Summary
Adult pancreatic beta cells can replicate during growth and after injury to maintain glucose homeostasis. Here we report that beta cells deficient in Dnmt1, an enzyme that propagates DNA methylation patterns during cell division, were converted to alpha cells. We identified the lineage determination gene aristaless related homeobox (Arx), as methylated and repressed in beta cells, and hypo-methylated and expressed in alpha cells and Dnmt1-deficient beta cells. We show that the methylated region of the Arx locus in beta cells was bound by methyl binding protein MeCP2 which recruited PRMT6, an enzyme that methylates histone H3R2 resulting in repression of Arx. This suggests that propagation of DNA methylation during cell division also ensures recruitment of enzymatic machinery capable of modifying and transmitting histone marks. Our results reveal that propagation of DNA methylation during cell division is essential for repression of alpha cell lineage determination genes to maintain pancreatic beta cell identity.
Introduction
Terminally differentiated pancreatic beta cells retain the capacity to proliferate during growth and after injury to maintain glucose homeostasis (Dor et al., 2004; Georgia and Bhushan, 2004; Teta et al., 2007; Zhong et al., 2007). This suggests that the gene expression pattern of beta cells needs to be propagated with high fidelity to ensure cell identity is maintained after cell division. The gene expression pattern of beta cells is established during development, as the fates of pancreatic progenitor cells are progressively restricted. Genetic studies have identified a number of genes that are important for restricting cell fate choice within the different endocrine lineages. In particular, two homeobox genes, Arx and Pax4 are exclusively expressed in alpha and beta cell lineages respectively. Mice lacking Arx display increased numbers of beta and delta cells at the expense of alpha cells (Collombat et al., 2003). Conversely, mice lacking Pax4 display reduced beta cells and increased in alpha cell numbers (Sosa-Pineda et al., 1997). Gain of function experiments show that forced misexpression of Pax4 or Arx in alpha and beta cells respectively, can lead to cell fate conversion (Collombat et al., 2007; Collombat et al., 2009). Furthermore, loss of another homeobox gene, Nkx2.2 results in increased epsilon cell mass at the expense of beta cells (Prado et al., 2004). These studies suggest that repression of lineage determination genes plays a prominent role in establishing cell fate during development. However, the mechanisms responsible for the stable propagation of the repressed state of these key lineage determination genes during cell division are not well understood.
It is generally believed that the assembly of a specific chromatin structure that can be propagated through DNA replication and cell division accompanies heritable gene repression. How lineage-specific transcription factor networks interact with different epigenetic marking systems to achieve stability of cell identity is not clear. One mechanism that ensures stable inheritance of repressed genes involves covalent modification of DNA by methyl groups. DNA methylation patterns are faithfully reproduced during cell division by DNA methyltranferase, Dnmt1, which recognizes hemi-methylated DNA to restore the symmetrical CpG methylation pattern, (reviewed in, (Goll and Bestor, 2005; Klose and Bird, 2006; Miranda and Jones, 2007)). However, the precise function of CpG methylation in gene repression has not been firmly established.
Here we report that pancreatic beta cells deficient in Dnmt1 were reprogrammed to alpha cells. Using genome-wide analysis we identified Arx to be methylated and repressed in beta cells but hypomethylated and expressed in Dnmt1 deficient beta cells. Methyl-specific binding proteins that recruit enzymatic machinery capable of locally altering histone modification bound the methylated region of Arx locus. Our results suggest that propagation of DNA methylation pattern forms the backbone for transmitting histone modifications and the assembly of Arx repressive chromatin structure that can be stably inherited through cell division to preserve beta cell identity.
Results
Deletion of Dnmt1 converts beta cells into alpha cells
To investigate the requirement for maintaining DNA methylation patterns during pancreatic beta cell replication, we crossed mice transgenic for Cre recombinase under the control of rat insulin promoter (RIP-cre) with DNA Methyltransferase (Dnmt1)fl/fl mice (Herrera, 2000; Jackson-Grusby et al., 2001) to selectively inactivate Dnmt1 in beta cells. Immunohistology confirmed cre recombinase activity specifically in beta cells (Figure S1A) and the absence of detectable Dnmt1 protein within the islets from pancreatic sections from RIP-Cre: Dnmt1fl/fl mice (denoted RC:Dnmt1fl/fl, conditional Dnmt1 mutant), while abundant Dnmt1 protein was observed within the islets of pancreatic tissue from Dnmt1fl/fl littermates (Figure 1A; Figure S1B). Staining of pancreatic sections from conditional Dnmt1 mutant mice with 5-methylcytosine antibody at different ages showed that islets from 3 month old pancreas had relative uniform staining of 5-methylcytosine, however islets from 8 months pancreas displayed patchy 5-methylcytosine staining (Figure S1C). Quantification of DNA methylation confirmed decrease in global DNA methylation in 8 month old islets isolated compared to isolated islets from 6 week old conditional Dnmt1 mutant mice, while no changes in global DNA methylation was observed in isolated islets from similarly aged control mice (Figure S1D). Beta cells have been shown to replicate in a homogenous fashion and as the animal ages, a greater proportion of beta cells within the islet would derive from replicating beta cells. Thus, the absence of Dnmt1 resulted in a passive loss of DNA methylation correlated with the rate of beta cell replication (Brennand et al., 2007; Teta et al., 2007) resulting in a gradual increase in the numbers of beta cells that lost cytosine methylation, as greater numbers of beta cell divide as the animal ages.
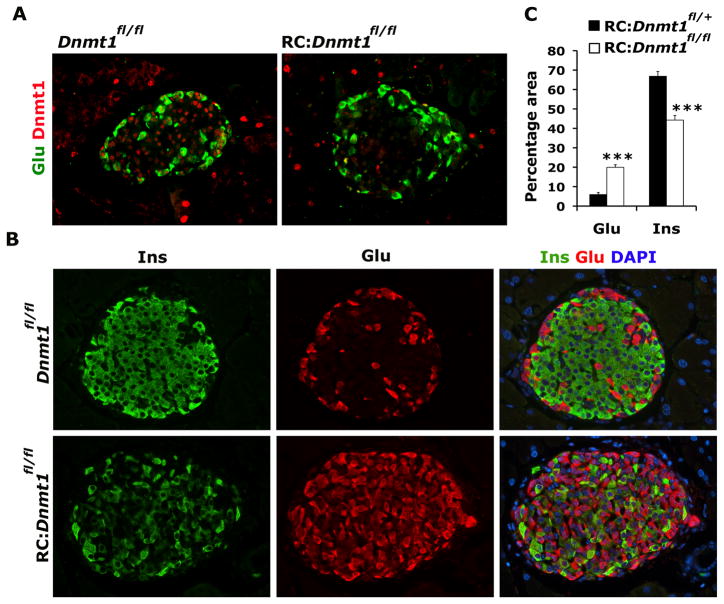
Figure 1. Loss of DNA methylation in beta cells results in an increase in the number of glucagon expressing cells.
