Abstract
Over the past few years, advances in biochemical and genetic studies of the structure and function of the Mediator complex have shed new light on its subunit architecture and its mechanism of action in transcription by RNA polymerase II (pol II). The development of improved methods for reconstitution of recombinant Mediator subassemblies is enabling more in-depth analyses of basic features of the mechanisms by which Mediator interacts with and controls the activity of pol II and the general initiation factors. The discovery and characterization of multiple, functionally distinct forms of Mediator characterized by the presence or absence of the Cdk8 kinase module have led to new insights into how Mediator functions in both Pol II transcription activation and repression. Finally, progress in studies of the mechanisms by which the transcriptional activation domains (ADs) of DNA binding transcription factors target Mediator have brought to light unexpected complexities in the way Mediator participates in signal transduction.
Introduction
Eukaryotic mRNA synthesis catalyzed by the multisubunit enzyme RNA polymerase II (pol II) is a critical site for the regulation of gene expression. Over the past thirty years, elegant biochemical and genetic studies have established that mRNA synthesis can be regulated at both the initiation and elongation stages of pol II transcription, by a seemingly endless collection of transcriptional regulatory proteins and mechanisms. Among these proteins are a core set of evolutionarily conserved transcription factors that function directly with pol II to control its recruitment to target genes and its activity in transcription initiation and elongation. These transcription factors include (i) the general initiation factors TFIIB, TFIID, TFIIE, TFIIF, and TFIIH, which are the minimum set of factors required to assist pol II to bind selectively to its promoters and initiate transcription, (ii) transcription elongation factors, such as SII/TFIIS, ELL, DSIF (Spt4/5), Elongin, and P-TEFb, which function by varied mechanisms to assist pol II to elongate its transcripts, and (iii) the Mediator complex, which can bind directly to pol II and appears to play important roles at each stage of transcription, from the recruitment of pol II to genes in response to many signals to controlling pol II activity during transcription initiation and elongation. Whereas there is now considerable information concerning the structures and activities of the general initiation and elongation factors, our understanding of the structure and mechanism of action of Mediator is less well-developed, owing in part to its enormous size and complexity.
The Mediator was discovered and first purified to near homogeneity from Saccharomyces cerevisiae as a more than 20 subunit complex required for activator-dependent transcription by pol II and the general initiation factors in vitro [1]. Orthologs of all or most of the S. cerevisiae Mediator subunits are found in higher eukaryotic Mediator complexes. Until recently, it was thought that metazoa and plant Mediator had acquired a set of additional subunits during evolution [2*, 3]; however, results of a recent bioinformatic analysis suggest that orthologs of subunits previously thought to be specific to metazoa and/or plants can be identified in some fungi, amoebae, and/or red algae, leading to the proposal that genes encoding nearly all Mediator subunits emerged very early in eukaryotic evolution, followed by loss or rearrangement of some peripheral subunits in various lineages [4*].
Evidence suggests that Mediator supports activator-dependent transcription in part by acting as a bridge between DNA binding transcription factors and pol II, the general initiation factors, and other components of the transcription apparatus. Mediator binds not only to pol II and its initiation factors, but also to the activation domains (ADs) of many DNA binding transcription factors [1, 5, 6, 7** and references therein]. Through these interactions, Mediator can enhance recruitment of pol II and initiation factors to activator-bound DNA in vitro. Suggesting it performs similar functions in cells, Mediator is detected by whole-genome ChIP at many activator-bound enhancer regions and at promoters, where it colocalizes with pol II [see, for example, 8, 9, 10, 11, 12**].
Mediator complexes from both yeast and higher organisms can also stimulate basal transcription in vitro, independent of DNA binding transcription factors [13, 14, 15]. Together with evidence from genetic experiments in yeast indicating that loss of certain key Mediator subunits affects transcription as severely as loss of Pol II [16], this observation suggests that the Mediator complex as a whole should properly be considered a component of the general transcription machinery. Nevertheless, results of both biochemical and genetic studies suggest that individual Mediator subunits can have remarkably gene-specific, and even tissue-specific functions.
Below we discuss recent findings, which are providing a deeper understanding of the function and regulation of the Mediator complex. We do not attempt a comprehensive review of the literature, but instead describe a collection of recent studies, which highlight various aspects of Mediator and its role in gene regulation.
