Abstract
SLUG is a transcriptional repressor protein implicated to have major role in the oncogenesis and metastasis of human breast cells. We previously have shown by chromatin immunoprecipitation assay that human SLUG (hSLUG) is co-localized with the co-repressor protein CtBP1 as bound to the BRCA2 gene silencer. hSLUG was predicted to be binding directly to CtBP1 because of an apparent presence of CtBP1 binding site in its amino acid sequences. Here, we provide evidence through yeast two-hybrid and in vitro co-immunoprecipitation analyses that hSLUG does not directly interacts with hCtBP1. This observation will help in the study of the mode of action of hSLUG in human cells.
Keywords: SLUG, CtBP1, Yeast 2-hybrid, Co-immunoprecipitation, Repressor, SNAG domain
SLUG is a member of a superfamily of zinc-finger transcriptional repressors [1-3]. SLUG and its family members are involved in tumor progression [1]. SLUG is implicated to induce epithelial to mesenchymal transition in many cells [2]. The different family members are now grouped into two families: Snail and Scratch. Vertebrates have three Snail members: SNAIL (new name SNAIL1), SLUG (now named SNAIL2), and SNAIL3 (formerly SMUC) [2,3]. Human cells have two SCRATCH proteins: SCRATCH1 and SCRATCH2. Human breast cells we have studied so far do not express SCRATCH transcripts (M.K. Tripathi and G. Chaudhuri, unpublished). Human SLUG gene is located at chromosome 8q11, has 3 exons and 2 introns, is transcribed into a ~2.1-kb mRNA (Accession No.NM_003068), and encodes a C2H2-type zinc finger transcription factor protein with 268 amino acids [4]. The encoded protein acts as a transcriptional repressor that binds to E2-box motif (5′-CACCTG-3′) [4,5].
Although many genes have the E2-box sequences at their promoters, only the expressions of few proteins are experimentally shown to be regulated by SLUG. In human mammary epithelial cells, SLUG is shown to negatively regulate the E-cadherin [5], aromatase [6], PUMA [7], BRCA2 [8], claudin-1 [9], integrin alpha3, beta1, and beta4 [10] and, cytokeratin 8 and 19 [11] gene expressions. Other genes that are potentially down regulated inside the human breast cells by SLUG may include those of VE-cadherin, other claudins, occludins, desmoplakin, and mucin-1 [2,3]. Majority of the genes that are down regulated by SLUG are directly or indirectly involved in cell–cell adhesion and their inhibition thus may induce dislodging of the cells and metastasis [3].
The exact mode of action of human SLUG is not known. While the C-terminal zinc-finger domains of SLUG are responsible for DNA binding, the N-terminal domain are predicted to be responsible for the recruitment of co-repressor molecules at the target gene promoter. SLUG has a domain with 20 amino acid residues at its N-terminus known as SNAG domain. The SNAG domain was originally characterized in the growth factor independence-1 (Gfi-1) oncoprotein, where it forms part of the first 20 amino acids that suffice for transcriptional repression [2,4]. Itis proposed that SLUG binds to the E2-box sequence of the DNA through its C-terminal zinc-finger domains and then recruits either CtBP1 [4,8] or a SNAG-domain binding protein (e.g., Sin3A) as a co-repressor. The co-repressor then recruits histone deacetylase (e.g., HDAC1) at the promoter to silent the target gene expression by chromatin remodeling [8]. Here we show evidence that hSLUG does not interact directly with hCtBP1.
