast cancer cell migration.
Confluent monolayers of MDA-MB-231 cells were treated with full-length recombinant sFRP-1 (5 µg/ml) and TSP1 (10 µg/ml). After 45 min, monolayers were scratched, and 1 h later the percent of recovered area was calculated on four wound edges per condition. The results are representative of three independent experiments (A). 100 µl cell suspension of 4×105 cells/ml MDA-MB-231 cells (B) or MDA-MB-468 cells (C) were added to the upper chamber of the CIM-Plate 16 in the presence or absence of full-length recombinant sFRP-1 (5 µg/ml) and TSP1 (10 µg/ml). After 30 min incubation, cell migration towards 10% FBS was monitored in real time using an impedance-based system and measurements were automatically collected by the analyzer every 5 min for up to 4 h. The results are presented as slope (changes in cell index/hour). The results are representative of two (B) to three (C) independent experiments performed in duplicate.
4. Discussion
The N-module of TSP1 interacts with multiple ligands including several cell surface β1-integrins [19–21, 78, 81], low-density lipoprotein (LDL)* receptor-related protein-1/calreticulin [82, 83], sulfated glycolipids and HSPG [23, 84], TSG-6 [26], and versican [27]. In this study, we have identified sFRP-1 and sFRP-2 but not sFRP-3 as additional ligands for the N-module of TSP1. sFRP-1 and sFRP-2 are closely related sFRPs (42% identical at the amino acid level), with conserved spacing of cysteine residues in their NTR domains, implying a similar pattern of protein folding, whereas the NTR domain of sFRP-3 displays a distinct pattern of disulfide bonds [38]. At pH 7.4 in the presence of physiological salt concentrations and divalent cations, the N-terminal region of TSP1 interacts with the NTR domain of sFRP-1. This suggests that sFRPs could bind to other proteins that contain a TSP N-module [84]. Previously, we noted that TSPEAR, which has a TSP N-module, also has the sFRP-1 binding motif identified in our peptide phage display analysis [60]. In contrast to TSP1, the peptide motif of TSPEAR is located in its TSP N-module, but the significance of this motif to predict binding remains unclear.
TSP1 plays an important role in hemostasis, wound repair, and inflammatory responses, in part, by its ability to modulate adhesion of many cell types [69, 85]. The N-module of TSP1 mediates adhesion and stimulates migration of breast cancer cells via α3β1 integrin [32, 33], and this activity is crucial for cancer progression and metastasis [8]. We demonstrate here that functional and physical interactions between TSP1 and sFRP-1 specifically disrupt MDA-MB-231 breast cancer cell adhesion and migration mediated by the N-module of TSP1 and this integrin. This may contribute to the reduced motility and metastatic potential of sFRP-1-expressing MDA-MB-231 cells [49].
In addition to binding TSP1, sFRP-2 was previously reported to co-immunoprecipitate with fibronectin and to enhance integrin-mediated adhesion of primary cells cultured from canine mammary gland tumors on a fibronectin substrate [50]. Similarly, we found that sFRP-2 increases α3β1 integrin-mediated adhesion of MDA-MB-231 human breast carcinoma cells on TSP1. Thus, we propose that the stimulatory effects of sFRP-2 on breast carcinoma cell adhesion are independent of its direct binding to TSP1. One possibility is that sFRP-2 signaling through the Wnt pathway activates a different integrin that recognizes TSP1.
Despite a general consensus that TSP1 limits tumor growth by inhibiting angiogenesis and independent evidence that TSP1 can limit tumor growth through its effects on anti-tumor immunity (reviewed in [17]), some clinical studies of TSP1 expression in breast cancer unexpectedly have revealed maintenance or up-regulation of TSP1 expression with disease progression [29, 30, 86, 87]. Consistent with these clinical studies, a murine model of breast cancer driven by the polyoma middle T antigen indicated that TSP1 in the mammary tumor microenvironment inhibits angiogenesis and primary tumor growth but promotes metastasis to the lung [31]. Our data suggests that, besides inhibiting tumor progression and metastasis by disrupting Wnt signaling, sFRP-1 plays an additional inhibitory role in breast cancer by blocking the pro-adhesive and chemotactic activities of TSP1. Because α3β1 integrin interactions with the N-module of TSP1 also regulate endothelial cell adhesion and chemotaxis in vitro and angiogenesis in the chicken chorioallantoic membrane [70], sFRP-1 could also be an important regulator of TSP1 in angiogenesis. sFRP-1 is reported to have both pro- and anti-angiogenic activities, depending on the context [88–90]. Thus, if sFRP-1 is present in a breast tumor we would predict that the inhibitory anti-angiogenic activity of TSP1 would be dominant, but in the absence of sFRP-1 expression the tumor promoting effects of the N-module of TSP1 may be unmasked. Furthermore, α3β1 integrin interactions with the N-module of TSP1 regulate adhesion, motility, and growth of melanoma, small cell lung carcinoma, neuroblastoma, and malignant astrocytoma cells [33, 91, 92], suggesting that sFRP-1 could have a broader role in regulating tumor responses to TSP1.
