Abstract
We present the 1.2 Å resolution X-ray crystal structure of a Ni-methyl species that is a proposed catalytic intermediate in methyl-coenzyme M reductase (MCR); the enzyme that catalyzes the biological formation of methane. The methyl group is situated 2.1 Å proximal of the Ni-atom of the MCR coenzyme F430. A rearrangement of the substrate channel has been posited to bring together substrate species, but Ni(III)-methyl formation alone does not lead to any observable structural changes in the channel.
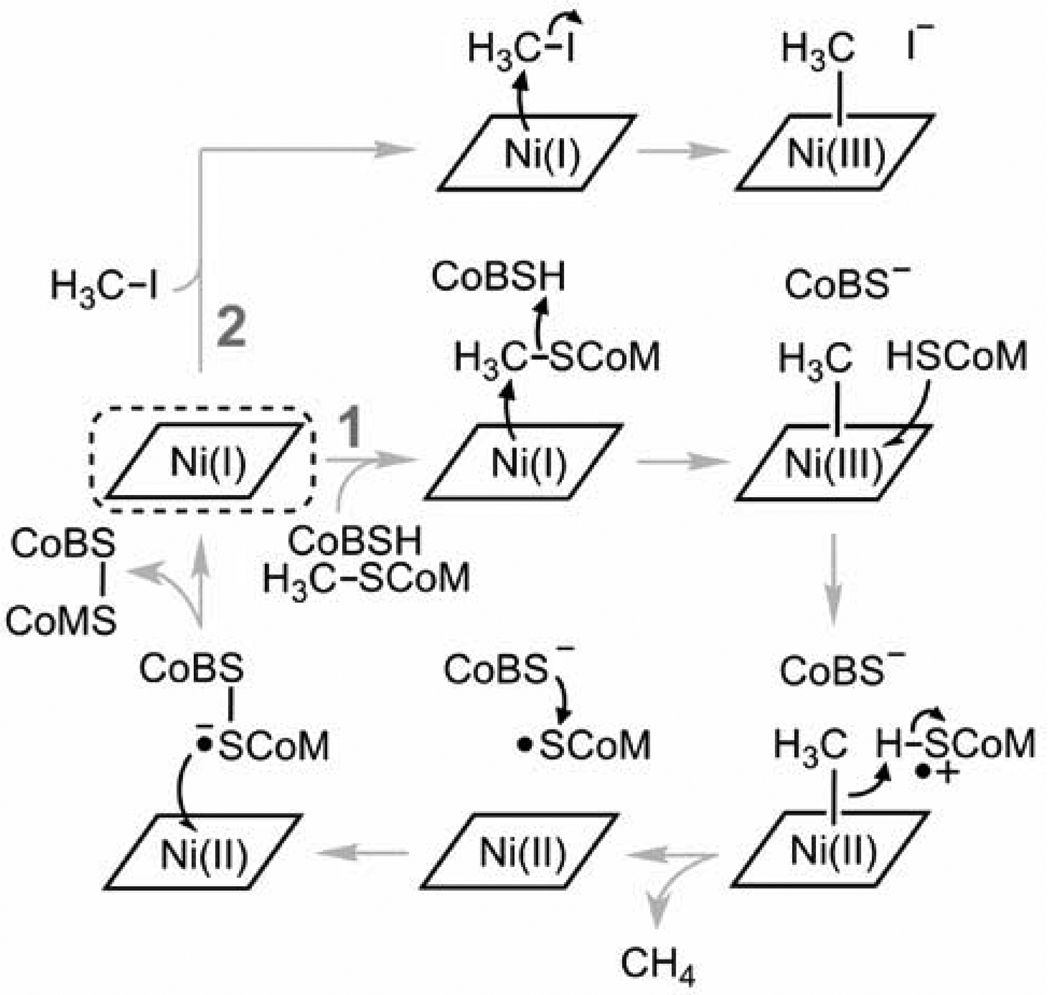
Methane has potential as an alternative fuel, but is also a potent greenhouse gas with surplus methanogenesis currently leading to a slow increase in methane concentrations within the atmosphere. The major source of methane produced in nature is the end product of the archaeal decomposition of organic matter in strict anaerobic environments such as lake sediments and the intestinal tract of animals.1 In the methanogenic archaea, methyl-coenzyme M reductase (MCR) catalyzes the final and rate-limiting step in methane biogenesis; the reduction of methyl-coenzyme M (methyl-SCoM, 2-(methylthio)ethanesulfonate) by coenzyme B (CoBSH, N-7-mercaptoheptanoylthreonine phosphate) to methane and a heterodisulfide (Supporting Information, Scheme S1). The 273 kDa MCR enzyme contains two deeply buried active sites, each containing a highly reduced Ni-tetrapyrrole cofactor, named coenzyme F430 (F430)2. The Ni atom in F430 is redox active and has been proposed to shuttle between oxidation states +1, +2 and +3 during turnover1,3,4 (Scheme 1.1), whilst other studies have suggested the involvement of only Ni oxidation states +1 and +2 (Supporting Information Scheme S2)5–7. The actual mechanism remains elusive since no true catalytic intermediate of MCR has ever been observed. However, a stable MCR-Ni(III)-methyl (MCRMe) state can be artificially produced by treating the resting active Ni(I) form of the enzyme (MCRred1) with methyl halide (Scheme 1.2).8,9 Treatment of MCRMe with coenzyme M (HSCoM, demethylated methyl-SCoM) generates MCRred1 along with the substrate methyl-SCoM, supporting this species as a possible catalytic intermediate.8
Scheme 1.
(1) Proposed MCR mechanism involving a Ni(III)-methyl intermediate. (2) Generation of a Ni(III)-methyl species using methyl iodide. The resting Ni(I) state is circled with a dashed line. For clarity the axial glutamine residue has been excluded
The geometric and electronic structure of MCRMe was recently characterized in solution by X-ray absorption spectroscopy (XAS) and density functional theory (DFT).10 Here we describe the X-ray crystal structure of the protein in this mechanistically relevant state. To confirm that MCRMe could form in crystals, a Ni K-edge XAS spectrum was obtained from a MCRred1 crystal treated anaerobically with methyl iodide, which demonstrated MCRMe formation in the crystal (Supporting Information Methods, Results and Figure S1).
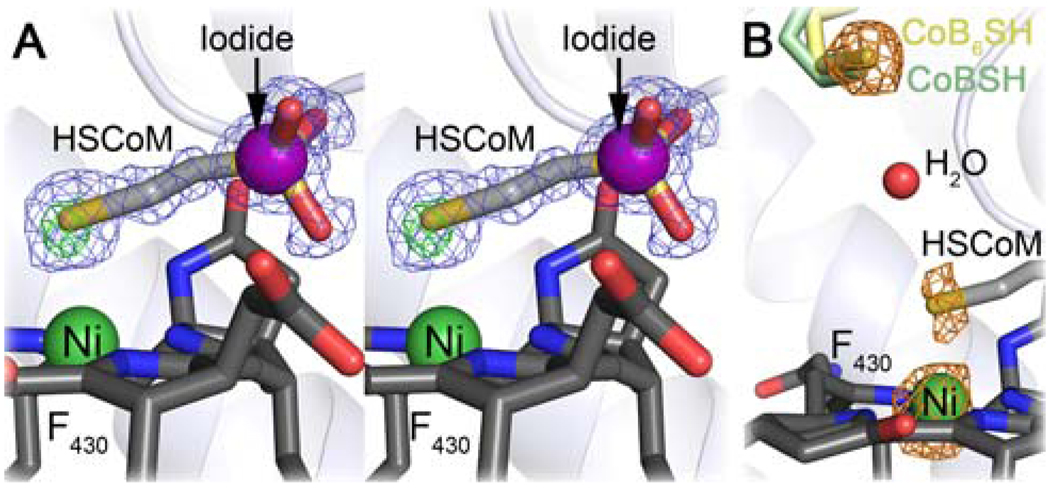
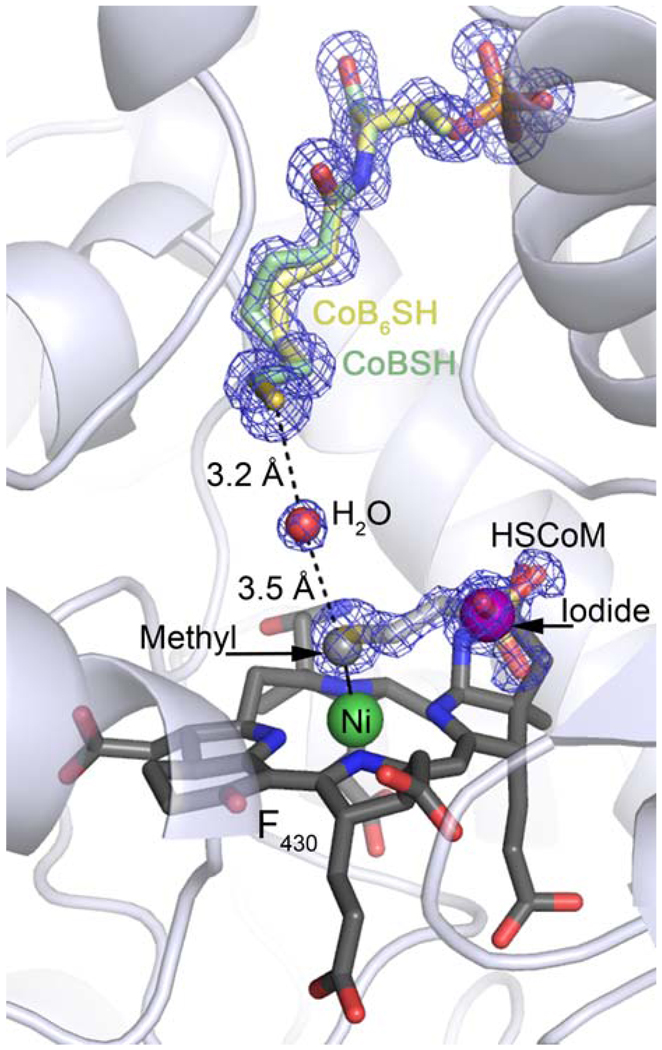
X-ray diffraction data were collected on a MCRMe crystal, and modeling and refinement led to final R-values are 13.2 % and 15.6 % for Rwork and Rfree respectively (Supporting Information Methods, Results and Table S1) The electron density map at 1.2 Å resolution contained two distinct species bound proximal to the Ni of F430. Besides a background of bound HSCoM (added during purification to stabilize MCR) there was clear Fo-Fc difference density consistent with a Ni-coordinated methyl group (Figure 1A). To deconvolute the multiple species from within the active site anomalous data generated from the native dataset (wavelength 0.98 Å) and at two additional