Abstract
The innate immune system relies on the recognition of pathogens by pattern recognition receptors as a first line of defense and to initiate the adaptive immune response. Substantial progress has been made in defining the role of Nod (nucleotide-binding oligimerization domain)-like receptors and AIM2 (absent in melanoma 2) as pattern recognition receptors that activate inflammasomes in macrophages. Inflammasomes are protein platforms essential for the activation of inflammatory caspases and subsequent maturation of their pro-inflammatory cytokine substrates and induction of pyroptosis. This paper summarizes recent developments regarding the function of Nod-like receptors in immunity and disease.
Keywords: inflammation, caspase-1, Nod-like receptor, pattern recognition, autoinflammatory disease
I. INTRODUCTION
In addition to the mechanical barrier of the skin, mammals are protected by two arms of the immune system, the innate and the adaptive immune systems, which cooperatively function to eradicate invading pathogens. The innate immune system is responsible for initial defense and also activates the adaptive immune system for long-lasting and highly specific immunity by antigen-specific, clonally expanded B and T lymphocytes. A key requirement in this process is the discrimination between self and non-self to specifically eliminate pathogens. Therefore, germ line-encoded pattern recognition receptors (PRRs) of innate immune cells sense pathogens directly at the infection site through recognition of pathogen-associated molecular patterns (PAMPs), which are conserved, and essential structures that are present on microorganisms but absent on host cells.1 PRRs can be classified into phagocytic PRRs, which directly bind pathogens and mediate their internalization, and sensor PRRs, which respond to recognition of PAMPs by initiating pro-inflammatory signaling pathways.2 Sensor PRRs, such as the membrane-bound Toll-like receptors (TLRs) recognize PAMPs on the cell surface and lumen of endosomes and lysosomes.3 More recently, PRRs that sense the cytosolic presence of PAMPs have been discovered, and these include the RIG-1 (retinoic acid-inducible gene-1)-like receptors (RLRs),4 the nucleotide-binding and oligomerization domain (Nod)-like receptors (NLRs),5–8 and AIM2 (absent in melanoma-2).9
Macrophages also express a number of phagocytic PRRs, including scavenger receptors, C-type lectins, collectins, and pentraxins.2 The broad activity of the innate immune system is provided by phagocytes such as monocytes, macrophages, neutrophils, dendritic cells, and epithelial cells, which engulf infectious agents and initiate an inflammatory host response. This response is mediated by the release of cytokines and chemokines that act on the surrounding tissue cells, as well as by up-regulation of adhesion and co-stimulatory molecules to promote recruitment of immune cells into the infected tissue. Two of these pro-inflammatory cytokines, the related interleukin (IL)-1β and IL-18, are produced early during host defense and are responsible for perpetuating this inflammatory response and exerting multiple effects during infection, tissue damage, inflammation, and autoimmune and autoinflammatory disease.10
II. IL-1β AND IL-18: TWO CASPASE-1-DEPENDENT PRO-INFLAMMATORY CYTOKINES
The processes that generate the biologically active cytokines IL-1β and IL-18 have a similar mechanism. In contrast to most cytokines, which are only transcriptionally induced and immediately secreted in response to inflammatory and infectious stimuli, IL-1β and IL-18 require two signals. First, activation of TLRs, RLRs, and other PRRs that induce the inflammatory response by activation of primarily NF-κB (nuclear factor kappa light-chain enhancer of activated B cells) and MAPK (mitogen-activated protein kinase) signaling pathways that collectively promote a transcription factor-mediated response, is required for the up-regulation of IL1B transcripts. In contrast, the IL18 transcript is constitutively produced in most cell types. Transcription/translation, however, only produces the intracellular, inactive precursors, pro-IL-1β (31 kDa) and pro-IL-18 (24 kDa). The second signal required for cytokine release causes the activation of caspase-1. Active caspase-1 proteolytically cleaves the prodomain to liberate the 17- and 18-kDa mature cytokines, which are then released by an atypical, leader peptide-independent mechanism, which is still controversial.11–13 This caspase-1-dependency of IL-1β and IL-18 maturation appears to be restricted to monocytes and macrophages.
While monocytes express constitutively active caspase-1, and therefore only require the priming step for IL-1β release,14 other cells such as neutrophils and mast cells release serine proteases that are frequently present in inflammatory fluids. These proteases, including proteinase-3, elastase, chymase, matrix metalloproteinases, and others, can mature pro-IL-1β and pro-IL-18, which can be released from short-lived and damaged cells.15 Mature IL-1β and IL-18 are recognized by their receptors, IL-1RI and IL-18Rα, respectively, and cause a conformational change that allows high-affinity binding in the complex with the IL-1R accessory protein (IL-1RAc or IL-1RIII) or the IL-18Rβ, respectively.10 Signal transduction is then mediated by the TIR (Toll/IL-1 receptor) domain, which is also present in TLRs, further emphasizing their link to innate immunity. Furthermore, both cytokines have a naturally occurring inhibitor, the IL-1R antagonist (IL-1Ra) and the IL-18 binding protein (IL-18BP), respectively.
In addition to its well-established role in maturing pro-IL-1β and pro-IL-18, caspase-1 has been implicated in unconventional protein secretion extending beyond cytokine release.16 Caspase-1 is able to also promote cell survival in response to microbial pore-forming toxins, which activate NLRP3 (NLR containing a PYD-3) and NLRC4 (NLR containing a CARD-4) inflammasomes, through activating lipid metabolic pathways to mediate membrane biogenesis and repair.17,18 In this capacity, caspase-1 activates the sterol regulatory element-binding proteins (SREBPs) 1 and 2, which in turn facilitate fatty acid metabolism and cholesterol and lipid biosynthesis.19 This might appear to contradict the better known role of caspase-1 in promoting an inflammatory form of cell death called pyroptosis, which causes loss of plasma membrane integrity and local inflammation.20–24 However, morphologically, pyroptosis is reminiscent of pyronecrosis, yet another form of inflammatory cell death, which is NLRP3, ASC (apoptosis-associated speck-like protein containing a CARD), and cathepsin B dependent, but independent of caspase-1.25,26 Thus, the consequence of caspase-1 activation might depend on the state of the infection. While maturation of proinflammatory cytokines is a first step in limiting the spread of a pathogen, the activation of membrane biogenesis could repair damage resulting from the microbial insult. There is now evidence that active caspase-1 is rapidly released from macrophages, which might prevent it from causing any further cell damage.27 However, if the infection persists and cannot be cleared, caspase-1 is capable of inducing pyroptosis to eliminate the pathogen through cell death.