Multiple Functionally Distinct Forms of Mediator
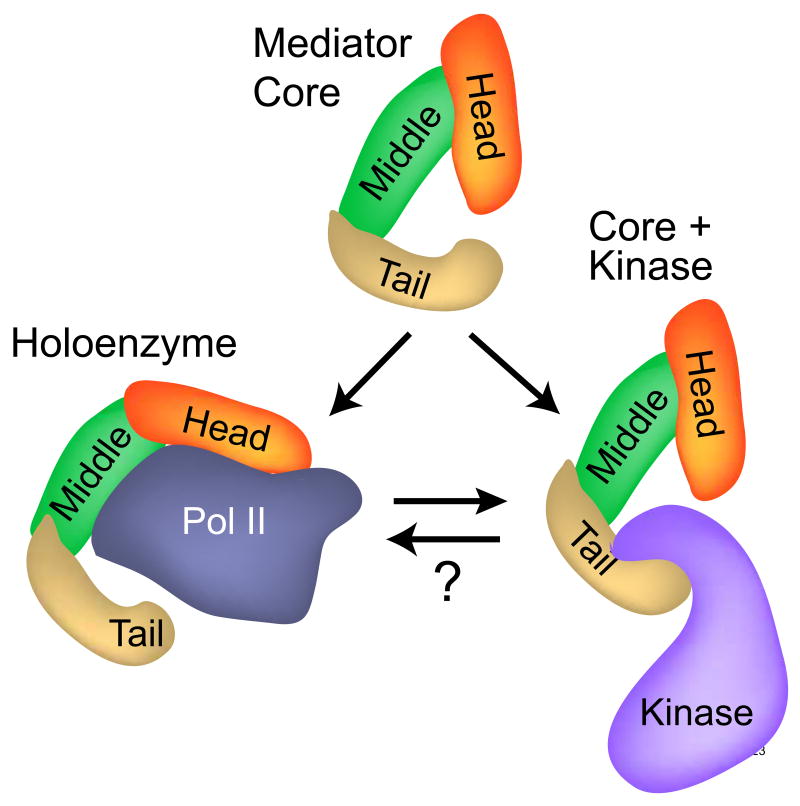
Shortly after the discovery of Mediator, a combination of biochemical, genetic, and EM structural studies revealed that Mediator exists in cells in at least two distinct forms [1, 6, 17]. In its simplest form, Mediator is composed of a set of core subunits organized into three modules referred to as the head, middle, and tail (Figure 1). Core Mediator can bind tightly to pol II to form the so-called holoenzyme; in higher eukaryotes, the holoenzyme is enriched for an additional subunit, Med26. An alternative form of Mediator is free of pol II but includes a fourth module referred to as the kinase module, which contains the cyclin-dependent kinase Cdk8, Cyclin C, and two additional subunits, Med12 and Med13. Intriguingly, higher chordates include multiple isoforms of one or more of the kinase module subunits, raising the possibility that association of core Mediator with alternative kinase modules provides opportunities to fine-tune tissue-, cell, or pathway-specific gene regulation [3, 4].
Fig. 1. Multiple Forms of Mediator.
Biochemical and EM structural studies have revealed that Mediator is present in cells in at least three forms: a Mediator core, which is composed of more than twenty subunits arranged in three modules referred to as head, middle, and tail; a holoenzyme, in which the Mediator core adopts a more open conformation that encircles and binds tightly to pol II; and a third form, in which the kinase module binds to the Mediator core in a manner that precludes Mediator binding to pol II. Understanding molecular mechanisms by which each form of Mediator contributes to transcriptional activation and repression, as well as the biochemical pathways leading to the assembly and possible interconversion of the different forms of Mediator, are key challenges for future investigations of the role Mediator plays in regulating gene expression.