wavelengths, 1.51 Å and 2.29 Å, were used (Figure 1B, Supporting Information Methods, Results, Table S2 and Figure S2). The Ni-S distance refines to 2.4 Å which is in line with the previous crystal structures containing HSCoM2,11–13 and the distance reported by XAS.10 The Ni-C distance refines to 2.07 Å and 2.10 Å in the two crystallographically independent active sites in the asymmetric unit (the distance determined by XAS for MCRMe in solution is 2.04 Å10). The substrate channel contains a mixture of coenzyme B analogue (CoB6SH (Figure S3); a slow substrate added in crystallization) and CoBSH (co-purifies with the enzyme: Figure 2). The respective thiols of CoB6SH and the substrate CoBSH are coincident and situated ~8.7 Å away from the Ni in F430.11 As CoBSH is a substrate, the long distance between the thiol and Ni(II) in the previous inactive MCR crystal structures has been puzzling.2,11–13 Thus, a conformational change has been proposed that would bring the CoBSH closer to the Ni,11,15 which is consistent with recent transient kinetic studies revealing substrate-triggered conformational changes in the catalytic resting state MCRred1.16
Figure 1.
Ni-methyl MCR electron density. A) Stereo figure showing positive Fo − Fc electron density (green mesh, contoured at 5σ) when methyl is not included in the structural model. 2Fo − Fc electron density (contoured at 1σ) is shown as blue mesh. B) Anomalous electron density map from X-ray diffraction data collected at wavelength 2.29 Å (orange mesh, contoured at 5σ) (f″ (electrons): sulfur 1.14; nickel 1.03). Coenzymes are shown in stick colored by atom (carbon: CoB6SH, pale yellow; CoBSH, pale green; HSCoM, light grey (faded); coenzyme F430, dark grey with the Ni atom shown as green sphere). Iodide as a purple sphere and a water molecule as a red sphere. Protein is shown as cartoon.