Caspase-1, the first member of the caspase family of cysteine proteases to be identified, belongs to the inflammatory caspase subfamily, which also includes caspase-4, caspase-5 (and their mouse paralogue, caspase-11), and caspase-12.28 The inflammatory caspases belong to the initiator caspases and contain a CARD as their pro-domain. Caspases are initially synthesized as inactive precursor proteins (zymogens) and require activation to achieve catalytic activity. CARD is essential for the clustering of two or more pro-caspases required for their autocatalytic transactivation by the induced proximity mechanism.29 The clustering of pro-caspases occurs in large protein platforms specific for each caspase, which is the inflammasome for the inflammatory caspases.27
III. INFLAMMASOMES: PROTEIN PLATFORMS TO ACTIVATE INFLAMMATORY CASPASES
While the activation of TLRs, RLRs, and some NLRs initiates signaling cascades that ultimately promote a transcriptional response to up-regulate pro-inflammatory mediators, the activation of several NLRs and AIM2 causes the formation and activation of inflammasomes, multi-protein platforms that mediate the activation of inflammatory caspases, including caspase-1 and caspase-5, by the induced proximity mechanism.27 For most characterized inflammasomes, ASC is the required adaptor for pro-caspase-1 activation.30,31 It binds to the CARD-containing pro-domain of caspase-1 to mediate its oligomerization. Accordingly, enforced oligomerization of ASC is sufficient to activate caspase-1.27,32–34 Therefore, ASC and caspase-1 are shifted into a high-molecular-weight protein complex following in vitro activation. ASC is the adaptor to bridge activated NLRs with pro-caspase-1, and consequently NLRP1 and caspase-5 are shifted into these large-molecular-weight complexes in response to in vitro activation.27 However, in response to MDP (muramyl dipeptide) activation of macrophages in vivo, only NLRP1, Nod2, and caspase-1 are shifted into high-molecular-weight complexes.35
Consistently, in vitro reconstituted NLRP1 inflammasomes are able to assemble in the absence of ASC, although ASC is able to enhance inflammasome activity.36 In contrast to this ASC-independent response of NLRP1 inflammasomes, immunodepletion of ASC is able to abrogate NLRP1 inflammasome activation in the human monocytic cell line THP-1 in response to lipopolysaccharide, which is reminiscent of the best-studied NLRP3 inflammasome.27 ASC directly interacts with NLRP3 by a PYD-PYD interaction, and recruits pro-caspase-1 by a CARD-CARD interaction.34,37 For NLRC4 inflammasomes, genetic evidence suggests that ASC is required for caspase-1 activation in spite of the direct interaction of NLRC4 with pro-caspase-1 by a CARD-CARD interaction.30,38,39 Thus, although ASC is required for caspase-1 activation by some inflammasomes, and ASC-deficient macrophages fail to respond to most tested stimuli with caspase-1 activation and release of IL-1β and IL-18,30,31,40 caspase-1 activation in response to MDP and perhaps some other stimuli is mostly uncoupled from ASC.35,36 These data suggest that while in some inflammasomes ASC is essential for assembly, ASC might play a less prominent role in other inflammasomes. Indeed, recent studies demonstrated that knockdown of ASC in THP-1 cells impaired the NF-κB-mediated up-regulation of several pro-inflammatory cytokines, including pro-IL-1α, in response to several TLR agonists and infection with Escherichia coli and Porphyromonas gingivalis, which could explain the partial requirement for ASC41: NLR oligomerization is required for their interaction with ASC.42
The NACHT domain of at least some NLRs has ATPase activity, and ATP binding is a prerequisite for NLR oligomerization.36,43,44 Several NLRs are complexed via their LRRs (leucine-rich repeats) with the ubiquitin ligase-associated protein SGT1 (suppressor of G2 allele of SKP1) and HSP90 (heat-shock protein 90) in unstimulated cells, which might keep the receptors in an inactive, but signaling ready state. In the absence of SGT1 or upon treatment of cells with geldanamycin to impair HSP90 chaperon activity, NLRs are degraded.45,46 However, upon inflammatory activation of cells, SGT1 and HSP90 dissociate from NLRs, followed by nucleotide binding and NLR oligomerization.45,46 Although 22 human NLRs and 33 mouse Nlr genes exist,47 the function of most NLRs has yet to be elucidated, and so far only a few NLRs have been linked to inflammasome activation.