Initial efforts to establish functions of the different forms of Mediator suggested that core Mediator and/or holoenzyme might be responsible for Mediator's activities in transcriptional activation, whereas Mediator associated with the kinase module might serve as a transcriptional repressor. Results of genetic analyses indicated that the kinase module negatively regulates a subset of genes in both budding and fission yeast [18, 19]. Core Mediator and holoenzyme were found to stimulate basal transcription and support activation of transcription in vitro by a variety of DNA binding transcription factors, whereas Mediator complexes containing kinase module did not [20, 21, 22, 23]; in addition, recent studies have shown that core Mediator can be converted to an inactive form by addition of purified kinase module [24]. Consistent with these biochemical observations, a repressive form of the transcription factor C/EBPβ appeared to block transcription of target genes in cells by recruiting Mediator with the kinase module to promoters, while C/EBPβ activated by phosphorylation through the RAS-MAPK pathway recruited Mediator that is free of kinase module and includes Med26 [25]. Similarly, kinase module subunits were found to be displaced from Mediator at a model promoter concomitant with VP16-dependent pol II recruitment and transcriptional activation both in vitro and in cells [26].
Although there is evidence that the kinase module can, in some cases, repress transcription via Cdk8-dependent phosphorylation of DNA binding transcriptional activators or general initiation factors [6], results of a combination of biochemical studies and single particle EM reconstructions indicate that kinase module can also repress transcription independent of Cdk8 kinase activity. In particular, binding of the kinase module to Mediator was found to block assembly of Mediator with pol II, either by occluding the pol II binding site on Mediator or by an allosteric mechanism [24, 27]. Core Mediator adopts a relatively compact structure, in which the middle and tail modules (or the middle module alone in fission yeast, which lacks the tail) are folded against the head in a way that blocks the pol II interaction surface [27, 28, 29]. When assembled with pol II into the holoenzyme, yeast Mediator undergoes a major conformational change to an elongated structure that allows extensive contact of each of the core Mediator modules with pol II [24, 28, 30*, 31, 32, 33]. EM studies performed with fission yeast Mediator suggest that the kinase module can interact with the head and middle modules in a way that locks the core into a closed conformation and, in so doing, sterically blocks the pol II binding site on Mediator [27].
Although it is clear that the kinase module can prevent Mediator binding to pol II and thereby block transcription, a growing body of evidence indicates that the kinase module can function by additional mechanisms not only to repress, but also to activate transcription. One intriguing new mechanism of kinase-independent transcriptional repression by the kinase module comes from the observation that Mediator, through the kinase module subunit Med12, can recruit the histone H3K9 methyltransferase G9a to genes regulated by the DNA binding transcriptional repressor, RE1 silencing transcription factor or REST [34**]. Recruitment of Mediator by REST in turn depends on REST binding to the Mediator subunit Med26, a somewhat surprising finding since Med26 (i) had been generally thought to function specifically in transcriptional activation and (ii) is present in only a small fraction of kinase module-associated Mediator [35].
Examples of roles for the kinase module in transcriptional activation have been accumulating. Cdk8 was found to be required for activation of transcription of a thyroid receptor-regulated gene [36*] and of several p53 target genes including p21 [6, 37*]. Phosphorylation of receptor-activated Smads by Cdk8 promotes recruitment of YAP and subsequent activation of transcription of genes involved in BMP suppression of neural differentiation of mouse ES cells [38*]. Cdk8 is also a positive regulator of transcription of several serum response genes; interestingly, Cdk8 was found to regulate these genes not during transcription initiation, but rather during elongation, where it is needed for recruitment of P-TEFb and BRD4 to transcribing pol II [39**]. Cdk8 has been shown to participate indirectly in activation of β-catenin-dependent transcription by phosphorylating and inactivating E2F1, an inhibitor of β-catenin; notably, the gene encoding Cdk8 was recently shown to be amplified in some colon cancers, with Cdk8 overexpression correlating with de-regulation of β-catenin [40]. Evidence that the kinase module plays a more direct role in transcriptional activation by β-catenin has come from the observations (i) that, in human cells, the β-catenin AD recruits Mediator to genes via interaction with Med12 and (ii) that, in Drosophila, the β-catenin cofactor Pygopus activates transcription by recruiting Mediator in a way that depends on both Med12 and Med13 [41*]. Med12 and/or Med13 have also been shown to interact with or to be required for transcriptional activation by other transcription factors, including Nanog [42], members of the GATA and RUNX families [43*], and yeast Pdr3 [44]. Some of these activities of Med12 and Med13 do not require Cdk8 and Cyclin C [41, 43], indicating that there are phosphorylation-independent functions of the kinase module in transcriptional activation, just as there are in repression.