(A) Representative pancreatic sections from 3 months old Dnmt1fl/fl (control) and RIP-Cre:Dnmt1fl/fl (RC:Dnmt1fl/fl) littermates were immunostained for glucagon (Glu; green) and Dnmt1 (red). (B) Immunostaining of representative pancreatic sections from 8 months old Dnmt1fl/fl and RC:Dnmt1fl/fl littermates showing insulin (green), glucagon (red) and overlay with DAPI (to counter-stain the nuclei; blue). (C) Quantification of glucagon and insulin area in 8 months old RIP-Cre:Dnmt1fl/+ (control) and RIP-Cre:Dnmt1fl/fl animals, shown as percentage of total islet area. N=4 animals. The error bars represent standard error (SEM) of the mean. See also Figure S1.
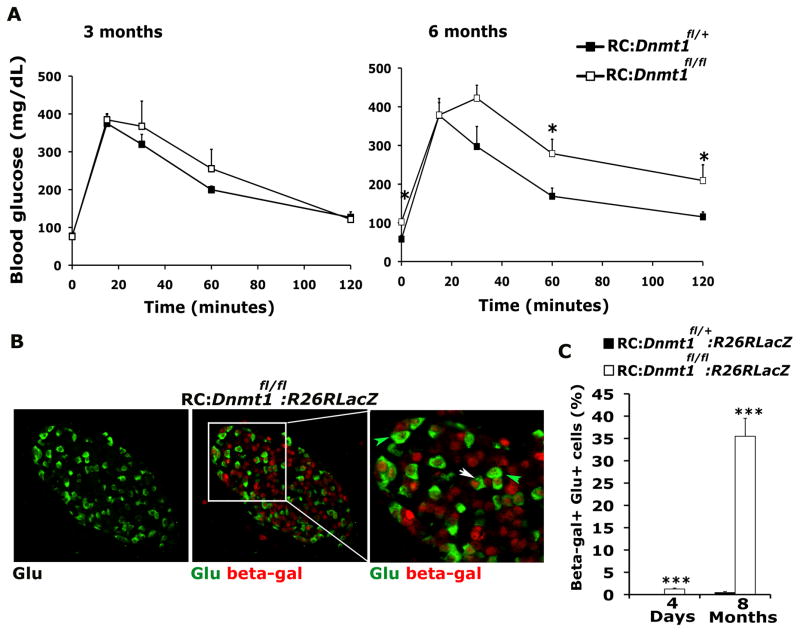
Pancreatic tissue isolated from mice at different ages was immunostained for insulin and glucagon, endocrine hormones that mark beta and alpha cells, respectively. Islets from wild-type and conditional Dnmt1 mutant littermates at birth did not show any differences and displayed a characteristic distribution of endocrine cells with beta cells forming the core and alpha cells at the periphery forming the mantle. Even though Dnmt1 is deleted in beta cells in young conditional Dnmt1 mutant mice (Figure S1B), DNA methylation is not significantly different in these beta cells (Figure S1C, D), consistent with the absence of an overt phenotype observed in the young conditional Dnmt1 mutant mice. However, with increasing age (and increased beta cell duplications along with passive demethylation), significant differences became apparent in conditional Dnmt1 mutants, which displayed reduced DNA methylation, increased number of glucagon-expressing cells throughout the islet while the number of insulin-expressing cells were diminished (Figure 1B; Figure S1C, D, E). Quantification of insulin and glucagon area in islets confirmed the increase in glucagon-expressing cells in conditional Dnmt1 mutants (Figure 1C). Glucose tolerance tests showed that with increasing age the conditional Dnmt1 mutants mice displayed abnormal glucose homeostasis, consistent with the reduction in beta cells (Figure 2A). To assess whether these excessive glucagon-expressing cells originated from beta cells, cells transcribing the insulin gene were heritably labeled using the R26R-LacZ, or R26R-YFP reporters to reveal that a number of glucagon-expressing cells observed in the conditional Dnmt1 mutant pancreas indeed derived from cells that had transcribed the insulin gene (Figure 2B; Figure S2A, B). The number of glucagon-expressing cells that also displayed enzymatic beta-galactosidase activity increased dramatically in older conditional Dnmt1 mutants (Figure 2C).
Figure 2. DNA methylation is required for maintenance of beta cell identity.
(A) Glucose tolerance test (GTT) for RC: Dnmt1fl/+ (control) and RC:Dnmt1fl/fl animals at ages 3 months (left panel) and 6 months (right panel) (n=5 for each genotype). (B) Representative pancreatic section from an 8 months old RC:Dnmt1fl/fl:R26RLacZ animal showing immunostaining for glucagon (green) in the left panel and an overlay of glucagon with beta galactosidase (b-gal; red), in the middle panel. The inset marks a representative area, magnified and shown in right panel. A number of beta-galactosidase and glucagon double labeled cells (white arrows) are evident in these RC:Dnmt1fl/fl:R26RLacZ animals. Green arrows indicate normal alpha cells, which express glucagon and do not stain for beta galactosidase. (C) Quantification of endocrine cells co-staining for glucagon and RIP-Cre driven beta-galactosidase in RC: Dnmt1fl/+:R26RLacZ (control) and RC:Dnmt1fl/fl:R26RLacZ animals at 4 days and 8 months of age. N=3 animals per group. The error bars represent standard error (SEM) of the mean. See also Figure S2.
Dnmt1 propagates DNA methylation of alpha cell lineage determinant Arx
We hypothesized that key determinants of the alpha cell lineage were methylated and repressed to maintain beta cell lineage stability and these determinants were de-repressed in the beta cells deficient in DNA methylation. To identify such methylated alpha cell lineage determinants, we carried out genome-wide mapping of DNA methylation patterns at 5’upstream promoter regions using the methylated DNA immunoprecipitation (MeDIP) method (Weber et al., 2005). Methylated CpG-enriched DNA fractions using anti-5-methyl cytosine antibody were collected from mouse alpha and beta cell lines (α-TC1 and Min6) and hybridized to high-resolution promoter microarrays. We screened for 5’upstream regulatory regions that showed higher methylation levels in Min6 cells when compared with α-TC1 cells. Analysis of the methylation profiles of endocrine cell fate determination genes led to the identification of two CpG-rich regions in the 5’regulatory region of Arx that were differentially methylated in Min6 and α-TC1 cells (Figure S3A). Overexpression of Arx in Min6 cells was sufficient to repress beta cell markers and induce alpha cell markers suggesting that Arx plays a crucial role in alpha cell fate (Figure S3B, C). This is consistent with previous studies showing that Arx is not expressed in the beta cells and mis-expression of Arx in beta cells can re-specify them to an alpha cell fate (Collombat et al., 2005; Collombat et al., 2007; Collombat et al., 2003). This suggested that methylation of Arx regulatory regions in beta cells may correlate to its repression in beta cells.