IV. NOD-LIKE RECEPTORS AND AIM2: SENSORS OF PATHOGEN INFECTION AND CELLULAR STRESS
A. NLRP1 and Nod2
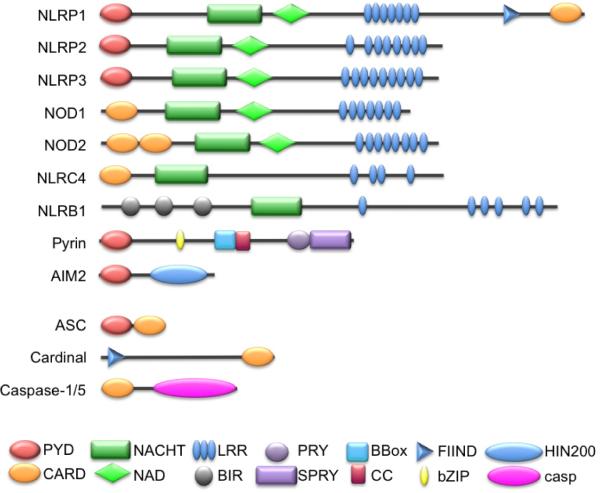
The first identified inflammasome was that of NLRP1.27 NLRP1 (also known as NALP1, DEFCAP, NAC, CARD7, and CLR17.1) is unique among NLRPs because, in addition to its PYD, NACHT, NACHT-associated domain (NAD), and LRRs, it contains a C-terminal function-to-find (FIIND) domain and CARD (Figs. 1 & 2).48,49 Although there is only one NLRP1, there are three paralogous Nlrp1 genes encoded in mice, all of which lack the PYD. The NLRP1 inflammasome contains caspase-1, which is recruited by ASC, and caspase-5, which directly interacts with the CARD of NLRP1.27 However, the roles of ASC and caspase-5 in the NLRP1 inflammasome are still uncertain. ASC interacts with NLRP1, immunodepletion of ASC impairs NLRP1 inflammasomes, and the presence of ASC enhances in vitro reconstituted NLRP1 inflammasomes that assemble in response to MDP.36,27 However, ASC was also found to be not essential for NLRP1 inflammasome function, and mouse Nlrp1 lacks the PYD and therefore cannot interact with ASC.35,36 Caspase-5, although part of the NLRP1 inflammasome, is lacking from other inflammasomes, including NLRP2 and NLRP3-containing inflammasomes, and thus might not have an essential role in all inflammasomes.37
FIGURE 1.
Domain structure of NLR and adaptor proteins involved in inflammasome activation. Proteins were analyzed using the simple modular architecture research tool (SMART) (http://smart.embl.de/)48,49 and conserved domains are indicated.
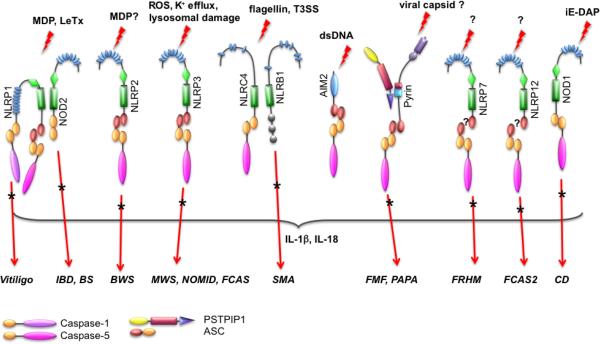
FIGURE 2.
Composition and activators of known inflammasomes. Known and putative inflammasomes are shown. Inflammatory diseases caused by mutations or polymorphisms of inflammasome proteins are indicated by an asterisk. A question mark indicates that the inflammasome agonist or precise inflammasome composition is not known.
MDP is also sensed by Nod2 (also known as NLRC2, CARD15, CD, BLAU, IBD1, PSORAS1, and CLR16.3), an NLRC that can directly interact with the CARD of pro-caspase-1. However, Nod2-mediated pro-caspase-1 activation and IL-1β maturation also require the presence of NLRP1 or NLRP3 (see below) (Figs. 1 & 2).35,50 This interaction does not require the adaptor protein RIP2 (receptor-interacting protein-2; also known as Cardiak, RICK, CCK, and Ripk2), which bridges Nod2 to the NF-κB activating IKK (IκB kinase) complex, although RIP2 itself can bind to caspase-1.51–54 Both NLRP1 and Nod2 form an MDP-sensing inflammasome, and in reconstitution assays, both proteins enhanced each other's interaction with caspase-1 and inflammasome activation, which is most likely explained by their ability to heterodimerize.35,55 However, this MDP-induced inflammasome does not contain NLRP3, although NLRP3 has also been linked to MDP-mediated inflammasome activation, which also required RIP2 and ASC.35,50,56
In addition to MDP, Nlrp1 is involved in sensing Bacillus anthracis infections. The lethal toxin (LeTx) is responsible for mortality in systemic anthrax infections; inbred mice show a variable degree of sensitivity to LeTx, and this trait has been mapped to Nlrp1b.57 LeTx activation of Nlrp1b depends on lysosomal membrane destabilization.58 Similar to MDP, recognition of LeTx also requires NLRP1 and Nod2, indicating that NLRP1/Nod2 heterodimers might be more prevalent.35 It is still not known if MDP or LeTx are directly bound by NLRP1 or Nod2, but in a minimal in vitro reconstitution system NLRP1 is still able to respond to MDP, which suggests that MDP could be directly recognized.36 LeTx, on the other hand, causes macrophage death through inactivation of p38α, which is required for macrophage survival, through up-regulation of the caspase-1 inhibitor PAI-2 (plasminogen activator inhibitor-2).59–61 Thus, it is more likely that LeTx is rather indirectly recognized by NLRP1/Nod2. As proposed for NLRP1, the Nod2 LRRs also engage with the CARD-NACHT region by intramolecular interaction, and this autoinhibitory interaction is reversed by MDP.35,36
B. NLRP2
NLRP2 (also known as NALP2, PYPAF2, NBS1, PAN1, and CLR19.9) interacts with ASC to form an inflammasome that also contains Cardinal (also known as TUCAN and CARD8) (Figs. 1 & 2).37,62 Cardinal is a FIIND and CARD-containing protein that can interact with caspase-1.63 The interaction of Cardinal with NLRP2 occurs via the NACHT and FIIND domains and requires an activated NLRP2 or deletion of the LRR (which can activate NLRs), but does not interact with full-length naive protein, due to its inactive conformation.37 Cardinal might provide the FIIND-CARD that is also present in NLRP1, but lacks in other NLRs, including NLRP2. However, unlike NLRP1, this interaction is insufficient to recruit caspase-5 to NLRP2.37 While cell rupture can activate NLRP2 inflammasomes, similar to NLRP1 and NLRP3, the physiological NLRP2 agonist is still elusive.