Complexities Arising in the Targeting of Mediator by Activation Domains
In light of Mediator's crucial role as an intermediary between ADs of DNA binding transcription factors and pol II and the general transcription machinery, a major goal has been to understand, at the molecular level, how ADs recruit and regulate the activities of Mediator. Early studies suggested that ADs might target Mediator via simple binary mechanisms, where a given Mediator subunit would be responsible for Mediator's physical and functional interactions with a particular transcription factor or class of transcription factors. For example, Med25 was found to be a target for the AD of the viral transactivator VP16 [45, 46], while MED15 has been shown to be targeted by the Smad2-Smad4 and Smad3-Smad4 transcriptional activators in response to transforming growth factor-β signaling [47] and by the sterol regulatory element binding protein (SREBP), which has a critical role in lipid homeostasis [48, 49] Similarly, it has long been known that Med1 binds in a ligand-dependent fashion, through its LXXLL motifs, to many nuclear receptors, including peroxisome proliferator-activated receptors (PPARs) and the thyroid, vitamin D, and retinoic acid receptors, and it was proposed that this interaction is critical for optimal recruitment of Mediator to nuclear receptor-regulated genes [50].
Recent studies have provided provocative new twists in our understanding of how ADs target Mediator and suggest that the same or closely related transcription factors can function through different Mediator subunits in different contexts. First, it has become clear that nuclear receptors target Mediator subunits in addition to Med1 and that different Mediator subunits can have dominant roles in regulation of different genes by the same nuclear receptor. For example, the nuclear receptor-like transcription factors Oaf1, Pdr1, and Pdr3 from yeast and NHR-49 from C. elegans have been shown to act through Med15 rather than Med1 [44, 49, 51]. In addition, the glucocorticoid receptor (GR) has been reported to use its ligand-dependent activation domain to target Med1, while its ligand-independent N-terminal activation domain targets Med14; notably, expression of some GR-regulated genes depends on Med14 but not Med1, while expression of others depends on both Med1 and Med14 [52]. Consistent with these observations, the N-terminal activation domain of nuclear receptor PPARγ binds Med14, and Med14 is required for Mediator-dependent activation of some but not all genes regulated by PPARγ [53].
Adding yet another degree of complexity, members of the same transcription factor family can target different Mediator subunits to activate transcription of the same gene, through the same promoter elements, in different cell types. Support for this idea came from the observation that Med23 is essential for expression of the Egr1 gene in mouse ES cells, but is dispensable for Egr1 expression in more differentiated cells. Further investigation revealed that in mouse ES cells, the Ets family member ELK-1 activates Egr1 transcription via binding to Med23. In contrast, in differentiated cells, different, but closely related, Ets family members activate Egr1 expression through interaction with one or more other Mediator subunits [7**].
Future Prospects
Over the past few years, biochemical and genetic studies have greatly improved our understanding of the structure and function of Mediator and have laid a solid foundation for progress in the near term. First, the definition of a complete set of Mediator subunits is enabling reconstitution of Mediator subcomplexes and investigation of their interactions with pol II and the general initiation factors and of their roles in promoting transcription initiation; indeed, the recent development of methods for preparing the seven subunit recombinant Mediator head module in sufficient quantities for high resolution structural studies [30*] gives hope that a view of Mediator at atomic resolution is forthcoming. Second, the discoveries of the diverse roles played by the Cdk8 kinase module in activating and repressing activities of Mediator during both initiation and elongation have paved the way for more in-depth investigations of Mediator function and regulation. It should be possible, in the next few years, to gain an understanding of how Mediator undergoes conversion to forms containing the kinase module or pol II and how the kinase module regulates Mediator activity. Finally, the emerging complexity of the mechanisms underlying the targeting of Mediator by the ADs of transcriptional activators provides an intriguing new story line and suggests that, though the going might be tough, the end result will be a deeper understanding of what is turning out to be a remarkably complex set of mechanisms that eukaryotes have evolved to fine-tune gene expression.
Acknowledgments
Work in the authors' laboratory is supported in part by National Institutes of Health Grant GM41628.