5. Conclusions
This study demonstrates direct high affinity binding of TSP1 to sFRP-1 and sFRP-2, but not sFRP-3. Binding to sFRP-1 is mediated by the N-terminal domain of TSP1 and the NTR domain of sFRP-1. This interaction and functional effects of sFRP-1 modulate the pro-adhesive and migratory activities of TSP1 for breast carcinoma cells and identifies an alternative mechanism by which sFRP-1 could regulate breast cancer progression and metastasis.
Acknowledgments
Grant support: this research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (DDR, JSR). Gema Martin-Manso was recipient of a grant BEFI from Instituto de Salud Carlos III (Spanish Ministry of Health).
We thank Deane Mosher, and Jack Lawler for providing reagents, Van Hoang and Young Kim from the Protein Chemistry Laboratory, SAIC/NCI-Frederick for mass spectroscopy and protein sequence analysis, respectively, Swaraj Sinha and David R. Soto-Pantoja for help with the wound scratch assays, and Thomas Miller for help with the migration assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: the abbreviations used are: sFRP, secreted frizzled-related protein; TSP1, thrombospondin-1; HSPG, heparan sulfate proteoglycan; CRD, cysteine-rich domain; NTR, netrin; TIMP, tissue inhibitor of metalloproteinases; pCOLCEs, procollagen C-proteinase enhancer proteins; GST, glutathione-S-transferase; vWC, von Willebrand factor type C; LDL, low-density lipoprotein; EGF, epidermal growth factor.
Contributor Information
Gema Martin-Manso, Email: mansog@mail.nih.gov.
Maria J. Calzada, Email: mcalzada.hlpr@salud.madrid.org.
Yoshiro Chuman, Email: chuman@sci.hokudai.ac.jp.
John M. Sipes, Email: sipesj@mail.nih.gov.
Charles P. Xavier, Email: Charles.Xavier@nih.gov.
Vladimir Wolf, Email: vladimir.wolf@qiagen.com.
Svetlana A. Kuznetsova, Email: kuznetsova4872@yahoo.co.uk.
Jeffrey S. Rubin, Email: rubinj@mail.nih.gov.
References
- 1.Baenziger NL, Brodie GN, Majerus PW. Proc Natl Acad Sci U S A. 1971;68:240–243. doi: 10.1073/pnas.68.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonnefoy A, Moura R, Hoylaerts MF. Cell Mol Life Sci. 2008;65:713–727. doi: 10.1007/s00018-007-7487-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isenberg JS, Romeo MJ, Yu C, Yu CK, Nghiem K, Monsale J, Rick ME, Wink DA, Frazier WA, Roberts DD. Blood. 2008;111:613–623. doi: 10.1182/blood-2007-06-098392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Good DJ, Polverini PJ, Rastinejad F, Le Beau MM, Lemons RS, Frazier WA, Bouck NP. Proc Natl Acad Sci U S A. 1990;87:6624–6628. doi: 10.1073/pnas.87.17.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taraboletti G, Roberts D, Liotta LA, Giavazzi R. J Cell Biol. 1990;111:765–772. doi: 10.1083/jcb.111.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstat-Saslow DL, Zabrenetzky VS, VanHoutte K, Frazier WA, Roberts DD, Steeg PS. Cancer Res. 1994;54:6504–6511. [PubMed] [Google Scholar]
- 7.Li Q, Ahuja N, Burger PC, Issa JP. Oncogene. 1999;18:3284–3289. doi: 10.1038/sj.onc.1202663. [DOI] [PubMed] [Google Scholar]
- 8.Roberts DD. Faseb J. 1996;10:1183–1191. [PubMed] [Google Scholar]