The Arx regulatory region is characterized by several CpG-rich sites; we focused on two of these, one within the proximal promoter and close to the transcription start site (TSS) referred to as UR1 and another one 2Kb upstream of TSS, termed UR2. Detailed characterization of the Arx upstream regulatory region methylation profiles by bisulfite sequencing analysis in FACS purified alpha and beta cells revealed that a vast majority of CpG dinucleotides in UR2 region were methylated in beta cells but unmethylated in alpha cells, while no difference in DNA methylation was observed in UR1 region (Figure 3A; Figure S3D). Quantification of DNA methylation of the UR2 region revealed significantly reduced DNA methylation in isolated DNA from sorted alpha cells compared to sorted beta cells (Figure 3B). A region 2.5 kb upstream of TSS, named UR3, was also identified in the MeDIP screens along with UR2 and showed differential methylation patterns similar to the UR2 region (Figure S3E, F). However, no significant differences in DNA methylation were observed in the 3’ conserved enhancer region identified in previous studies (Figure S3G), suggesting that regulation of Arx expression by this downstream enhancer region is not dependent on DNA methylation (Collombat et al., 2005). Furthermore, bisulfite analysis of a CpG-rich region +1292 to +1400 bp region within intron 1 showed no significant differences in alpha and beta cells (Figure S3H).
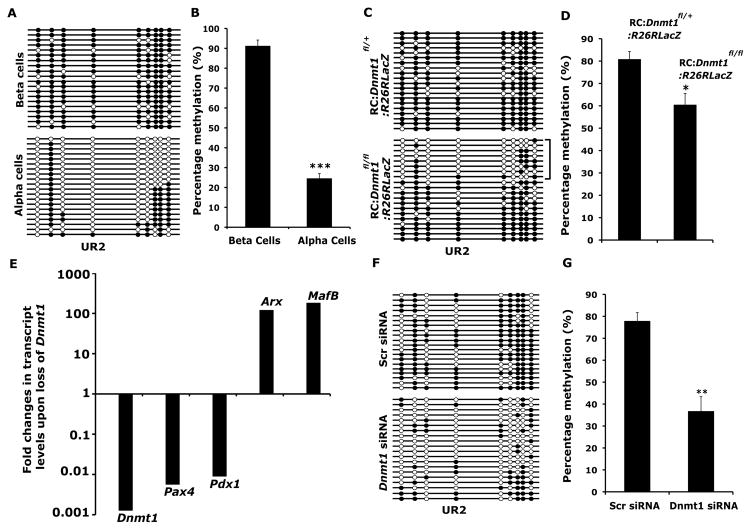
Figure 3. Dnmt1 methylates and leads to the repression of Arx locus in beta cells.
(A) Bisulfite sequencing analysis and (B) quantification of the bisulfite sequencing data, shown as percentage DNA methylation (N=3) for the CpG rich UR2 region of Arx locus (−2103 to −1992 bp) in isolated beta and alpha cells. (C) Bisulfite sequencing analysis and (D) quantification of the bisulfite sequencing data, shown as percentage DNA methylation (N=3), for the UR2 region of Arx locus in cells marked with beta-galactosidase activity from RIP-Cre:Dnmt1fl/fl:R26RLacZ (RC:Dnmt1fl/fl:R26RLacZ) animals. Littermate RC:Dnmt1fl/+:R26RLacZ mice were used as controls. Each horizontal line with dots is an independent clone and 20 clones are shown here. The UR2 region is almost fully DNA methylated (filled circles) in beta cells, but largely hypomethylated (open circles) in alpha cells. Cells isolated using beta-galactosidase activity from RC:Dnmt1fl/fl:R26RLacZ animals showed a subset of clones that were hypomethylated, as marked by the bracket. (E) Fold changes in the levels of Dnmt1, Pax4, Pdx1, MafB and Arx transcripts determined by real-time RT-PCR in the cells marked with beta-galactosidase activity from RC:Dnmt1fl/fl:R26RLacZ and control littermates, with the levels in control set as one arbitrary unit. The levels are depicted on a logarithmic scale. (F) Bisulfite sequencing analysis and (G) quantification of the bisulfite sequencing data, shown as percentage DNA methylation (N=3), for the UR2 region of Arx promoter from Min6 cells treated with control, scrambled (Scr) or Dnmt1 siRNAs. The error bars represent standard error (SEM) of the mean. See also Figure S3.
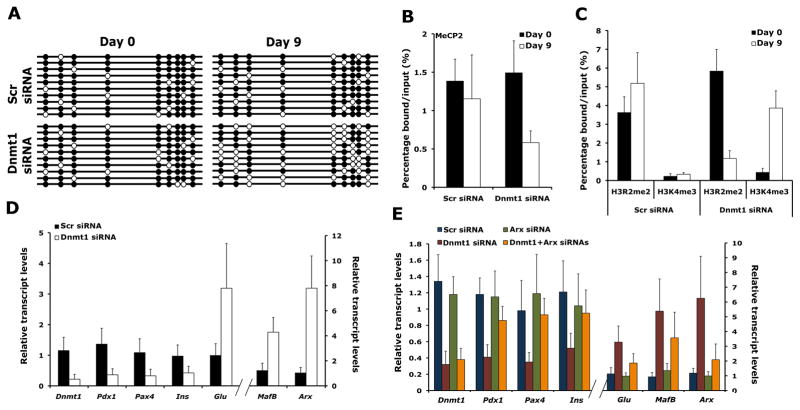
To investigate whether beta cells deficient in Dnmt1 displayed changes in DNA methylation at these CpG-rich regions of the Arx locus, we used the R26R-LacZ reporter to isolate heritably labeled cells that transcribed the insulin gene, using beta-galactosidase activity to cleave a fluorescent substrate, followed by FACS. Bisulfite sequencing analysis on beta-galactosidase positive cells isolated from control mice showed a heavily methylated pattern in the UR2 region similar to the pattern observed in purified beta cells. In contrast, beta-galactosidase positive cells isolated from conditional Dnmt1 mutant littermates revealed two distinct patterns of methylation on the UR2 region of the Arx gene, indicating clonal heterogeneity. One set of clones was methylated, characteristic of beta cells, while the other set of clones was predominately unmethylated and characteristic of alpha cells (Figure 3C). The heterogeneity is consistent with the fact that isolated cells from Dnmt1 deficient mice would be a mixed population consisting not only of undivided beta cells that retained genomic patterns of cytosine methylation ,but also the progeny of divided cells that lost cytosine methylation. These results indicate that Arx gene was unmethylated in the progeny of divided beta cells of conditional Dnmt1 mutant mice (Figure 3D). In addition, isolated beta cells from conditional Dnmt1 mutant mice showed massive increase in the expression of Arx compared to beta cells isolated from wildtype littermates, along with another alpha cell marker MafB and significant reduction in the levels of beta cell markers Pdx1 and Pax4 (Figure 3E). To determine whether DNA methylation was directly correlated with Arx repression, we performed knockdown of Dnmt1 in Min6 cells using specific siRNA. Bisulfite sequencing analysis of Dnmt1 deficient Min6 cells showed that loss of Dnmt1 resulted in loss of methylation at the UR2 region of Arx, resulting in a methylation profile similar to purified alpha cells (Figure 3F,G). Furthermore, Dnmt1 deficient Min6 cells displayed increase in the expression of Arx (Figure S3I). These experiments indicate that repression of Arx in beta cells correlates with methylation of the Arx promoter. In the absence of Dnmt1, the loss of Arx promoter methylation results in de-repression of the Arx locus.