Abbreviations
- mRNA
messenger RNA
- pol II
RNA polymerase II
- AD
activation domain
- TFII
pol II transcription factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Myers LC, Kornberg RD. Mediator of Transcriptional Regulation. Annu Rev Biochem. 2000;69:729–749. doi: 10.1146/annurev.biochem.69.1.729. [DOI] [PubMed] [Google Scholar]
- *2.Backstrom S, Elfving N, Nilsson R, Wingsle G, Bjorklund S. Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]; This paper describes the first purification and characterization of Mediator from plants. The authors' findings illustrate the remarkable evolutionary conservation of plant and animal Mediator complexes.
- 3.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- *4.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides an in-depth bioinformatic comparison of Mediator subunits across species. The author's findings reveal the evolutionary conservation and ancient origin of Mediator in eukaryotes.
- 5.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **7.Balamotis MA, Pennella MA, Stevens JL, Wasylyk B, Belmont AS, Berk AJ. Complexity in transcription control at the activation domain-mediator interface. Sci Signal. 2009;2:ra20. doi: 10.1126/scisignal.1164302. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the intriguing observation that different, but closely related, members of the Ets family of transcription factors regulate activation of expression of the Egr1 gene in different cell types through interactions with different Mediator subunits. The authors' findings bring to light an additional level of complexity of Mediator function in fine-tuning gene expression.
- 8.Andrau JC, van de PL, Lijnzaad P, Bijma T, Koerkamp MG, van de PJ, Werner M, Holstege FC. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell. 2006;22:179–192. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 9.Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell. 2006;22:169–178. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 10.Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci U S A. 2009;106:16734–16739. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **12.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports evidence that Mediator functions together with cohesin to promote activation of transcription of genes in mouse embryonic stem cells. The authors' findings support the model that DNA looping created by the Mediator-cohesin interaction provides the chromosomal architecture required for gene transcription.
- 13.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 14.Mittler G, Kremmer E, Timmers HTh, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/Mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAFIIs. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci U S A. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 19.Samuelsen CO, Baraznenok V, Khorosjutina O, Spahr H, Kieselbach T, Holmberg S, Gustafsson CM. TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution. Proc Natl Acad Sci U S A. 2003;100:6422–6427. doi: 10.1073/pnas.1030497100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PC's. Mol Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 21.Spahr H, Khorosjutina O, Baraznenok V, Linder T, Samuelsen CO, Hermand D, Makela TP, Holmberg S, Gustafsson CM. Mediator influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro. J Biol Chem. 2003;278:51301–51306. doi: 10.1074/jbc.M306750200. [DOI] [PubMed] [Google Scholar]
- 22.Akoulitchev S, Chuikov S, Reinberg D. TFIIH is negatively regulated by cdk8-containing complexes. Nature. 2000;407:102–106. doi: 10.1038/35024111. [DOI] [PubMed] [Google Scholar]
- 23.Taatjes DJ, Naar AM, Andel F, Nogales E, Tjian R. Structure, function, and activator-induced conformations of the CRSP coactivator. Science. 2002;295:1058–1062. doi: 10.1126/science.1065249. [DOI] [PubMed] [Google Scholar]
- 24.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo X, Kowenz-Leutz E, Xu H, Leutz A. Ras induces mediator complex exchange on C/EBPβ. Mol Cell. 2004;13:241–250. doi: 10.1016/s1097-2765(03)00521-5. [DOI] [PubMed] [Google Scholar]
- 26.Uhlmann T, Boeing S, Lehmbacher M, Meisterernst M. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J Biol Chem. 2007;282:2163–2173. doi: 10.1074/jbc.M608451200. [DOI] [PubMed] [Google Scholar]
- 27.Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci U S A. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis JA, Takagi Y, Kornberg RD, Asturias FJ. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 29.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci U S A. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of mediator interactions. Mol Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]; This paper reports the complete reconstitution of a recombinant yeast seven subunit Mediator head module that can interact stably with RNA polymerase II in the presence of the general initiation factor TFIIF. The authors' work provides the foundations for systematic investigation of the function of the Mediator head module and for high resolution structural studies.
- 31.Cai G, Imasaki T, Takagi Y, Asturias FJ. Mediator structural conservation and implications for the regulation mechanism. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator head module structure and functional interactions. Nat Struct Mol Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II and adopts a specific conformation. Genes Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, Boyer TG. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports the discovery that Mediator subunit Med12 orchestrates silencing of neuronal genes in non-neuronal cells by recruiting REST and the G9a methyltransferase to silence those genes. Notably, Med12 mutations found in the X-linked mental retardation (XLMR) disorders FG syndrome and Lujan syndrome interfere with its activity in silencing.