- 9.Kazerounian S, Yee KO, Lawler J. Cell Mol Life Sci. 2008;65:700–712. doi: 10.1007/s00018-007-7486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown LF, Guidi AJ, Schnitt SJ, Van De Water L, Iruela-Arispe ML, Yeo TK, Tognazzi K, Dvorak HF. Clin Cancer Res. 1999;5:1041–1056. [PubMed] [Google Scholar]
- 11.Tuszynski GP, Smith M, Rothman VL, Capuzzi DM, Joseph RR, Katz J, Besa EC, Treat J, Switalska HI. Thromb Haemost. 1992;67:607–611. [PubMed] [Google Scholar]
- 12.Volpert OV, Lawler J, Bouck NP. Proc Natl Acad Sci U S A. 1998;95:6343–6348. doi: 10.1073/pnas.95.11.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bocci G, Francia G, Man S, Lawler J, Kerbel RS. Proc Natl Acad Sci U S A. 2003;100:12917–12922. doi: 10.1073/pnas.2135406100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isenberg JS, Hyodo F, Ridnour LA, Shannon CS, Wink DA, Krishna MC, Roberts DD. Neoplasia. 2008;10:886–896. doi: 10.1593/neo.08264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Manso G, Galli S, Ridnour LA, Tsokos M, Wink DA, Roberts DD. Cancer Res. 2008;68:7090–7099. doi: 10.1158/0008-5472.CAN-08-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxhimer JB, Soto-Pantoja DR, Ridnour LA, Shih HB, Degraff WG, Tsokos M, Wink DA, Isenberg JS, Roberts DD. Sci Transl Med. 2009;1:3ra7. doi: 10.1126/scitranslmed.3000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isenberg JS, Martin-Manso G, Maxhimer JB, Roberts DD. Nat Rev Cancer. 2009;9:182–194. doi: 10.1038/nrc2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson CB, Lawler J, Mosher DF. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-007-7484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calzada MJ, Sipes JM, Krutzsch HC, Yurchenco PD, Annis DS, Mosher DF, Roberts DD. J Biol Chem. 2003;278:40679–40687. doi: 10.1074/jbc.M302014200. [DOI] [PubMed] [Google Scholar]
- 20.Calzada MJ, Zhou L, Sipes JM, Zhang J, Krutzsch HC, Iruela-Arispe ML, Annis DS, Mosher DF, Roberts DD. Circ Res. 2004;94:462–470. doi: 10.1161/01.RES.0000115555.05668.93. [DOI] [PubMed] [Google Scholar]
- 21.Staniszewska I, Zaveri S, Del Valle L, Oliva I, Rothman VL, Croul SE, Roberts DD, Mosher DF, Tuszynski GP, Marcinkiewicz C. Circ Res. 2007;100:1308–1316. doi: 10.1161/01.RES.0000266662.98355.66. [DOI] [PubMed] [Google Scholar]
- 22.Asch AS, Liu I, Briccetti FM, Barnwell JW, Kwakye-Berko F, Dokun A, Goldberger J, Pernambuco M. Science. 1993;262:1436–1440. doi: 10.1126/science.7504322. [DOI] [PubMed] [Google Scholar]
- 23.Roberts DD. Cancer Res. 1988;48:6785–6793. [PubMed] [Google Scholar]
- 24.Murphy-Ullrich JE, Schultz-Cherry S, Hook M. Mol Biol Cell. 1992;3:181–188. doi: 10.1091/mbc.3.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg PJ. Thromb Haemost. 1994;72:787–792. [PubMed] [Google Scholar]
- 26.Kuznetsova SA, Day AJ, Mahoney DJ, Rugg MS, Mosher DF, Roberts DD. J Biol Chem. 2005;280:30899–30908. doi: 10.1074/jbc.M500701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuznetsova SA, Issa P, Perruccio EM, Zeng B, Sipes JM, Ward Y, Seyfried NT, Fielder HL, Day AJ, Wight TN, Roberts DD. J Cell Sci. 2006;119:4499–4509. doi: 10.1242/jcs.03171. [DOI] [PubMed] [Google Scholar]
- 28.Byrne GJ, Hayden KE, McDowell G, Lang H, Kirwan CC, Tetlow L, Kumar S, Bundred NJ. Int J Oncol. 2007;31:1127–1132. doi: 10.3892/ijo.31.5.1127. [DOI] [PubMed] [Google Scholar]