Materials and methods
cDNA amplifications and cloning
The ORF of hSLUG was amplified with Pfu DNA polymerase (Stratagene, La Jolla, CA) from its cDNA (8) using 5′CAACATATGATGCCGCGCTCC-3′ and 5′-CAAGTCGACGTGTGCTACACAGCAGCC-3′ which contained NdeI and SalI sites at the 5′-ends, respectively. The PCR product (831 bp) was purified and cloned into pCR-4-TOPO (Invitrogen, Carlsbad, CA). Purified pCR-4-TOPO-hSLUG plasmid DNA was digested with NdeI and SalI and cloned into the NdeI/SalI sites of the bait vector pGBKT7 (BD Biosciences Clontech, Palo Alto, CA). This gave us pGBK-hSLUG clone. Human CtBP1 (hCtBP1) cDNA was a gift from Prof. G. Chinnadurai of Saint Louis University Medical Center, St. Louis, MO. The ORF of hCtBP1 was amplified using following primers: 5′CAACATATGATGGGCAGCTCG-3′ and 5′CAAATCGATCAACTGGTCACT-3′. The PCR product (1.3 kb) was purified and cloned into pCR-4-TOPO (Invitrogen) and cut out of this plasmid with NdeI and EcoR I. Next, the CtBP1 fragment was cloned into the NdeI and EcoRI sites in the multiple cloning site of the prey vector pGADT7 (BD Biosciences Clontech). This gave us pGAD-hCtBP1. The inserts of the pGAD-hCtBP1 and pGBK-hSLUG constructs were sequenced to confirm that the ORFs are in frame with their respective fusion partners.
Yeast 2-hybrid analysis
Saccharomyces cerevisiae strain AH109 (BD Biosciences Clontech) was co-transformed with pair wise combinations of bait and prey vectors with lithium acetate (Matchmaker System 3; BD Biosciences Clontech), as described in the manufacturer’s protocol. The yeast strain AH109 was co-transformed with Gal4 DBD-hSLUG fusion construct in pGBKT7 together with a Gal4 activation domain-hCtBP1 fusion construct in pGADT7. Interactions between the proteins, which tethered both domains of GAL4 together, were identified by growth of plasmid-carrying cells on minimal medium without leucine, tryptophan, histidine and adenine and by α-galactosidase activity.
Co-immunoprecipitation experiments
Epitope-tagged proteins (myc-hSLUG and HA-hCtBP1) were expressed from the bait (hSLUG-pGBKT7) and the prey (hCtBP1-pGADT7) vectors using the reagents from the rabbit reticulocyte extract-based TNT T7 Quick-coupled Transcription/ Translation System; (Promega, Madison, WI). Equal volumes of radiolabeled (35S-methionine) proteins, myc-hSLUG and HA-hCtBP1, were mixed together and incubated at 25 °C for 1 h and were subjected to immunoprecipitation with either rabbit polyclonal HA antibodies or mouse c-Myc monoclonal antibodies using the MATCHMAKER Co-IP kit (BD Biosciences Clontech) according to the manufactures protocols. Immunoprecipitated proteins were resolved on NuPAGE 4–12% Bis-Tris gradient gels (Invitrogen) and analyzed by autoradiography. In some experiments, we preincubated the hSLUG with 20 fmols of human BRCA2 gene silencer DNA (8) or a pRL-Null (Promega) plasmid construct containing a 641-bp human claudin 7 gene promoter that has seven E2-box sequences before the binding reaction. We also performed the binding reaction at 37 °C for 1 h.
Results and discussion
Human SLUG has the potential consensus CtBP1 binding site
CtBP1 belongs to the group of proteins that comprise a distinct class of corepressors that interact with a subset of transcription factors through a short sequence motif, PXDLSX[R/K], where X is any amino acid [4,12]. Human SLUG protein has a sequence element [PSDTSSK] similar to this CtBP1 binding consensus sequences at 91–97 positions (Fig. 1). While characterizing the mode of hSLUG in mediating cell cycle stage specific silencing of human BRCA2 gene expression we investigated whether hCtBP1 is associated with this repression [8]. Chromatin immunoprecipitation analysis suggested that hSLUG is co-localized at the 221 bp BRCA2 gene silencer with hCtBP1 [8]. We postulated that hSLUG directly or with the help of an adapter protein binds to hCtBP1 at the promoter of SLUG-regulated genes. Apart from the putative CtBP1 binding motif hSLUG also have a N-terminal SNAG domain (Fig. 1), which may bind to co-repressor molecules like Sin3A [2,3]. We have not yet explored whether hSLUG directly binds to Sin3A.