C. NLRP3
NLRP3 (also known as cryopyrin, cold-induced autoinflammatory syndrome-1 [CIAS1], NALP3, PYPAF1, and CLR1.1) is perhaps the best-studied NLRP and requires the adaptor ASC to bridge to caspase-1 (Figs. 1 & 2).30 Like NLRP2, NLRP3 inflammasomes also contain Cardinal, although the precise role of Cardinal in inflammasomes is elusive, given the fact that it is absent from the mouse genome.37 The interaction with ASC is induced by enforced NLRP3 oligomerization, which occurs upon ligand binding or upon expression of disease-associated mutations in the NLRP3 protein, indicating that inflammatory disease-linked mutants are constitutively active and uncoupled from ligand sensing.42 Accordingly, disease-associated NLRP3 mutants cause enhanced oligomerization of ASC.64 Because the LRRs are the ligand-sensing/-binding domain and keep the NLR inactive, deletion of the LRR renders the protein constitutively active.42
A first insight into the physiological role of NLRP3 came from macrophages isolated from NLRP3-deficient mice, which demonstrated that NLRP3 is activated by a large number of diverse agonists, including PAMPs, DAMPs (danger-associated molecular patterns), pathogens, and synthetic substances that cause potassium efflux, lysosomal damage, and/or reactive oxygen species (ROS) generation.18,40,65,66 PAMPs and other exogenous ligands that are recognized by NLRP3 include MDP, viral and bacterial dsRNA, poly(I:C), or the imidazoquinolines R848/R837, mainly in conjunction with ATP.56,65,67,68 The precise mechanism of how diverse PAMPs activate NLRP3 is not clear, but potentially promotes macrophage priming to up-regulate pro-IL-1β, while ATP potentially provides the second signal for inflammasome activation and IL-1β maturation. Activation and opening of the pannexin-1 channel may allow entry of PAMPs into the cytosol. In fact, pannexin-1 channel opening occurs as a consequence of binding of exogenous ATP, which is a danger signal released from stressed and dying cells following tissue injury to the purinergic P2X7 receptor (P2X7R) and subsequent potassium efflux.69–72 Indeed, exogenous ATP is a potent activator of the NLRP3 inflammasome, which causes production of ROS and activation of PI3K (phosphoinositide 3-kinase).18,72,73 Exogenous triggers appear to require ATP/P2X7R/K+ efflux for inflammasome activation, while intracellular pathogens, such as Salmonella or Listeria and intracellular flagellin, do not require this step.69 Furthermore, while mouse macrophages require exogenous ATP for IL-1β release in response to certain stimuli, human macrophages release ATP upon treatment with inflammatory or infectious stimuli, which then acts through an autocrine mechanism on P2X7R to promote IL-1β release.74
In support of these findings, the action of ATP can be mimicked with microbial pore-forming toxins such as streptolysin O or pneumolysin.72 Similarly, maitotoxin and nigericin, two K+/H+ antiport ionophores, cause intracellular K+ ion depletion and promote NLRP3 activation.18 Preventing K+ depletion, for example by culturing of macrophages in medium containing 130 mM KCl, also reduces inflammasome activation and emphasizes the significance of K+ efflux for NLRP3 activation.22,75,76 In the case of Staphylococcus aureus MDP, phagocytosis and lysozyme-based cell wall degradation are essential to make MDP accessible for inflammasome activation, while preventing degradation also diminishes inflammasome activation.77 However, caspase-1 activation can also occur in the absence of infectious stimulation during a sterile inflammatory response. Caspase-1 can be activated in macrophages and dendritic cells by incubation with inflammatory cytokines such as TNFα and IL-1β, but not with CD40L, Fas, IFNγ, or RANKL in combination with ATP or particulates (see below).78
Crystalline and polymeric structures, including environmental pollutants and danger signals, are also potent activators of NLRP3. Asbestos, silica, calcium pyrophosphate dehydrate crystals, dextran sulfate sodium polymers, aluminum salt, monosodium urate crystals, cholesterol crystals, and amyloid-β fibrillar peptides all activate NLRP3.66,79–87 These NLRP3 agonists undergo similar phagocytosis that leads to subsequent inflicted phagolysosomal damage, causing the release of lysosomal proteases, most prominently cathepsin B and L, suggesting that NLRP3 might be activated downstream of a proteolytic pathway. Cathepsin B- and L-deficient mice display impaired IL-1β processing, and short-interfering RNA (siRNA)-mediated knockdown of cathepsin B abrogates caspase-1 activation in response to nigericin or dextran sulfate sodium.87–89 However, siRNA-mediated knockdown of cathepsin B did not affect inflammasome activation in response to LeTx, which, however, is an Nlrp1b agonist.58 The same study also pointed out that CA-074Me, a previously considered cathepsin B-specific inhibitor, blocks LeTx-induced Nlrp1b activation independently from acting on cathepsin B through inhibiting alternative protease activity induced or released by lysosomal membrane permeabilization.58 The NLRP3-agonist role of such crystals can be mimicked by direct lysosomal rupture using pharmacological or mechanical mechanisms, which cause NLRP3 activation, supporting the essential role of lysosomal destabilization for NLRP3 activation.81,83,90
NLRP3 is also responsible for sensing diverse pathogens, including the gram-positive bacteria S. aureus and Listeria monocytogenes18,91 and the gram-negative V. vulnificus and V. cholerae.90S. aureus is recognized by NLRP3 through α-hemolysin, a pore-forming virulence factor responsible for pneumonia.92 Similarly, V. vulnificus and V. cholerae-mediated NLRP3 activation occurs by hemolysins and MARTX (multifunctional repeat-in toxins).90 NLRP3 also senses infections by C. pneumoniae, which is dependent on potassium efflux, lysosomal damage, and cathepsin B release.93Chlamydia trachomatis also activates NLRP3, which requires Syk (spleen tyrosine kinase), potassium efflux, and ROS generation.94 NLRP3 further senses infections by DNA and RNA viruses, including Sendai virus and influenza virus, and recognizes the adenoviral capsid.67,68,95–97 The influenza proton-selective ion channel M2 is localized to the Golgi, and its acidification is required for influenza M2-induced NLRP3 inflammasome activation, which is caused by imbalances in H+ and other cations.97,98 However, caspase-1 activation by viral RNA occurs independently of NLRP3 by a complex of the RNA helicase RIG-I and ASC.99 Parasites such as Plasmodium chabaudi adami DS are recognized by NLRP3 through its metabolic waste hemozoin, which has a crystalline appearance, and deficiency of NLRP3 augments survival in a mouse model of P. chabaudi adami DS infection.100