- 29.Bertin N, Clezardin P, Kubiak R, Frappart L. Cancer Res. 1997;57:396–399. [PubMed] [Google Scholar]
- 30.Linderholm B, Karlsson E, Klaar S, Lindahl T, Borg AL, Elmberger G, Bergh J. Eur J Cancer. 2004;40:2417–2423. doi: 10.1016/j.ejca.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 31.Yee KO, Connolly CM, Duquette M, Kazerounian S, Washington R, Lawler J. Breast Cancer Res Treat. 2009;114:85–96. doi: 10.1007/s10549-008-9992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taraboletti G, Roberts DD, Liotta LA. J Cell Biol. 1987;105:2409–2415. doi: 10.1083/jcb.105.5.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandrasekaran S, Guo NH, Rodrigues RG, Kaiser J, Roberts DD. J Biol Chem. 1999;274:11408–11416. doi: 10.1074/jbc.274.16.11408. [DOI] [PubMed] [Google Scholar]
- 34.Hyder SM, Liang Y, Wu J, Welbern V. Endocr Relat Cancer. 2009;16:809–817. doi: 10.1677/ERC-08-0311. [DOI] [PubMed] [Google Scholar]
- 35.Hyder SM, Liang Y, Wu J. Int J Cancer. 2009;125:1045–1053. doi: 10.1002/ijc.24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones SE, Jomary C. Bioessays. 2002;24:811–820. doi: 10.1002/bies.10136. [DOI] [PubMed] [Google Scholar]
- 37.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. J Cell Sci. 2008;121:737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 38.Chong JM, Uren A, Rubin JS, Speicher DW. J Biol Chem. 2002;277:5134–5144. doi: 10.1074/jbc.M108533200. [DOI] [PubMed] [Google Scholar]
- 39.Klaus A, Birchmeier W. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 40.Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 41.Schlange T, Matsuda Y, Lienhard S, Huber A, Hynes NE. Breast Cancer Res. 2007;9:R63. doi: 10.1186/bcr1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leyns L, Bouwmeester T, Kim SH, Piccolo S, De Robertis EM. Cell. 1997;88:747–756. doi: 10.1016/s0092-8674(00)81921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, Krinks M, Lin K, Luyten FP, Moos M., Jr Cell. 1997;88:757–766. doi: 10.1016/s0092-8674(00)81922-4. [DOI] [PubMed] [Google Scholar]
- 44.Finch PW, He X, Kelley MJ, Uren A, Schaudies RP, Popescu NC, Rudikoff S, Aaronson SA, Varmus HE, Rubin JS. Proc Natl Acad Sci U S A. 1997;94:6770–6775. doi: 10.1073/pnas.94.13.6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubin JS, Barshishat-Kupper M, Feroze-Merzoug F, Xi ZF. Front Biosci. 2006;11:2093–2105. doi: 10.2741/1952. [DOI] [PubMed] [Google Scholar]
- 46.Klopocki E, Kristiansen G, Wild PJ, Klaman I, Castanos-Velez E, Singer G, Stohr R, Simon R, Sauter G, Leibiger H, Essers L, Weber B, Hermann K, Rosenthal A, Hartmann A, Dahl E. Int J Oncol. 2004;25:641–649. [PubMed] [Google Scholar]
- 47.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- 48.Gumz ML, Kreinest PA, Zou H, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon B, Sinha M, Parker AS, Sun L-Z, Ahlquist DA, Wood CG, Copland JA. Clin Cancer Res. 2007 doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 49.Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. Breast Cancer Res. 2009;11:R32. doi: 10.1186/bcr2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JL, Lin CT, Chueh LL, Chang CJ. J Biol Chem. 2004;279:14602–14609. doi: 10.1074/jbc.M309008200. [DOI] [PubMed] [Google Scholar]