Figure 2.
Final structural model and electron density. The final model contains a 60:40 mixture of coenzyme B6 (CoB6SH) and coenzyme B (CoBSH), 50% coenzyme M (HSCoM), 50 % methyl and 35% iodide. Coenzymes are shown in stick colored by atom (carbon: CoB6SH, pale yellow; CoBSH, pale green; HSCoM, light grey (faded); coenzyme F430, dark grey with the Ni atom shown as green sphere). Methyl is displayed as a grey sphere, iodide as a purple sphere and a water molecule as a red sphere. Protein is shown as cartoon. 2Fo − Fc electron density (contoured at 1σ) is shown as blue mesh.
Our data suggest that if Ni(III)-methyl is an intermediate in methane formation, its presence alone does not affect the architecture of the MCR substrate channel, which appears unchanged compared to the structure of the channel in the Ni(II) structures. The release of methane involves a protonation step, which has been postulated to either be provided by CoBSH via a CoM species (Scheme 1.1), or possibly via a water molecule (Figure 2), two Tyr residues2 (Tyrα333 and Tyrβ367, whose hydroxyls point towards the Ni) or the C-ring nitrogen of F4303. Since no catalytic intermediates of MCR have been observed thus far by fast spectroscopic techniques, any Ni(III)-methyl catalytic intermediate formed from the native substrates must be fleeting during turnover. Recently transient kinetic methods using the natural substrate, methyl-SCoM, and the slow substrate CoB6SH, enabled observation of a short-lived Ni(III)-alkyl intermediate and its subsequent decay at catalytically competent rates.16 In contrast, when Ni(III)-methyl is artificially generated using methyl iodide, as in this study, it is relatively stable enabling spectroscopic and biochemical characterization.8,9
Interestingly, MCR is also implicated in “reverse” methanogenesis in the anaerobic oxidation of methane within microbial mats found at methane seeps in the deep ocean.17,18 The dissociation energy of the C-H bond (+439 kJ/mol) is larger than in any other organic compound except benzene. Thauer and Shima have proposed that in “reverse” methanogenesis only the strong electrophile F430-Ni(III) of MCR is capable of breaking the C-H bond in a modified “reversed” version of the Ni(III)-methyl mechanism proposed for methanogenesis (Scheme 1.1).19 The recent evidence of a Ni-(σ-alkane) intermediate in the reaction of MCR is consistent with this proposal.20
The crystal structure of MCRMe is the first structure resembling a postulated intermediate in one of the proposed mechanisms of MCR. The high resolution of the X-ray diffraction data, coupled with anomalous diffraction at several wavelengths, has enabled us to extract the relevant Ni-methyl structure from confounding species. The active Ni(I) enzyme, its complex with methyl-SCoM, substrate channel conformational changes and true catalytic intermediates trapped during turnover still wait to be structurally defined.
Supplementary Material
Acknowledgement
This work was supported by a Department of Energy grant DE-FG02-08ER15931 to S.W.R and supplement to C.M.W. and a Minnesota Partnership for Biotechnology and Medical Genomics grant SPAP-05-0013-P-FY06. We thank Stephan L. Ginell, Ph.D. and the staff at the SBC-CAT beamline 19-ID-D at the Advanced Photon Source (APS), Argonne National Laboratory for valuable assistance during X-ray data collection. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. SSRL operations are funded by the Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology program is supported by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the Department of Energy, Office of Biological and Environmental Research.
Abbreviations
- MCR
Methyl-coenzyme M reductase
- F430
coenzyme F430
- methyl-SCoM
methyl-coenzyme M
- CoBSH
coenzyme B
- HSCoM
coenzyme M
- CoB6SH
coenzyme B analogue
Footnotes
Supporting Information Available: methods, supporting results, X-ray crystallographic data table, anomalous signal table, single crystal X-ray absorption spectra, anomalous electron density figure, drawing of CoBSH and analogue CoB6SH, EPR spectrum, catalytic reaction scheme and enzymatic mechanism scheme. This material is available free of charge via the Internet at http://pubs.acs.org. The structure is deposited in the Protein Data Bank with PDB entry 3POT.