In agreement with its crystalline structure, hemozoin recognition depends on phagocytosis, lysosomal damage, potassium efflux, ROS generation, and cathepsin B release.100 Moreover, hemozoin causes activation of the tyrosine kinases Syk and Lyn (a Yamaguchi sarcoma viral oncogene homolog) and subsequently activation of PI3K and ERK (extracellular signal-regulated kinase), providing evidence for the link between inflammasome activation and kinase signaling.100 Fungal infections, such as by Candida albicans and Aspergillus fumigatus, are sensed by NLRP3 inflammasomes in concert with Syk-coupled C-type lectin receptors with an immunoreceptor tyrosine-based activation motif, such as dectin-1 and dectin-2.75,101C. albicans-mediated NLRP3 activation is independent of lysosomal damage and cathepsin B release, but dependent on ROS generation, which is in agreement with an essential role of Syk in the generation of ROS in response to fungal infections.75,102 Similarly, monosodium urate and other particulates that activate NLRP3 have been shown to activate Syk. Therefore, the generation of ROS is a common requirement for most, if not all, of the NLRP3 activators.
Further support for the importance of ROS comes from experiments with ROS scavengers such as N-acetyl cysteine.70,73,75,76,85,100 Recently, the link between ROS and NLRP3 has been elucidated. The LRRs of NLRP3, but not other NLRs, interact specifically with the thioredoxin (TRX)-interacting protein (TXNIP). While TXNIP is bound to TRX during homeostasis, elevated ROS levels induce the dissociation of TXNIP from TRX, allowing TXNIP to interact and activate NLRP3 in a ROS-dependent manner.103 However, not all stimuli causing the generation of ROS also cause activation of NLRP3 and maturation of IL-1β, suggesting some form of regulation downstream of ROS. Thus, there might be several distinct mechanisms responsible for NLRP3 activation, including ATP/P2X7R/pannexin-1/K+ depletion, lysosomal destabilization and potentially release of proteases such as cathepsin B and L, and the generation of ROS.
D. NLRC4 and NLRB1
NLRC4 (also known as CARD12, NOD27, CLAN, IPAF, and CLR2.1) is an NLR containing a CARD, which can directly recruit caspase-1, but also interacts with ASC (Figs. 1 & 2).104–106 NLRC4 is activated by the intracellular gram-negative S. typhimurium (but not by bacteria lacking the salmonella invasion protein B), Legionella pneumophilia, Shigella flex-neri, and Pseudomonas aeruginosa.30,38,39,107–109 Although NLRC4 can interact directly with caspase-1, ASC is also required for NLRC4 inflammasome activation in response to S. typhimurium, S. flexneri, and P. aeruginosa.30,38,108,109 The common PAMP that is recognized by NLRC4 is flagellin.38,39,107 Accordingly, a L. pneumophilia mutant lacking flagellin is unable to activate NLRC4 and avoids clearance due to a defective lysosomal fusion of phagosomes, and thus efficiently replicates in NLRC4-deficient macrophages.107 Also the flagellin-deficient S. typhimurium fliC-fljB or flhCD mutants fail to activate NLRC4, whereas cytosolic delivery of flagellin is sufficient.38,39 NLRC4 recognizes a conserved C-terminal 35-amino-acid peptide in flagellin in addition to another region.110
Flagellin is not the only PAMP recognized. P. aeruginosa recognition by NLRC4 is independent of flagellin, since the flagellin-deficient P. aeruginosa mutant PAKΔfliC is still capable of activating caspase-1 by an NLRC4-dependent mechanism.109 Similarly, the non-flagellated S. flexneri is sensed by NLRC4, indicating that other structures are also recognized by NLRC4.108 NLRC4 recognition of L. pneumophilia depends on a competent type IV secretion system.107 Similarly, S. typhimurium- and S. flexneri-mediated caspase-1 activation requires a functional type III secretion system39,108,111–113 The S. typhimurium type III secretion system rod structure encoded by PrgJ from the pathogenicity island-1 (SPI1), but not the rod SsaI from SPI2, resembles the hollow flagellin tube, which is injected into the host cell, and both share a conserved C-terminal peptide sequence sufficient for NLRC4 detection and inflammasome activation. This demonstrates that in addition to flagellin, the type III secretion system itself is also recognized by NLRC4.114 Other rod proteins recognized by NLRC4 are BsaK from Burkholderia pseudomallei, EprJ and EscI from E. coli, MxiI from S. flexneri, and PscI from P. aeruginosa, indicating a conserved mechanism of NLRC4 recognition.114
Activation of NLRC4 also induces pyroptosis of macrophages. Depending on the pathogen, macrophage death is either beneficial for pathogen spread and selectively induced, or is a mechanism to suppress or subvert macrophage-mediated inflammatory host responses.115,116S. flexneri and P. aeruginosa-induced pyroptosis is blocked in cells lacking caspase-1 or NLRC4, but not in cells lacking ASC, although caspase-1 activation requires both NLRC4 and ASC, suggesting two distinct mechanisms.108,109
A PRR role for NLRB1 (NLR containing a baculovirus inhibitor of apoptosis domain-1; also known as NAIP5, BIRC1e, and CLR5.1) has been proposed due to its ability to restrict L. pneumophilia infection in vitro and in vivo.117–120 Although NLRB1 does not have a CARD or PYD, its signaling is dependent on caspase-1 (Figs. 1 & 2). While ASC is not required for L. pneumophilia growth restriction by NLRB1, it does require NLRC4.118 NLRB1 also recognizes flagellin and detects the same conserved region in flagellin as NLRC4.110,119 While NLRB1, but not NLRC4, is essential to restrict L. pneumophilia infections via recognition of flagellin, flagellin-dependent recognition of P. aeruginosa and S. typhimurium is mediated by both NLRC4 and NLRB1.110 Similarly, L. monocytogenes infection activates several NLRs, including NLRC4, NLRP3, and AIM2.121–123 While humans encode one NLRB1, mice encode seven paralogous genes (Naip1–7).124