- 51.Lee JL, Chang CJ, Wu SY, Sargan DR, Lin CT. Breast Cancer Res Treat. 2004;84:139–149. doi: 10.1023/B:BREA.0000018412.83348.ff. [DOI] [PubMed] [Google Scholar]
- 52.Courtwright A, Siamakpour-Reihani S, Arbiser JL, Banet N, Hilliard E, Fried L, Livasy C, Ketelsen D, Nepal DB, Perou CM, Patterson C, Klauber-Demore N. Cancer Res. 2009;69:4621–4628. doi: 10.1158/0008-5472.CAN-08-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulte G, Bryja V. Trends Pharmacol Sci. 2007;28:518–525. doi: 10.1016/j.tips.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Lin K, Wang S, Julius MA, Kitajewski J, Moos M, Jr, Luyten FP. Proc Natl Acad Sci U S A. 1997;94:11196–11200. doi: 10.1073/pnas.94.21.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bafico A, Gazit A, Pramila T, Finch PW, Yaniv A, Aaronson SA. J Biol Chem. 1999;274:16180–16187. doi: 10.1074/jbc.274.23.16180. [DOI] [PubMed] [Google Scholar]
- 56.Banyai L, Patthy L. Protein Sci. 1999;8:1636–1642. doi: 10.1110/ps.8.8.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williamson RA, Panagiotidou P, Mott JD, Howard MJ. Mol Biosyst. 2008;4:417–425. doi: 10.1039/b717901d. [DOI] [PubMed] [Google Scholar]
- 58.Uren A, Reichsman F, Anest V, Taylor WG, Muraiso K, Bottaro DP, Cumberledge S, Rubin JS. J Biol Chem. 2000;275:4374–4382. doi: 10.1074/jbc.275.6.4374. [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Rios J, Esteve P, Ruiz JM, Bovolenta P. Neural Dev. 2008;3:19. doi: 10.1186/1749-8104-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chuman Y, Uren A, Cahill J, Regan C, Wolf V, Kay BK, Rubin JS. Peptides. 2004;25:1831–1838. doi: 10.1016/j.peptides.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 61.Roberts DD, Cashel J, Guo N, Tissue Cult J. Methods. 1994;16:217–222. [Google Scholar]
- 62.Akiyama SK, Yamada KM. J Biol Chem. 1985;260:4492–4500. [PubMed] [Google Scholar]
- 63.Misenheimer TM, Huwiler KG, Annis DS, Mosher DF. J Biol Chem. 2000;275:40938–40945. doi: 10.1074/jbc.M007022200. [DOI] [PubMed] [Google Scholar]
- 64.Anilkumar N, Annis DS, Mosher DF, Adams JC. J Cell Sci. 2002;115:2357–2366. doi: 10.1242/jcs.115.11.2357. [DOI] [PubMed] [Google Scholar]
- 65.Calzada MJ, Kuznetsova SA, Sipes JM, Rodrigues RG, Cashel JA, Annis DS, Mosher DF, Roberts DD. Matrix Biol. 2008;27:339–351. doi: 10.1016/j.matbio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Legrand C, Thibert V, Dubernard V, Begault B, Lawler J. Blood. 1992;79:1995–2003. [PubMed] [Google Scholar]
- 67.Wolf V, Endo Y, Rubin JS. Methods Mol Biol. 2008;468:31–44. doi: 10.1007/978-1-59745-249-6_3. [DOI] [PubMed] [Google Scholar]
- 68.Todaro GJ, Fryling C, De Larco JE. Proc Natl Acad Sci U S A. 1980;77:5258–5262. doi: 10.1073/pnas.77.9.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson KE, Li Z, Kara M, Gardner KL, Roberts DD. J Immunol. 1999;163:3621–3628. [PubMed] [Google Scholar]
- 70.Chandrasekaran L, He CZ, Al-Barazi H, Krutzsch HC, Iruela-Arispe ML, Roberts DD. Mol Biol Cell. 2000;11:2885–2900. doi: 10.1091/mbc.11.9.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Munson PJ, Rodbard D. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 72.Roszmusz E, Patthy A, Trexler M, Patthy L. J Biol Chem. 2001;276:18485–18490. doi: 10.1074/jbc.M100100200. [DOI] [PubMed] [Google Scholar]