Recruitment of PRMT6 at methylated region of Arx facilitates repression
We next sought to establish the mechanism by which DNA methylation facilitated the transcriptional repression of the Arx locus, by screening for methyl-DNA binding (MDB) proteins that bound the UR2 region of the Arx locus in beta cells. Chromatin immunoprecipitation (ChIP) analysis showed that one member of the methyl binding proteins family (Bird, 2002), MeCP2 was bound to the UR2 region of the Arx promoter in beta cells but not in alpha cells (Figure 4A). Binding of MeCP2 to the UR2 regions of Arx locus was also significantly reduced in sorted beta-galactosidase positive cells isolated from conditional Dnmt1 mutant mice (Figure S4A). In addition to the MDB domain, MeCP2 has a transcriptional repressive domain that recruits enzymatic machinery capable of altering histone modification and regulating chromatin structure (Jones et al., 1998; Nan et al., 1998). To identify the binding partners of MeCP2, we transfected a Flag-tagged MeCP2 into Min6 cells and used an anti-Flag antibody to precipitate MeCP2-associated proteins. This analysis revealed several specific bands that co-immunoprecipitated with MeCP2, including one around 42 kDa (Figure S4B). Mass spectrometry analysis of this band identified PRMT6 (protein arginine methyltransferase 6) to be one of the proteins interacting with MeCP2, among others. PRMT6 resides predominantly in the nucleus and acts as a H3R2 methyltransferase and represses transcription by counteracting H3K4 trimethylation (Hyllus et al., 2007). To confirm the specificity of the above interaction, immunoprecipitates of Flag-tagged MeCP2 transfected Min6 cells subjected to Western blotting using the PRMT6 antibody revealed that PRMT6 co-precipitated with MeCP2 (Figure 4B). These data were verified by immuno-precipitations using a myc-tagged PRMT6 over-expression construct to complex with MeCP2 (Figure 4C).
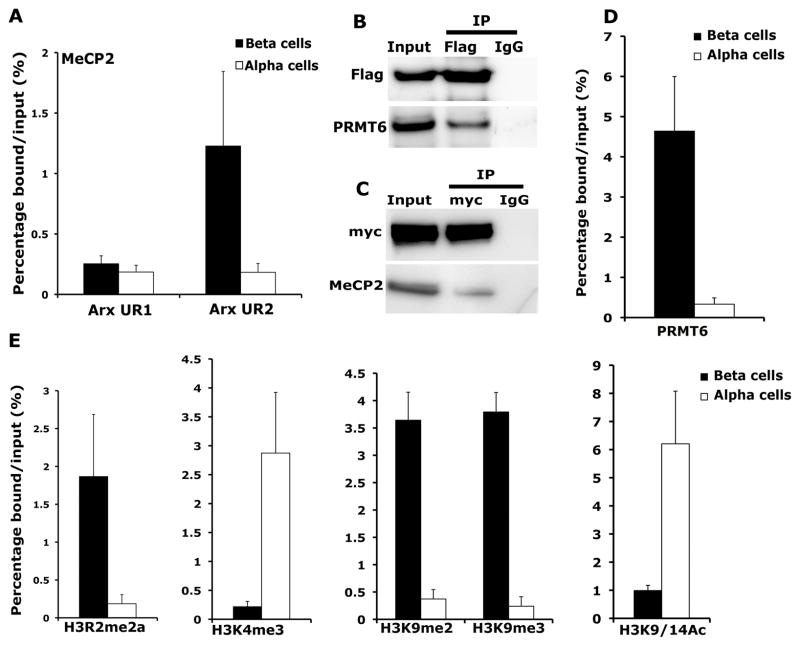
Figure 4. MeCP2 binds to the Arx locus in beta cells and recruits H3R2 methyltransferase PRMT6 to repress Arx expression.
(A) Chromatin immunoprecipitation (ChIP) analysis showing the binding of MeCP2 to the two CpG rich regions, UR2 (−2111 to −1960) and UR1 (−184 to −254) of the Arx locus in beta and alpha cells. (B) Co-immunoprecipitation analysis examining the interaction of MeCP2 and PRMT6. Cell extracts from Min6 cells transfected with Flag-MeCP2 construct were used as input for immunoprecipitation (IP) with anti-Flag antibody and analyzed by immunoblotting with Flag and PRMT6 antibodies. (C) Co-immunoprecipitation analysis for the interaction of PRMT6 with MeCP2 using immunoprecipitation with myc-tag antibody on Min6 cells transfected with myc-PRMT6 construct, and Western blotting with myc-tag and MeCP2 antibodies. (D) ChIP analysis comparing the recruitment of PRMT6 to the UR2 region of Arx locus in beta and alpha cells. (E) ChIP analysis comparing the levels of asymmetric H3R2 di-methylation (H3R2me2a), H3K4 trimethylation (H3K4me3), H3K9 di- and tri-methylation (H3K9me2 and H3K9me3) and H3K9/14 acetylation (H3K9/14Ac) at the differentially methylated UR2 region of the Arx locus in beta and alpha cells purified from 5 months old wildtype mice. These analyses indicate enrichment of histone modification associated with repression of gene expression, namely, H3R2me2a, H3K9me2 and H3K9me3 at the Arx locus in pancreatic beta cells. The error bars represent standard error (SEM) of the mean. See also Figure S4.
To further analyze whether PRMT6 bound to the methylated UR2 region of the Arx promoter, we measured the binding of PRMT6 by ChIP assays. ChIP analysis showed that PRMT6 was bound to the Arx locus in beta cells where the UR2 region is methylated, while no binding of PRMT6 to the Arx locus was observed in alpha cells where the UR2 region is unmethylated (Figure 4D). The UR3 region, which is differentially methylated in beta cells as well, is also specifically bound by MeCP2 and PRMT6 (Figure S4C), while the downstream regions that were not differentially methylated were not bound by these proteins (Figure S4D). Taken together, these data indicate that a complex containing MeCP2 and PRMT6 is recruited to the methylated regions of the Arx promoter and could play a role in the assembly and propagation of repressive chromatin structure of Arx locus.