E. Nod1
Like Nod2, Nod1 (also known as NLRC1, CARD4, and CLR7.1) contains a CARD and has a well-characterized role in activation of NF-κB and MAPKs via its adaptor RIP2 in protein platforms called nodosomes (Figs. 1 & 2).125,126 Also similar to Nod2, Nod1 detects breakdown products from peptidoglycans, specifically meso-diaminopimelic acid from gram-negative bacteria.127,128 There is very limited evidence for an involvement of Nod1 in inflammasome activation. However, based on an overexpression study, Nod1 interacts with pro-caspase-1 by CARD interaction, causing its activation and maturation of IL-1β.129 There is also indirect evidence for this interaction, since meso-diaminopimelic acid-induced caspase-1 activation and IL-1β release correlates with Nod1 expression in uveitis.130
F. NLRP12
Little is known about NLRP12 (also known as Monarch-1, NALP12, PYPAF7, RNO2, PAN6, and CLR19.3), which can inhibit NF-κB.131–133 NLRP12 also has ATPase activity, which is required for its NF-κB-inhibitory function.44 However, based on overexpression studies, NLRP12 can also interact with ASC to form a caspase-1 activating inflammasome (Fig. 2).34
G. AIM2
AIM2 belongs to the HIN-200 (hematopoietic interferon-inducible nuclear protein with a 200-amino acid repeat; also known as p200 or PYHIN) family of interferon-response genes, which consists of four human (MNDA, IFI16, IFIX, and AIM2) and six mouse proteins, with AIM2 being the most conserved member.134 HIN-200 proteins share a conserved domain architecture consisting of an N-terminal PYD (except p202a and p202b) and one or two copies of the HIN-200 domain (Figs. 1 & 2). Although HIN-200 members were initially considered to be transcriptional regulators of cell proliferation and differentiation, it has been recently established that AIM2 functions as a PRR by directly binding to cytosolic double-stranded DNA (dsDNA). The DNA-binding capacity of the HIN-200 proteins was recognized early on, and is thought to be responsible for their multiple functions in transcriptional regulation.135–139 DNA binding occurs by the HIN-200 domain, which consists of two consecutive oligonucleotide-/oligosaccharide-binding domains.140
AIM2 is the only cytosolic HIN-200 protein and the sole HIN-200-binding partner of ASC.141,142 Based on these requirements, AIM2 has been identified as a cytosolic PRR that activates an inflammasome upon recognition of cytosolic dsDNA but not single-stranded DNA.141–144 In agreement, reconstitution of AIM2 inflammasomes in human embryonic kidney (HEK) 293 cells resulted in a fully functional inflammasome upon transfection of dsDNA.141,145 AIM2 recognizes cytosolic dsDNA of any origin and size, including synthetic DNA (poly dA:dT), DNA isolated from vaccinia virus, plasmid DNA, bacterial DNA, and genomic DNA. AIM2 oligomerizes upon DNA binding and recruits ASC to activate caspase-1.142 Accordingly, knockdown of AIM2 abrogates caspase-1 activation and IL-1β maturation in response to dsDNA, but not other inflammasome agonists.141–144
The essential role of AIM2 in dsDNA-induced inflammasome activation has been validated in macrophages from AIM2-deficient mice, which fail to activate caspase-1 and fail to release IL-1β in response to transfected dsDNA, but not to other inflammasome activators.122,145 Also, infection of AIM2−/− macrophages with the bacterial pathogens Francisella tularensis and L. monocytogenes, as well as the DNA viruses vaccinia virus, herpes simplex virus-1, and mouse cytomegalovirus, results in impaired caspase-1 activation and IL-1β release.122,145 As discussed above, L. monocytogenes is also sensed by other PRRs such as NLRC4 via flagellin, while a flagellin-deficient mutant is primarily sensed by AIM2 inflammasomes.123 AIM2-deficient mice are also more susceptible to infection by F. tularensis, which is not sensed by other inflammasomes, and lack IL-18 production in response to mouse cytomegalovirus infection resulting in higher virus titers than controls.122,145 Analogous to other inflammasomes, the AIM2 inflammasome also activates pyroptosis, and AIM2−/− macrophages are protected from caspase-1-induced cell death.122,145 The dsDNA inflammasome response in mice is also regulated by p202, another HIN-200 member, which has no human ortholog. Because p202 binds dsDNA similarly to AIM2 (p210), but lacks the PYD, it cannot interact with ASC to assemble an inflammasome, and thus blocks dsDNA-induced caspase-1 activation. Therefore which causes knockdown of p202, increasing caspase-1 activation in response to dsDNA.144
H. Pyrin
Pyrin is the founding member of the PYD family. Like AIM2, it has a different domain architecture from NLRs. It consists of a PYD, a central BBox/ Coiled Coil (CC), and a C-terminal PRY/SPRY domain (Figs. 1 & 2). Pyrin interacts with several inflammasome proteins, but its role in inflammasome activation is still controversial. Evidence for its inflammasome-activating function came recently from the discovery that pyrin specifically interacts with PSTPIP1 (proline serine threonine phosphatase-interacting protein 1), also known as cluster of differentiation-2 (CD2) binding protein-1 (CD2BP1).146 This association is mediated by the BBox/CC in pyrin and by the SH3 (Src homology-3)/CC region in PSTPIP1, and is markedly increased in PSTPIP1 harboring pyogenic arthritis, pyoderma gangrenosum, and acne (PAPA) syndrome-associated mutations such as A230T and E250Q.146 These PSTPIP1 mutations also cause increased IL-1β levels in monocytes. Pyrin can form ASC and caspase-1 containing inflammasomes to promote IL-1β release in reconstituted inflammasomes.33,64,147 On endogenous level, PSTPIP1 binding to pyrin promotes the unmasking of its PYD and the interaction with ASC, to recruit caspase-1, which is enhanced by the presence of PSTPIP1 proteins harboring the PAPA-linked mutations.148 Any pathogens or PAMPs that are potentially sensed by pyrin remain elusive.