- 73.Ishida T, Horiike K, Tojo H, Nozaki M. J Theor Biol. 1988;130:49–66. doi: 10.1016/s0022-5193(88)80163-2. [DOI] [PubMed] [Google Scholar]
- 74.Adams JC, Lawler J. Int J Biochem Cell Biol. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang YW, Su Y, Volpert OV, Vande Woude GF. Proc Natl Acad Sci U S A. 2003;100:12718–12723. doi: 10.1073/pnas.2135113100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Michaud M, Poyet P. Anticancer Res. 1994;14:1127–1131. [PubMed] [Google Scholar]
- 77.Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, Galm O, Camara O, Durst M, Kristiansen G, Huszka C, Knuchel R, Dahl E. Oncogene. 2006;25:3479–3488. doi: 10.1038/sj.onc.1209386. [DOI] [PubMed] [Google Scholar]
- 78.Krutzsch HC, Choe BJ, Sipes JM, Guo N, Roberts DD. J Biol Chem. 1999;274:24080–24086. doi: 10.1074/jbc.274.34.24080. [DOI] [PubMed] [Google Scholar]
- 79.Calzada MJ, Annis DS, Zeng B, Marcinkiewicz C, Banas B, Lawler J, Mosher DF, Roberts DD. J Biol Chem. 2004;279:41734–41743. doi: 10.1074/jbc.M406267200. [DOI] [PubMed] [Google Scholar]
- 80.Dihlmann S, von Knebel Doeberitz M. Int J Cancer. 2005;113:515–524. doi: 10.1002/ijc.20609. [DOI] [PubMed] [Google Scholar]
- 81.Li Z, Calzada MJ, Sipes JM, Cashel JA, Krutzsch HC, Annis DS, Mosher DF, Roberts DD. J Cell Biol. 2002;157:509–519. doi: 10.1083/jcb.200109098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pallero MA, Elzie CA, Chen J, Mosher DF, Murphy-Ullrich JE. Faseb J. 2008;22:3968–3979. doi: 10.1096/fj.07-104802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan Q, Murphy-Ullrich JE, Song Y. Biochemistry [Google Scholar]
- 84.Tan K, Duquette M, Liu JH, Zhang R, Joachimiak A, Wang JH, Lawler J. Structure. 2006;14:33–42. doi: 10.1016/j.str.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jurk K, Clemetson KJ, de Groot PG, Brodde MF, Steiner M, Savion N, Varon D, Sixma JJ, Van Aken H, Kehrel BE. Faseb J. 2003;17:1490–1492. doi: 10.1096/fj.02-0830fje. [DOI] [PubMed] [Google Scholar]
- 86.Wang-Rodriguez J, Urquidi V, Rivard A, Goodison S. Breast Cancer Res. 2003;5:R136–R143. doi: 10.1186/bcr620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gasparini G, Toi M, Biganzoli E, Dittadi R, Fanelli M, Morabito A, Boracchi P, Gion M. Oncology. 2001;60:72–80. doi: 10.1159/000055300. [DOI] [PubMed] [Google Scholar]
- 88.Dufourcq P, Couffinhal T, Ezan J, Barandon L, Moreau C, Daret D, Duplaa C. Circulation. 2002;106:3097–3103. doi: 10.1161/01.cir.0000039342.85015.5c. [DOI] [PubMed] [Google Scholar]
- 89.Dufourcq P, Descamps B, Tojais NF, Leroux L, Oses P, Daret D, Moreau C, Lamaziere JM, Couffinhal T, Duplaa C. Stem Cells. 2008;26:2991–3001. doi: 10.1634/stemcells.2008-0372. [DOI] [PubMed] [Google Scholar]
- 90.Hu J, Dong A, Fernandez-Ruiz V, Shan J, Kawa M, Martinez-Anso E, Prieto J, Qian C. Cancer Res. 2009;69:6951–6959. doi: 10.1158/0008-5472.CAN-09-0541. [DOI] [PubMed] [Google Scholar]
- 91.Guo N, Templeton NS, Al-Barazi H, Cashel JA, Sipes JM, Krutzsch HC, Roberts DD. Cancer Res. 2000;60:457–466. [PubMed] [Google Scholar]
- 92.Pijuan-Thompson V, Grammer JR, Stewart J, Silverstein RL, Pearce SF, Tuszynski GP, Murphy-Ullrich JE, Gladson CL. Exp Cell Res. 1999;249:86–101. doi: 10.1006/excr.1999.4458. [DOI] [PubMed] [Google Scholar]