- 35.Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem. 2009;284:2648–2656. doi: 10.1074/jbc.M806514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *36.Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol. 2010;30:2437–2448. doi: 10.1128/MCB.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation [39**]
- *37.Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation [39**]
- *38.Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massague J. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation [39**]
- **39.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]; These four papers by Belakavadi and Fondell [36*], Donner et al [37*], Alarcon et al [38*], and Donner et al [39**] describe diverse mechanisms by which the Cdk8 kinase can participate in transcriptional activation and support the notion that Cdk8 and the Mediator kinase module can function not only in transcriptional repression but also in activation. In addition, Donner et al [39**] and Belakavadi and Fondell [36*] provide evidence for an intriguing link between the transcription elongation factor P-TEFb and the Mediator kinase module.
- 40.Morris EJ, Ji JY, Yang F, Di SL, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *41.Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci U S A. 2008;105:6644–6649. doi: 10.1073/pnas.0709749105. [DOI] [PMC free article] [PubMed] [Google Scholar]; See annotation [43*].
- 42.Tutter AV, Kowalski MP, Baltus GA, Iourgenko V, Labow M, Li E, Kadam S. Role for Med12 in regulation of Nanog and Nanog target genes. J Biol Chem. 2009;284:3709–3718. doi: 10.1074/jbc.M805677200. [DOI] [PubMed] [Google Scholar]
- *43.Gobert V, Osman D, Bras S, Auge B, Boube M, Bourbon HM, Horn T, Boutros M, Haenlin M, Waltzer L. genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/ RUNX-activated transcription in Drosophila. Mol Cell Biol. 2010;30:2837–2848. doi: 10.1128/MCB.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; These two papers by Carerra et al [41*] and Gobert et al [43*] provide evidence that the kinase module can contribute to gene activation via phosphorylation-independent mechanisms.
- 44.Shahi P, Gulshan K, Naar AM, Moye-Rowley WS. Differential roles of transcriptional mediator subunits in regulation of multidrug resistance gene expression in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:2469–2482. doi: 10.1091/mbc.E09-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mittler G, Stühler T, Santolin L, Kremmer E, Lottspeich F, Berti L, Meisterernst M. A novel docking site on Mediator is critical for activation by VP16 in mammalian cells. EMBO J. 2003;22:6494–6504. doi: 10.1093/emboj/cdg619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang F, DeBeaumont R, Zhou S, Naar AM. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci U S A. 2004;101:2339–2344. doi: 10.1073/pnas.0308676100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato Y, Habas R, Katsuyama Y, Naar AM, He X. A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- 48.Yang F, Vought BW, Satterlee JS, Walker AK, Jim Sun ZY, Watts JL, DeBeaumont R, Saito RM, Hyberts SG, Yang S, Macol C, Iyer L, Tjian R, van den HS, Hart AC, Wagner G, Naar AM. An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis. Nature. 2006;442:700–704. doi: 10.1038/nature04942. [DOI] [PubMed] [Google Scholar]
- 49.Taubert S, Hansen M, Van Gilst MR, Cooper SB, Yamamoto KR. The Mediator subunit MDT-15 confers metabolic adaptation to ingested material. PLoS Genet. 2008;4:e1000021. doi: 10.1371/journal.pgen.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- 51.Thakur JK, Arthanari H, Yang F, Chau KH, Wagner G, Naar AM. Mediator subunit Gal11p/MED15 is required for fatty acid-dependent gene activation by yeast transcription factor Oaf1p. J Biol Chem. 2009;284:4422–4428. doi: 10.1074/jbc.M808263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Rogatsky I, Garabedian MJ. MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol Endocrinol. 2006;20:560–572. doi: 10.1210/me.2005-0318. [DOI] [PubMed] [Google Scholar]
- 53.Grontved L, Madsen MS, Boergesen M, Roeder RG, Mandrup S. MED14 tethers mediator to the N-terminal domain of peroxisome proliferator-activated receptor gamma and is required for full transcriptional activity and adipogenesis. Mol Cell Biol. 2010;30:2155–2169. doi: 10.1128/MCB.01238-09. [DOI] [PMC free article] [PubMed] [Google Scholar]