Fig. 1.
Amino acid sequence of human SLUG protein showing the putative domains. The N-terminal 30 amino acid residues constitute the SNAG domain (shown in green). The putative CtBP1 binding sequence is shown in blue. Underscored amino acid residues indicate the zinc-finger domains. The cysteine and histidine residues of the zinc-fingers are shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Yeast 2-hybrid analysis suggests that hSLUG does not bind directly to hCtBP1
To verify whether hSLUG indeed binds directly with hCtBP1 we utilized the yeast two-hybrid techniques (Fig. 2). hSLUG bait constructs were co-transformed into yeast with a prey construct encoding hCtBP1 fused to the Gal4 activation domain. As a control, the bait constructs were co-transformed with the empty vector to confirm that the hSLUG bait constructs were unable to activate reporter genes in the absence of an interaction. A positive interaction was assessed by the ability of the yeast to grow on SD medium lacking tryptophan, leucine, adenine, and histidine (Fig. 2). Unexpectedly, hSLUG did not interact with hCtBP1 whereas the positive control proteins (SV40 T4 antigen and a P53 protein fragment) did show binding (Fig. 2). Verification of the nucleotide sequences of the bait and the prey constructs, reversion of the bait and prey, and the evaluation of the expressions of the hybrid mRNAs and proteins in the transfected yeast cells were performed to ensure the experimental procedures.
Fig. 2.
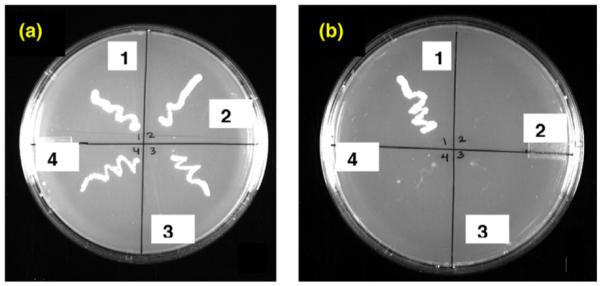
Yeast-2-hybrid analysis data. Growth of yeast cells on minimal agar plate containing selective dropout medium is shown. The left agar plate (a)is with selective medium (SD medium without leucine and tryptophan) for the growth of the transformants (no insert in the vectors are necessary for the growth). The plate in the right (b) the selective medium (SD medium without leucine, tryptophan, histidine, and adenine) only allowed growth of the yeast cells if the cloned bait protein and the prey protein physically interact. The bait and prey protein pairs in different segments in the photograph are as follows: (1) pGADT7-SV40 T-antigen/pGBKT7-P53 (positive control); (2) blank vectors (negative control); (3 and 4) pGBKT7-hSLUG/pGADT7-hCtBP1.
In vitro co-IP analysis confirms that hSLUG does not physically interact with hCtBP1
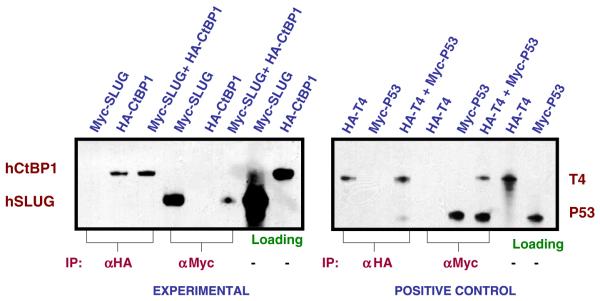
To further evaluate whether Myc-tagged hSLUG can directly bind with HA-tagged hCtBP1, we synthesized these proteins as 35S-methionine labeled in an in vitro transcription and translation system (see Materials and methods). The proteins were mixed together and incubated for 1 h at 25 or 37 °C. The proteins were then precipitated with HA or Myc antibody. The autoradiogram showed no apparent direct binding of hSLUG with hCtBP1 (Fig. 3). Under the conditions of the experiment two proteins known to bind to each other, SV40 T4 antigen and a P53 protein fragment, showed binding (Fig. 3). hSLUG did not show any binding to hCtBP1 either at 25 °C or at 37 °C.