Pyrin is transcriptionally up-regulated in response to pro-inflammatory stimuli and inhibited by anti-inflammatory cytokines in myeloid cells.149 Furthermore, type I interferons and retroviral infection induces expression of pyrin, raising the possibility that pyrin is involved in the antiviral host response.148 Pyrin is also known as TRIM20 (tripartite motif-containing protein 20). Other TRIM family members, most notably TRIM5α, function to restrict retroviral replication via SPRY domain-mediated viral capsid recognition. Furthermore, TRIM22 mediates its antiviral activity by directly interacting with the HIV core.150–152 Zebrafish contain several hybrid proteins comprised of NLRs and the SPRY of pyrin, further suggesting a role of pyrin in pattern recognition. A significant number of familial Mediterranean fever (FMF)-linked mutations in pyrin are located inside the SPRY domain, further emphasizing the importance of this domain in promoting inflammasome activation.153
In addition to its role in assembling an inflammasome, there is also evidence for an inflammasome-antagonistic role of pyrin. Macrophages expressing a truncated pyrin protein retaining only the PYD express increased levels of active caspase-1 and are hyperactive with regard to secreting IL-1β.154 The PYD of pyrin is still expressed in these cells. Because pyrin interacts with ASC through their PYDs to modulate downstream signaling pathways, expression of the pyrin-PYD could potentially affect IL-1β maturation.64,155,156 This result was confirmed by the observation that knockdown of pyrin in the human THP-1 monocytic cell line augmented release of IL-1β, providing further evidence that pyrin potentially negatively regulates inflammasomes.157,158 Mechanistically, pyrin interacts via its SPRY domain with several inflammasome components, including the NACHT domains of NLRP1, NLRP2, and NLRP3; the caspase domain of caspase-1; and the pro-domain of pro-IL-1β, suggesting that pyrin potentially complexes with these proteins to prevent their activation. Furthermore, the commonly FMF-associated pyrin mutations M694V and M680I impair the interaction of pyrin with caspase-1, thus providing a possible explanation for the excessive IL-1β production reported in FMF patients.158 However, in another study, knockdown of pyrin in the same THP-1 cell line and in primary human monocytes impaired maturation of IL-1β, suggesting that the limited pyrin expression in macrophages is the cause of their reduced ability to release IL-1β.147 In addition, pyrin itself is cleaved by caspase-1, and the N-terminal fragment comprised of the PYD and the bZIP (basic leucine zipper) domain activates NF-κB.159 This fragment directly interacts with p65 and potentially acts as a transcription factor, and it also binds to IκBα (inhibitor of NF-κB-alpha), which induces calpain-mediated degradation of IκBα and activation of NF-κB.159 Certainly, activation of NF-κB is essential for transcription of IL-1β. Consequently, the precise role of pyrin in IL-1β maturation is still controversial and is likely to be dependent on the context of cellular activation.
V. NOD-LIKE RECEPTORS: LINK TO AUTOINFLAMMATORY AND AUTOIMMUNE DISEASES
Because IL-1β and IL-18 play such essential and prominent roles in inflammation and innate immunity, it is not surprising that excessive production of both cytokines is closely linked to an expanding number of periodic fever syndromes and autoinflammatory diseases.160 The term “systemic autoinflammatory disease” was first described for the TNFR1 (tumor necrosis factor receptor-1)-associated periodic syndromes,161 and has since been expanded to include an increasing number of self-directed tissue-inflammatory conditions that are generally characterized by the absence of high-titer autoantibodies and antigen-specific T cells, thus having limited autoimmune origin.162 The more details emerge on inflammasomes and their activation, the more evidence also points to a direct contribution of inflammasome defects, such as hereditary mutations in inflammasome activators leading to exaggerated immune responses in, IL-1β inflammasomopathies.