We then assessed whether recruitment of PRMT6 to the methylated UR2 region of the Arx locus could play a role in the assembly and propagation of repressive chromatin state of Arx affected histone modification at the Arx locus. We first measured levels of asymmetric di-methylation of H3R2, (H3R2me2a) at the Arx promoter in FACS purified beta and alpha cells. ChIP analysis using anti-H3R2me2a antibody showed high levels of enrichment at the Arx promoter in beta cells and low binding in alpha cells (Figure 4E). As recent studies have shown that H3K4 methylation is antagonized by PRMT6-mediated H3R2me2a (Guccione et al., 2007; Hyllus et al., 2007; Kirmizis et al., 2007), we sought to determine the methylation levels of H3K4 by ChIP analysis using an anti-H3K4me3 antibody. These experiments revealed high levels of H3K4me3 at the Arx locus in alpha cells, while H3K4 was unmethylated at the Arx promoter in beta cells (Figure 4E). Sorted beta-galactosidase positive cells isolated from conditional Dnmt1 mutant mice displayed high levels of H3K4me3 and low levels of H3R2me2a similar to sorted alpha cells (Figure S4E). Analysis of the levels of H3K9 di- and tri-methylation (H3K9me2 and H3K9me3) at the Arx locus by ChIP showed high levels of these two histone modifications in beta cells, confirming a repressed chromatin state of Arx locus (Figure 4E). ChIP analysis using anti-H3K9/14ac and HDAC1 antibodies showed very low levels of histone acetylation at the Arx locus (Figure 4E), which correlated with recruitment of HDAC1 to the Arx locus in beta cells compared to alpha cells (Figure S4F).
DNA methylation propagates the transcriptional repression of Arx during beta cell replication
To test whether DNA methylation is involved in maintaining Arx chromatin structure for many generations of actively dividing cells, we treated Min6 cells with Dnmt1 siRNA to assess cell numbers, DNA methylation, and histone modifications on Arx locus. Bisulfite sequencing analyses showed progressive loss of methylation within the UR2 region of the Arx locus with increase in cell duplications (Figure 5A; Figure S5A). ChIP analysis revealed that decreased methylation within the UR2 region of Arx locus upon loss of Dnmt1 correlated with reduced binding of MeCP2 (Figure 5B). We also observed a reduction in H3R2me2a levels and an increase in H3K4me3 levels suggesting that histone modifications profile of the Arx locus was altered (Figure 5C). Furthermore, reduced levels of H3K9me2 and H3K9me3 and increased levels of H3K9Ac confirmed the open chromatin structure of Arx locus in the absence of Dnmt1 (Figure S5B). These results suggest that loss of DNA methylation over several cell divisions can lead to de-repression of the Arx locus. We then asked whether changes in Arx chromatin structure that resulted in de-repression of the Arx locus led to changes in gene expression profile of the cell. Expression analysis of a number of endocrine markers by quantitative PCR analysis revealed that Dnmt1 siRNA treated Min6 cells after several cell divisions had reduced levels of beta cells markers Pdx1, Pax4 and Insulin. In contrast, the expression of alpha cell markers, Arx, Mafb and Glucagon was increased suggesting that Min6 cells no longer retained resemblance to beta cells but instead expressed markers characteristic of alpha cells (Figure 5D). A combined knockdown of Dnmt1 and Arx in Min6 cells, however, did not result in any reduction in beta cell markers or lead to induction of glucagon expression, indicating that de-repression of Arx is critical to the cell fate switch upon loss of Dnmt1 in beta cells (Figure 5E).
Figure 5. Loss of DNA methylation affects the inheritance of the repressive histone modifications on the Arx locus in successive cell generations.
Min6 cells transfected with scrambled or Dnmt1 siRNAs were grown in the exponential phase for 9 days in culture before cells were harvested. (A) Bisulfite sequencing of the UR2 region of Arx locus in Min6 cells transfected with Dnmt1 siRNA, harvested at day 0 and day 9. The UR2 region of the Arx locus that is almost fully DNA methylated (filled circles) at the start of the experiment and is hypomethylated (open circles) after 9 days of exponential growth. (B, C) ChIP analyses in Min6 cells transfected with scrambled or Dnmt1 siRNAs, 0 and 9 after transfection. The binding of MeCP2 to the UR2 region of Arx locus was reduced in Dnmt1 siRNAs transfected Min6 cells at D9 (B). The levels of H3R2me2a were reduced and H3K4me3 were increased in Dnmt1 siRNAs transfected Min6 cells at D9 (C). (D) Real-time RT-PCR analyses comparing the transcript levels of Dnmt1, Pdx1, Pax4, Insulin (Ins), Glucagon (Glu), MafB and Arx in Min6 cells after 0 and 9 days of transfection with Dnmt1 or control, scrambled (Scr) siRNAs. (E) Transcript levels of Dnmt1, Pdx1, Pax4, Insulin (Ins), Glucagon (Glu), MafB and Arx determined by real-time RT-PCRs in Min6 cells, after 9 days of transfection with scrambled (Scr), Dnmt1, Arx or Dnmt1+Arx siRNAs, showing a requirement for Arx in cell fate conversion upon loss of Dnmt1. n=3 for each experiment. The error bars represent standard error (SEM) of the mean. See also Figure S5.
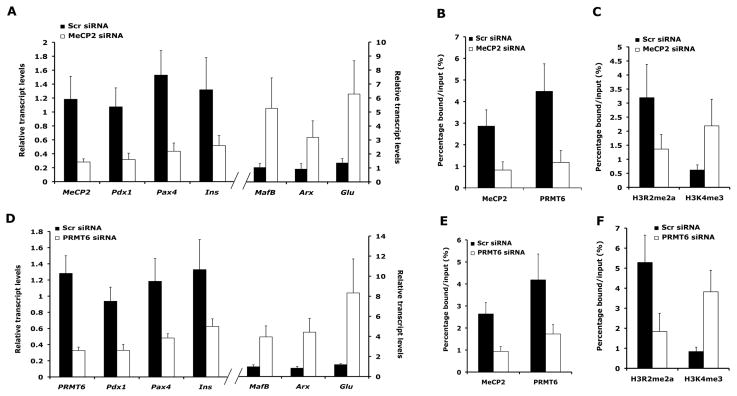
Our analysis suggested that a complex containing MeCP2 and PRMT6 was recruited to the methylated regions of Arx locus to repress its expression in beta cells. To test whether loss of MeCP2 or PRMT6 would mimic loss of Dnmt1, we performed loss-of-function studies for MeCP2 and Prmt6 in Min6 cells using specific siRNAs. Loss of MeCP2 in Min6 cells resulted in de-repression of the Arx locus, with concomitant up-regulation of other alpha cell markers glucagon (Glu) and MafB, and a reduction in the levels of beta cell markers, such as Pdx1, Pax4 and Insulin (Ins) (Figure 6A). These changes corresponded with reduced binding of MeCP2 and PRMT6 to the Arx locus, which also reflected in reduced levels of H3R2me2a and increased levels of H3K4me3 (Figure 6B, C). Similarly, loss of Prmt6 resulted in expression of Arx in Min6 cells, accompanied by induction of other alpha cell markers and loss of beta cell markers (Figure 6D). This was accompanied by loss of PRMT6 and MeCP2 binding to the Arx locus, leading to reduced H3R2me2a and increased H3K4me3 enrichment (Figure 6E, F). Thus, loss of MeCP2 and Prmt6 in Min6 cells was able to phenocopy the loss of Dnmt1, highlighting the functional importance of DNA methylation in regulation of Arx. This requirement for DNA methylation was further confirmed by treating Min6 cells with 5-aza-dC, a chemical which leads to DNA demethylation. Loss of DNA methylation due to 5-aza-dC treatment also resulted in loss of MeCP2 and PRMT6 binding at the Arx locus, concomitant with the induction of Arx expression, along with other alpha cell markers Glucagon and MafB, and reduction in the expression of beta cell markers, such as Pdx1, Pax4, Insulin (Figure S6A–D). This suggests that DNA methylation can influence the covalent histone modification patterns to perpetuate a repressed chromatin state of the Arx locus and maintain beta cell identity. Taken together, these results indicate that DNA methylation plays an active role in the assembly of chromatin structure to represses transcription of alpha cell determination gene Arx to maintain beta cell identity.
