This case represents the first report on the isolation of a difficult-to-identify clinical isolate of Mycobacterium elephantis from Asia.
Case presentation
A 72-year-old woman was admitted to hospital because of chronic respiratory disease for approximately 2 months. Her major symptoms included dry cough and nocturnal fever (to 38°C). Her outpatient record indicated that she was prescribed amoxicillin by her general practitioner but no improvement was obtained. She had no history of infection with mycobacteria or contact with an elephant. Results of physical examination were unremarkable. The patient's tuberculin test was negative. Routine laboratory testing on admission revealed an elevated C-reactive protein (CRP) of 34 mg/L, and an erythrocyte sedimentation rate of 45 mm/hr. The chest X-ray examination showed a diffuse shadow in the right apex suggestive of pulmonary tuberculosis. The initial microbiological examination of bronchial lavage by the hospital laboratory revealed the presence of small coccobacillary acid-fast organisms. The bronchial lavage specimen was cultured on Löwenstein-Jensen medium for mycobacterial isolation. Since the growth of the isolate on Löwenstein-Jensen medium was slower than that for the majority of rapidly growing mycobacteria, and due to apical pulmonary lesions the patient was misdiagnosed as having tuberculosis and entered into the TB Register. After one month of antituberculosis therapy, the patient discontinued treatment due to extreme weakness and severe abdominal pain. Meanwhile the isolate was referred to our laboratory for definitive identification. Further investigation on repeated specimens revealed that the infecting micro-organism was a rapidly growing mycobacteria. The patient's treatment was changed from the antituberculosis regimen to a combination therapy consisting of amikacin and ciprofloxacin. After two months of treatment she recovered and has remained well ever since.
The conventional identification and drug susceptibility of the Iranian isolate, namely, ‘M202’ was achieved according to the standard procedures described previously.1,2 The conclusive identification included molecular testing, i.e. the PCR restriction fragment length polymorphism analysis (PRA algorithm) of the hsp653 and direct sequencing analysis of almost full length of 16S rDNA4 and 16S–23S internal transcribed spacer (ITS)5 as well as partial sequencing of hsp653 and rpoB genes.6 The GenBank accession numbers of M. elephantis determined in this work are as follow: GU142921, HM229788-90 for 16S rDNA, rpoB, hsp65 and ITS genes, respectively.
Based on the phenotypic characteristics, M202 was a scotochromogenic rapidly growing mycobacterial species which grew at 37°C and 45°C as well as on Löwenstein-Jensen medium with 5% NaCl. It was positive for the key biochemical tests including catalase, nitrate, urease, tween hydrolysis and tellurite reduction tests. It was resistant to rifampicin but susceptible to amikacin, clarithromycin, ciprofloxacin, ethambutol, isoniazid, doxicyclin, streptomycin, cefoxitin, sulfamethoxazol and imipenem. The main microbiological traits of the Iranian isolate resembled those of the human clinical strains of M. elephantis characterized by Turenne et al.7 and Tortoli et al.8
In PRA method the isolate exhibited a unique pattern that was distinct from the previously published patterns3 as well as from those in our in-house library.
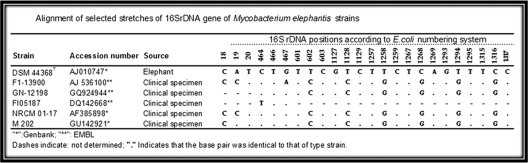
The 16S rDNA, rpoB and hsp65 gene sequences of the isolate showed 100%, 98.7% and 96.2% similarities with those of the validly published reference strains of M. elephantis, respectively. The hypervariable signature sequences of 16S rDNA of M202 were identical to those of the type strain of M. elephantis (Figure 1). The ITS sequence of M202 was unique compared with those of other validly published mycobacteria.
Figure 1.
Alignment of selected stretches of 16S rDNA gene of Mycobacterium elephantis strains
Discussion
Further to the phenotypic features, all molecular tests used in the current study provided the evidence that the Iranian isolate belongs to M. elephantis species.
M. elephantis is a rapidly growing mycobacteria first isolated from a lung abscess in a Sri Lankan elephant that died from chronic respiratory disease.4 Human isolation of this species has been reported from the developed countries including Canadian, Italian and Belgian patient specimens.7–9 It belongs to a group of clinical isolates of mycobacteria which has been evaluated as difficult-to-identify organisms by Springer et al.10
Our case represents the first report of isolation of M. elephantis from the human clinical specimen in a developing country. The elderly patient had no apparent underlying diseases. However, the risk factors for mycobacterial infection could not completely be excluded in the patient due to age.
The etiologic role of the isolate M202 might be inferred from the fact that acid fast bacilli were microscopically observed in two different bronchial lavage specimens of the patient. Furthermore, the isolate was recovered from the pure culture. Also, no such isolate was made in our laboratory in the past and during the same period of time. As for 15 previously reported clinical strains,7–9 the clinical relevance of M. elephantis remains difficult to ascertain since the isolate is very rare and hard to identify. Increased awareness of this species might help to distinct this species from closely related mycobacteria to more exactly determine the role of M. elephantis in human infection.
In conclusion, this case re-affirms the fact stated by Tortoli et al.8 that the environment is the most probable reservoir of M. elephantis for either human or animal infections. It is not limited to a certain geographic area. Furthermore, the key molecular markers when combined with the major microbiological traits provide a conclusive evidence for identification of the rare clinically significant non-tuberculous mycobacteria.
DECLARATIONS
Competing interests
None declared
Funding
The Office of Vice-chancellor for Research, Isfahan University of Medical Sciences
Ethical approval
Written informed consent to publication was obtained from the patient or next of kin
Guarantor
HS
Contributorship
All authors contributed equally
Acknowledgements
The authors are grateful to the Office of Vice-chancellor for Research, Isfahan University of Medical Sciences for the financial support of current study
Reviewer
Buddha Basnyat
References
- 1.Kent PT, Kubica GP Public health mycobacteriology: a guide for the level III laboratory. Atlanta, GA: CDC, 1985 [Google Scholar]
- 2.National Committee for Clinical Laboratory Standards Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; Approved Standard. Wayne, PA: NCCLS, 2003 [PubMed] [Google Scholar]
- 3.Kim H, Kim SH, Shim TS, et al. PCR restriction fragment length polymorphism analysis (PRA)-algorithm targeting 644 bp Heat Shock Protein 65 (hsp65) gene for differentiation of Mycobacterium spp. J Microbiol Methods 2005;62:199–209 [DOI] [PubMed] [Google Scholar]
- 4.Shojaei H, Magee JG, Freeman R, et al. Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic Mycobacterium isolated from an elephant. Int J Syst Evol Microbiol 2000;50:1817–20 [DOI] [PubMed] [Google Scholar]
- 5.Roth A, Fischer M, Hamid ME, et al. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol 1998;36:139–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adekambi T, Colson P, Drancourt M .rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol 2003;41:5699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turenne C, Chedore P, Wolfe J, et al. Phenotypic and molecular characterization of clinical isolates of Mycobacterium elephantis from human specimens. J Clin Microbiol 2002;40:1230–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tortoli E, Rindi L, Bartoloni A, et al. Mycobacterium elephantis: not an exceptional finding in clinical specimens. Eur J Clin Microbiol Infect Dis 2003;22:427–30 [DOI] [PubMed] [Google Scholar]
- 9.Potters D, Seghers M, Muyldermans G, et al. Recovery of Mycobacterium elephantis from sputum of a patient in Belgium. J Clin Microbiol 2003;41:1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer B, Stockman L, Teschner K, et al. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol 1996;34:296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]