Fig. 3.
Co-immunoprecipitation data. Autoradiograms containing the 35S-labeled proteins immunoprecipitaed from the binding reactions are shown. Labeling at the top of the autoradiograms indicates the 35S-labeled proteins in the binding reaction. The Myc-hSLUG is ~31 kDa and HA-hCtBP1 is ~50 kDa. The positive control panel shows interactions between the HA-tagged SV40 T4 antigen (65 kDa) and the Myc-tagged P53 fragment (35 kDa) as was analyzed similarly as the experimental proteins. Loading panels shows 1/10 of the respective proteins used in each binding assay.
These data suggests that despite the presence of the consensus hCtBP1 binding sequence in the hSLUG protein, they fail to bind each other. The possibilities for the mediation of hSLUG action thus may be through a adapter protein that bridges between hSLUG and hCtBP1. Direct recruitment of other co-repressor protein by the SNAG domain of hSLUG (Fig. 1) is another likely possibility. Our data are consistent with the observation that the N-terminal 20–30 amino acid residues of hSLUG is sufficient to mediate its repressor function [4]. Further detail analysis of the protein–protein interfaces in the hSLUG repressor complex will help us to understand the mode of action of this critical regulator of cell growth and metastasis.
Acknowledgments
We thank Ms. Holly Smith for technical assistance. Supported by the DOD Grant #DAMD17-00-1-0341 and the MMC/VICC cancer partnership Grant #1U54CA091408-010003 from NCI to G.C. C.K.B. is supported by a graduate fellowship from NHLBI (Grant #5 T32 HL007737).
References
- [1].Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, Aikou T. Slug Expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin. Cancer Res. 2005;11:1174–1180. [PubMed] [Google Scholar]
- [2].Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- [3].Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell. Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- [4].Hemavathy K, Guru SC, Harris J, Chen JD, Ip YT. Human Slug is a repressor that localizes to sites of active transcription. Mol. Cell. Biol. 2000;20:5087–5095. doi: 10.1128/mcb.20.14.5087-5095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- [6].Chen S, Ye J, Kijima I, Kinoshita Y, Zhou D. Positive and negative transcriptional regulation of aromatase expression in human breast cancer tissue. J. Steroid Biochem. Mol. Biol. 2005;95:17–23. doi: 10.1016/j.jsbmb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- [7].Wu WS, Heinrichs S, Xu D, Garrison SP, Zambetti GP, Adams JM, Look AT. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–665. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- [8].Tripathi MK, Misra S, Khedkar SV, Hamilton N, Irvin-Wilson C, Sharan C, Sealy L, Chaudhuri G. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J. Biol. Chem. 2005;280:17163–17171. doi: 10.1074/jbc.M501375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Martinez-Estrada OM, Culleres A, Soriano FX, Peinado H, Bolos V, Martinez FO, Reina M, Cano A, Fabre M, Vilaro S. The transcription factors Slug and Snail act as repressors of Claudin-1 expression in epithelial cells. Biochem. J. 2006;394:449–457. doi: 10.1042/BJ20050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Turner FE, Broad S, Khanim FL, Jeanes A, Talma S, Hughes S, Tselepis C, Hotchin NA. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J. Biol. Chem. 2006;281:21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- [11].Tripathi MK, Misra S, Chaudhuri G. Negative regulation of the expressions of cytokeratins 8 and 19 by SLUG repressor protein in human breast cells. Biochem. Biophys. Res. Commun. 2005;329:508–515. doi: 10.1016/j.bbrc.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]