Figure 6. Effect of loss of MeCP2 and PRMT6 mimics on Arx expression the loss of DNA methylation.
(A) Transcript levels of MeCP2, Pdx1, Pax4, Insulin (Ins), Glucagon (Glu), MafB and Arx determined by real-time RT-PCRs, (B) binding of MeCP2 and PRMT6 to the UR2 region of Arx locus and (C) levels of H3R2me2a and H3K4me3 at the UR2 region of the Arx locus in Min6 cells, after 9 days of transfection with scrambled (Scr) and MeCP2 siRNAs. (D) Transcript levels of Prmt6, Pdx1, Pax4, Insulin (Ins), Glucagon (Glu), MafB and Arx determined by real-time RT-PCRs, (E) binding of MeCP2 and PRMT6 to the UR2 region of Arx locus and (F) levels of H3R2me2a and H3K4me3 at the UR2 region of the Arx locus in Min6 cells, after 9 days of transfection with scrambled (Scr) and Prmt6 siRNAs. See also Figure S6.
Discussion
It has been generally assumed that the identity of adult differentiated cells is fixed, although there is emerging evidence that adult cells can be manipulated to change identity (Takahashi and Yamanaka, 2006; Vierbuchen et al., 2010; Zhou et al., 2008) or regain plasticity (Collombat et al., 2009; Thorel et al., 2010). Understanding the mechanisms that preserve cell identity would enable successful manipulation of cell plasticity in clinical settings. Our work reveals that Arx, a key determination gene for alpha cells is methylated and repressed in pancreatic beta cells. Dnmt1 propagates this methylation during cell division to ensure repression of Arx in the progeny and preservation of beta cell identity. Deletion of Dnmt1 in beta cells results in the de-repression of Arx and a gradual conversion to alpha cell-like identity. There are several reasons why the switch in cell identity occurs gradually. First, Dnmt1 primarily maintains methylation patterns during cell division and the effects of Dnmt1 deletion are only apparent in the progeny of cells that lack Dnmt1. Deletion of Dnmt1 leads to passive demethylation that is linked to the rates of beta cell replication. BrdU-labeling studies demonstrate that all beta cells are uniformly capable of cell division and newly divided beta cells are prevented from reentering the cell cycle after cell division (Brennand et al., 2007; Teta et al., 2007). Secondly, beta cell replication declines with age, resulting in fewer beta cell undergoing successive rounds of cell division (Dhawan et al., 2009; Rankin and Kushner, 2009; Tschen et al., 2009).Thirdly, cells displaying co-staining of insulin and glucagon are observed in mice with Dnmt1 deletion in beta cells, and could correspond to cells displaying intermediate stages in the cell identity switch. This implies that the switch in between beta and alpha cell-like identity is not immediate but gradual and is reminiscent of somatic cell reprogramming during which cells in different stages of reprogramming are observed (Hanna et al., 2009). All of these factors can contribute to the slow kinetics of cell conversion observed in mice that lack Dnmt1 in beta cells.
It is noteworthy that loss of Dnmt1 in pancreatic beta cells did not lead to large-scale de-repression of silent tissue specific genes. Our results suggests that DNA methylation regulated repression of a small set of genes involved in lineage specification of cell types that are closely related in developmental history. It is likely that a larger set of genes is regulated by DNA methylation but only genes that have appropriate transcriptional activator present will be derepressed in Dnmt1 deleted cells. Studies in ES cells also show that DNA methylation regulation of a single transcription factor plays a role in the separation of embryonic and trophoblast lineages. Elf5 is hypomethylated and expressed in trophoblast cells but DNA methylation stably represses Elf5 in the embryonic lineage. Deletion of Dnmt1 allows for efficient differentiation of methylation-deficient ES cells into the trophoblast lineage (Ng et al., 2008). Other studies also suggest that methylation of a lineage determination gene regulates astrocyte differentiation in the developing brain and T cell development, suggesting that regulation of a limited number of genes is likely to be a general feature of DNA methylation-dependent gene repression (Lee et al., 2001; Takizawa et al., 2001).
Our study highlights the crucial role of DNA methylation in the epigenetic inheritance of a cell differentiation state. Our work here provides a molecular link by which gene repression by DNA methylation is accompanied by the alterations in histone modifications and the assembly of a specific chromatin structure that can be stably inherited through cell division to preserve cell identity. Several studies suggest that methylated CpG binding proteins recruit enzymatic machinery capable of locally altering histone modifications, can maintain chromatin structure through many cell cycles. Histone modifiers such as HDACs and Sin3A associate with MeCP2, potentially linking DNA methylation dependent silencing with changes in chromatin modifications (Jones et al., 1998; Nan et al., 1998). In this study, we have identified PRMT6, an H3R2 methyltransferase, as a interaction partner of MeCP2, which results in CpG methylation directed asymmetric di-methylation of H3R2, a histone modification associated with repression of transcription (Guccione et al., 2007; Hyllus et al., 2007). Our data establishes a direct correlation between DNA methylation and establishment of a repressive chromatin structure dependent on DNA methylation. This supports a model in which propagation of methylation pattern DNA forms the backbone for transmitting histone modifications that regulate the assembly of repressive chromatin structure to the next cell generation (Martin and Zhang, 2007). Finally our results show that inheritance of epigenetic information can regulate pancreatic endocrine cell identity, suggesting that epigenetic reprogramming of cell types with shared developmental history could be an effective strategy for pancreatic beta cell replacement therapies for diabetes. Taken together, this study can form a basis for how inheritance of epigenetic information during cell division can be exploited for cell replacement therapies.