REFERENCES
- 1.Thauer RK. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 2.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 3.Duin EC, McKee ML. J. Phys. Chem. B. 2008;112:2466–2482. doi: 10.1021/jp709860c. [DOI] [PubMed] [Google Scholar]
- 4.Horng YC, Becker DF, Ragsdale SW. Biochemistry. 2001;40:12875–12885. doi: 10.1021/bi011196y. [DOI] [PubMed] [Google Scholar]
- 5.Chen SL, Pelmenschikov V, Blomberg MR, Siegbahn PE. J. Am. Chem. Soc. 2009;131:9912–9913. doi: 10.1021/ja904301f. [DOI] [PubMed] [Google Scholar]
- 6.Pelmenschikov V, Blomberg MRA, Siegbahn PEM, Crabtree RH. J. Am. Chem. Soc. 2002;124:4039–4049. doi: 10.1021/ja011664r. [DOI] [PubMed] [Google Scholar]
- 7.Pelmenschikov V, Siegbahn PE. J. Biol. Inorg. Chem. 2003;8:653–662. doi: 10.1007/s00775-003-0461-8. [DOI] [PubMed] [Google Scholar]
- 8.Dey M, Telser J, Kunz RC, Lees NS, Ragsdale SW, Hoffman BM. J. Am. Chem. Soc. 2007;129:11030–11032. doi: 10.1021/ja074556z. [DOI] [PubMed] [Google Scholar]
- 9.Yang N, Reiher M, Wang M, Harmer J, Duin EC. J. Am. Chem. Soc. 2007;129:11028–11029. doi: 10.1021/ja0734501. [DOI] [PubMed] [Google Scholar]
- 10.Sarangi R, Dey M, Ragsdale SW. Biochemistry. 2009;48:3146–3156. doi: 10.1021/bi900087w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cedervall PE, Dey M, Pearson AR, Ragsdale SW, Wilmot CM. Biochemistry. 2010;49:7683–7693. doi: 10.1021/bi100458d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabarse W, Mahlert F, Duin EC, Goubeaud M, Shima S, Thauer RK, Lamzin V, Ermler U. J. Mol. Biol. 2001;309:315–330. doi: 10.1006/jmbi.2001.4647. [DOI] [PubMed] [Google Scholar]
- 13.Grabarse W, Mahlert F, Shima S, Thauer RK, Ermler U. J. Mol. Biol. 2000;303:329–344. doi: 10.1006/jmbi.2000.4136. [DOI] [PubMed] [Google Scholar]
- 14.Dey M, Kunz RC, Lyons DM, Ragsdale SW. Biochemistry. 2007;46:11969–11978. doi: 10.1021/bi700925n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebner S, Jaun B, Goenrich M, Thauer RK, Harmer J. J. Am. Chem. Soc. 2010;132:567–575. doi: 10.1021/ja906367h. [DOI] [PubMed] [Google Scholar]
- 16.Dey M, Li X, Kunz RC, Ragsdale SW. Biochemistry. 2010;49:10902–10911. doi: 10.1021/bi101562m. [DOI] [PubMed] [Google Scholar]
- 17.Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF. Appl. Environ. Microbiol. 2003;69:5483–5491. doi: 10.1128/AEM.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheller S, Goenrich M, Boecher R, Thauer RK, Jaun B. Nature. 2010;465:606–608. doi: 10.1038/nature09015. [DOI] [PubMed] [Google Scholar]
- 19.Thauer RK, Shima S. Ann. N.Y. Acad. Sci. 2008;1125:158–170. doi: 10.1196/annals.1419.000. [DOI] [PubMed] [Google Scholar]
- 20.Scheller S, Goenrich M, Mayr S, Thauer RK, Jaun B. Angew. Chem. Int. Ed. Engl. 2010;49:8112–8115. doi: 10.1002/anie.201003214. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.