Experimental procedures
Animals, metabolic testing and cell lines
We used Cre/loxP system to conditionally delete Dnmt1 in pancreatic beta cells. Dnmt1fl/fl mice with loxP sites flanking exons 4 and 5 of the Dnmt1 gene and RIP-Cre mice expressing Cre-recombinase from rat insulin promoter have been described previously (Herrera, 2000; Jackson-Grusby et al., 2001). The RIP-Cre mice were bred into the Rosa26R-LacZ or Rosa26R-YFP background to indelibly mark all cells that were derived from insulin expressing cells (Jackson Labs)(Soriano, 1999). All animal experiments were performed in accordance with NIH policies on the use laboratory animals and approved by the Animal Research Committee of the Office for the Protection of Research Subjects at UCLA. Glucose tolerance test was performed following overnight fasting of the animals, as previously described (Zhong et al., 2007). Min6 cells were maintained in DMEM containing 10%FBS and 25 mM glucose at 37 °C in 5% CO2 environment. α-TC1 cells were cultured in DMEM containing 16.8 mM glucose, 4 mM L-glutamine, 17.8 mM NaHCO3 and 10% FBS at 37 °C in 5% CO2 environment.
Immunofluorescence staining and morphometric analysis
Standard immunofluorscence protocol was used for immuno-detection of various proteins in pancreatic sections(Zhong et al., 2007). Briefly, pancreatic tissue was dissected in PBS, fixed in 4% formaldehyde for 4 hours to overnight, dehydrated in grades of ethanol, and stored in −20°C until processed for paraffin embedding. Primary antibodies were diluted in the blocking solution at the following dilutions: Mouse anti-Glucagon (diluted 1:1000, Sigma-Aldrich G2654-.2ML); rabbit anti-Glucagon (diluted 1:500, Immunostar 20076); guinea pig anti-Insulin (diluted 1:400, Dako A0564); mouse anti-Dnmt1 (diluted 1:5000, Imgenex IMG-261A); chicken anti-beta galactosidase (1:1500, Abcam ab9361), mouse anti 5-methyl cytosine (1:200, Aviva Systems Biology AMM99021), chicken anti-GFP (1:500, Aves Labs Inc. 1020). Donkey- and goat-derived secondary antibodies conjugated to FITC or Cy3 were diluted 1:500 (Jackson ImmunoResearch Laboratories). Slides were viewed using a Leica DM6000 microscope and images were acquired using Openlab software (Improvision). For measurement of insulin and glucagon positive cell areas, 30 randomly chosen islets per animal (4 animals per group) from pancreatic sections immuno-stained for insulin and glucagon expression were imaged as above. Islets were defined as individual regions of interest, and total area along with the area of insulin and glucagon staining was quantified. Measurements of insulin and glucagon staining are expressed as a percentage of individual islet area. For calculation of glucagon cells co-expressing RIP-Cre driven beta-galactosidase lineage trace, total beta-galactosidase positive cells were counted along with cells double positive for beta-galactosidase and glucagon in 30 randomly chosen islets per animal (3 animals per group) and the data was expressed as percentage of double positive cells.
Islet isolation and cell sorting, beta galactosidase staining in islet cells for sorting
Islets were isolated using the Liberase enzyme blend (Roche Diagnostics) as described before (Zhong et al., 2007). For alpha cell isolation, islets from wildtype mice were digested into a single cells suspension and immunostained for glucagon, following brief fixation with BD cytofix reagent (BD Pharmingen), using rabbit anti-glucagon antibody, followed by incubation with a Cy3 conjugated secondary antibody, and sorted by FACS to an average percentage purity of 80–85%. Islet cells processed similarly, but without primary antibody were used as a negative control for FACS gating. For purification of beta cells, islets from MIP-GFP transgeneic mice were digested into single cells and sorted for GFP by FACS to an average percentage purity of 85–95%. Cells from wldtype islets were used as negative control for FACS gating. Beta cell-derived endocrine cells lineage traced with beta-galactosidase activity driven by RIP-Cre were purified after cleavage of fluorescent substrate fluorescein di-b-D-galactopyranoside (FDG; Sigma), by FACS sorting. For FACS sorting of these beta galactosidase positive cells, single cell suspension was prepared from isolated islets in ice-cold FACS sorting buffer (1X HBSS with 2% serum) and divided into 100 μl aliquots. 100 μl aliquots of 2mM FDG in amber tubes were pre-warmed at 37°C for 10 minutes along with cell aliquots. The cells were transferred to tubes with FDG and incubated at 37°C for one minute. The reaction was stopped by the addition of 2 ml ice-cold sorting buffer, followed by incubation on ice for 30 minutes and subsequently at room temperature for one hour. Cells were spun down, washed, suspended in sorting buffer and sorted.
siRNA based knockdown, RNA isolation, cDNA synthesis, real-time PCRs
Knockdown of Dnmt1, Arx and Dnmt1+Arx in Min6 cells were performed by transfection with a pool of specific targeting small inhibitory RNAs, or scrambled controls (siRNA, purchased from Dharmacon Research Inc.) using Lipofectamine-2000 (Invitrogen), according to manufacturer’s instructionsusing OPTI-MEM medium. Min6 cells were transfected with appropriate siRNAs or scrambled controls every three days (average transfection efficiency 65–80%) and samples were harvested at days post-transfection, as indicated. RNA was isolated from dissected pancreatic tissue or islets using TRI Reagent (MRC) and treated with DNase (Ambion) according to manufacturers’ instructions. RNA from sorted cells was prepared using RNeasy micro kit (Qiagen). 1 μg RNA was used for preparation of single stranded cDNA using Superscript III Reverse transcriptase (Invitrogen) by the oligodT priming method. Real-time RT-PCRs were performed using the LightCycler FastStartPLUS DNA SYBR Master kit (Roche) and the Light Cycler PCR equipment (Roche). The expression levels of each transcript were normalized to the housekeeping gene Cyclophilin. Each real-time PCR experiment shown is a representative from at least three independent experiments; for each experiment, islets were pooled from 3–4 mice per specified group. Real time PCR primers for RT-PCR are listed in Supplemental information.
Co-immunoprecipitation analyses
2.5 μg. of antibody (or control IgG) was immobilized onto Protein-A or Protein G Sepharose (Millipore) and washed with cold non-denaturing buffer (20 mM Tris-HCl pH=8, 137 mM NaCl, 10% glycerol, 1% NP-0, 2mM EDTA). The antibody coated beads were bound with cellular extracts prepared from Min6 cells (either un-transfected or transfected with the appropriate expression construct), in the non-denaturing buffer and incubated 5–16 hrs. at 4 °C with shaking. Beads were washed three times in ice-cold non-denaturing buffer and eluted into SDS-PAGE gel loading buffer. The immunoprecipitates were analyzed by Western blotting with appropriate antibodies. Western blots with the indicated antibodies were performed as described before (Zhong et al., 2007). Liquid chromatrography-mass spectrometry analysis was used to identify proteins co-immunoprecipitating with MeCP2, in Min6 cells, as described in Supplemental information.
DNA methylation analyses and ChIP analyses
Bisulfite conversion of DNA was performed as described previously (Millar et al., 2002). DNA samples were incubated with sodium bisulfite for 4–5 hours. Bisulfite treated DNA was then desalted and precipitated. We used 1/10 of precipitated DNA for each PCR using primers described in Supplemental information, to generate PCR products. Primers were designed using the MethPrimer software (Urogene). PCR products were gel purified and used for TOPO-TA cloning (Invitrogen), followed by sequencing. Primers used for generating bisulfite PCR products are described in Supplemental information, under “Experimental Procedures” section. MeDIP was performed according to standard methods (Weber et al., 2005). For MeDIP, 4 μg of sonicated genomic DNA (size: 300–1000 bp) was immuno-precipitated with 5 μg of antibody against 5-methyl cytidine (5mC, Aviva Systems Biology AMM99021) and collected for 4 h at 4°C with constant agitation using Protein G magnetic beads (Dynabeads, Invitrogen). A portion of the sonicated DNA was kept aside as input control. DNA was recovered from input (INP) and immunoprecipitated (IP) by Proteinase-K+ digestion, followed by phenol extraction and ethanol precipitation. 200 ng of immunoprecipitated and input DNA was prepared for hybridization on the as per manufacture’s instruction for Mammalian ChIP-on-chip hybrization to the mouse whole genome promoter array (Agilent). The arrays were scanned using the Agilent DNA microarray scanner. Data extraction and analyses were performed using the Agilent Feature Extraction software (version 9.1.3.1). Probe signals were normalized with Lowess normalization and then extracted in 16 step-wise 500 bp windows covering −5.5 Kb to +2.5 Kb promoter regions, with reference to the transcription start site (TSS) for each gene (Xie et al., 2009). ChIP experiments with the purified alpha and beta cells were carried out using the micro-ChIP protocol (Dahl and Collas, 2008). The primers used to amplify the Arx locus are listed in Supplementary Table 3. ChIP analyses on cell lines (Min6 and α-TC1) were performed using the Millipore chromatin immunoprecipitation kit (Cat. No. 17–295, Millipore, Bedford, MA) according to manufacturer’s instructions, with minor modifications described before (Dhawan et al., 2009).
Statistical methods
All data were expressed as Mean±S.E. Mean and s.e.m. values were calculated from atleast triplicates of a representative experiment. The statistical signifacnce of differences was measured by unpaired Student’s t-test. A P value < 0.05 indicated statistical significance. *P<0.05, **P<0.01, ***P<0.005.
Antibodies used for co-immunoprecipitation and ChIP, procedures for quantification of global DNA methylation and identification of proteins by liquid chromatography, mass-spectrometry and primers used for real-time PCRs for RNA expression analysis, quantification of ChIPs and for bisulfite sequencing analysis are presented in Supplemental experimental procedures.
Supplementary Material
Acknowledgments
We are grateful to Shaun Fouse for help with the DNA methylation analyses, Bing Li (Kurdistani lab), Wei Xei (Grunstein lab) for advice on MeDIP-ChIP experiments and Murtaza Kanji, Lendy Le for technical help. We also thank the Greenberg lab, Harvard Medical School, for the Flag-MeCP2 construct. The mass spectrometry analysis was performed in the W. M. Keck Proteomic Facility at the UCLA Molecular Instrumentation Center, which was established with a grant from the W. M. Keck Foundation. S. G. is supported by an NIDDK Career Development Award. This work was supported by grants from NIDDK, Juvenile Diabetes Research Foundation and the Helmsley Trust to A.B.
Footnotes
Author contributions:
S.D and S.G performed the experiments and analyses. G.F provided the Dnmt1lox/loxmice. A.B, S.D and S.G conceived and planned the experiments and interpreted data. S.T provided technical assistance. A.B and S.D wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Brennand K, Huangfu D, Melton D. All beta cells contribute equally to islet growth and maintenance. PLoS Biol. 2007;5:e163. doi: 10.1371/journal.pbio.0050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P, Mansouri A. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–2980. doi: 10.1242/dev.01870. [DOI] [PubMed] [Google Scholar]
- Collombat P, Hecksher-Sorensen J, Krull J, Berger J, Riedel D, Herrera PL, Serup P, Mansouri A. Embryonic endocrine pancreas and mature beta cells acquire alpha and PP cell phenotypes upon Arx misexpression. J Clin Invest. 2007;117:961–970. doi: 10.1172/JCI29115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Mansouri A, Hecksher-Sorensen J, Serup P, Krull J, Gradwohl G, Gruss P. Opposing actions of Arx and Pax4 in endocrine pancreas development. Genes Dev. 2003;17:2591–2603. doi: 10.1101/gad.269003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collombat P, Xu X, Ravassard P, Sosa-Pineda B, Dussaud S, Billestrup N, Madsen OD, Serup P, Heimberg H, Mansouri A. The ectopic expression of Pax4 in the mouse pancreas converts progenitor cells into alpha and subsequently beta cells. Cell. 2009;138:449–462. doi: 10.1016/j.cell.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Collas P. MicroChIP--a rapid micro chromatin immunoprecipitation assay for small cell samples and biopsies. Nucleic Acids Res. 2008;36:e15. doi: 10.1093/nar/gkm1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J Clin Invest. 2004;114:963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Luscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127:2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bahler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Millar DS, Warnecke PM, Melki JR, Clark SJ. Methylation sequencing from limiting DNA: embryonic, fixed, and microdissected cells. Methods. 2002;27:108–113. doi: 10.1016/s1046-2023(02)00061-0. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, Hemberger M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. 2008;10:1280–1290. doi: 10.1038/ncb1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101:2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin MM, Kushner JA. Adaptive beta-cell proliferation is severely restricted with advanced age. Diabetes. 2009;58:1365–1372. doi: 10.2337/db08-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sosa-Pineda B, Chowdhury K, Torres M, Oliver G, Gruss P. The Pax4 gene is essential for differentiation of insulin-producing beta cells in the mammalian pancreas. Nature. 1997;386:399–402. doi: 10.1038/386399a0. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Teta M, Rankin MM, Long SY, Stein GM, Kushner JA. Growth and regeneration of adult beta cells does not involve specialized progenitors. Dev Cell. 2007;12:817–826. doi: 10.1016/j.devcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Thorel F, Nepote V, Avril I, Kohno K, Desgraz R, Chera S, Herrera PL. Conversion of adult pancreatic alpha-cells to beta-cells after extreme beta-cell loss. Nature. 2010;464:1149–1154. doi: 10.1038/nature08894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell. 2009;33:417–427. doi: 10.1016/j.molcel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Georgia S, Tschen SI, Nakayama K, Bhushan A. Essential role of Skp2-mediated p27 degradation in growth and adaptive expansion of pancreatic beta cells. J Clin Invest. 2007;117:2869–2876. doi: 10.1172/JCI32198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.