A. Pyrin
FMF is a recessively inherited, prototypic autoinflammatory disease characterized by repeated and recurrent episodes of fever, which can be asymptomatic or coincide with abdominal pain; signs of peritonitis; joint pain in primarily one large joint; pain in the chest with pleuritis and pericarditis, myalgia, or erysipeloid; and elevated acute-phase reactions that can lead to secondary amyloidosis.163 Positional cloning identified the FMF-linked gene, which was called MEFV (Mediterranean fever gene) and which encodes pyrin.164,165 There have been 193 hereditary mutations in MEFV linked to FMF, with the majority being single-nucleotide substitutions that cluster in exon 2, which contains the bZIP, and exon 10, which contains PRY/SPRY (Fig. 2). Fewer mutations are found in exon 3 (containing the BBox) and exon 5 (see Infevers, an online database for autoinflammatory mutations, at http://fmf.igh.cnrs.fr/ISSAID/infevers).166–168 As discussed above, the precise inflammatory role of pyrin is still controversial, and it is feasible that it has a dual physiological role. If pyrin has a predominantly anti-inflammatory role, then the recessively inherited FMF mutations could render the protein defective, causing a loss-of-function phenotype. Alternatively, if a pro-inflammatory role with inflammasome assembly is the main function of pyrin, then the dominant inherited FMF mutations could manifest as a gain-of-function phenotype.169
Notably, some of the most common mutations are located inside the PRY/SPRY domain, including M694V, V726A, M680I, M694I, together with E148Q in exon 2, which account for more than 70% of all cases.170 This high frequency can only be explained by a positive selection mechanism, which possibly acts by conferring resistance to infection.169 This hypothesis is supported by the observations that some of the FMF mutations inside the PRY/SPRY domain, including M680I and V726A, are found in wild-type primate alleles, indicating that some FMF mutations recapitulate an ancestral state and that primates might not be exposed to this putative pathogen.171 Furthermore, the PRY/SPRY domain is missing in rodents, and therefore is a recent evolutionary development, further emphasizing a human-specific function.172 Several of the severe FMF mutations localize close to a putative binding pocket in the PRY/SPRY domain, suggesting that pyrin could potentially directly recognize a pathogen.153
B. NLRP3
NLRP3, initially known as cryopyrin and encoded by the CIAS1 gene, was initially cloned based on its frequent mutation in the hereditary fever syndromes familial cold autoinflammatory syndrome (FCAS, also known as familial cold urticaria or FCU) and Muckle-Wells syndrome (MWS) (Fig. 2).173 Subsequently, mutations in NLRP3 have also been identified in patients with neonatal-onset multisystem inflammatory disease (NOMID, also known as chronic infantile neurological cutaneous and articular syndrome or CINCA).174,175 NLRP3 inflammasome-linked disorders are now collectively referred to as cryopyrinopathies or CIAS1-associated periodic syndromes (CAPS), and are caused by autosomal dominant or de novo mutations in NLRP3, which are generally linked to excessive production and maturation of IL-1β.153 These disorders usually develop during childhood, and are characterized by unexplained episodes of fever and severe localized inflammation. Monocytes from cryopyrinopathy patients also display a deranged redox homeostasis, characterized by elevated ROS levels and thus a more accelerated IL-1β maturation, which might partially explain the episodic nature of this disorder.176 NOMID is characterized by recurrent episodes of fever, urticarial rash beginning in the neonatal period, chronic meningitis, uveitis, loss of hearing, and growth retardation.177 FCAS displays recurrent episodes of rash, cold-induced urticaria, fever, arthralgia, and conjunctivitis frequently triggered by cold exposure.177 MWS patients have fever, progressive loss of hearing, urticaria, arthralgia, and rare amyloidosis.177 At least 120 mutations are described in NLRP3, although not all are clearly linked to cryopyrinopathies. Most of these mutations are located in exon 3, which encodes the NACHT domain and the flanking sequences, and represent single-nucleotide substitutions.166–168 NLRP3 mutations cause spontaneous oligomerization, recruitment of ASC, inflammasome activation, and elevated production of IL-1β.42,64
Recently, mouse models of cryopyrinopathies that reflect the human IL-1β-dependent diseases have been generated by introducing FCAS and MWS mutations (A350V and L351P, corresponding to humans A352V and L353P), or by using the shared R258W mutation (corresponding to human R260W) into Nlrp3, and also show a skewed Th17 response.178,179 Most importantly, the mutations do not cause spontaneous IL-1β release, but macrophages isolated from these mice display significantly higher sensitivity to TLR ligands and do not require exogenous ATP for IL-1β release.
NLRP3 polymorphisms are also linked to a number of autoimmune conditions. Gout and pseudogout are common autoinflammatory arthritides, which are caused by deposition of monosodium urate and calcium pyrophosphate dihydrate crystals in articular and periarticular tissues. Monosodium urate is a danger signal and a potent activator of the NLRP3 inflammasome.66 Cholesterol crystals are also recognized by NLRP3 inflammasomes, and the inflammasome contributes to atherosclerosis in mice.84 Fibrillar β-amyloid also activates the NLRP3 inflammasome, which is linked to the pathology of Alzheimer's disease, and silicosis and fibrosis are at least partially dependent on the NLRP3 inflammasome.83,85,86,180 Schnitzler syndrome is a rare disease characterized by a chronic urticarial eruption and monoclonal gammopathy, and the symptoms include bone pain, fever, arthralgia, lymphadenopathy, leucocytosis, hepatomegaly and/or splenomegaly. NLRP3 (V198M) predisposes to Schnitzler syndrome and dysregulated NLRP3 inflammasomes contribute to the pathogenesis of Schnitzler syndrome.181,182
Crohn's disease is a multifactorial, chronic inflammatory bowel disease mostly affecting colon and terminal ileum, which is characterized by excessive production of NF-κB-dependent proinflammatory cytokines, including IL-1β. Mutations and polymorphisms in NLRP3 (Q705K) alone and in combination with Cardinal (C10X), contribute to increased IL-1β production and the susceptibility to Chrohn's disease, and polymorphisms in both genes also influence the severity of rheumatoid arthritis.183–186 Two NLRP3 single-nucleotide polymorphisms (rs4612666 and rs10754558) are significantly associated with the susceptibility to food allergy, food-induced anaphylaxis, and aspirin-induced asthma, and also predispose to type-1 diabetes and celiac disease, indicating that derailed NLRP3 inflammasomes affect the pathogenesis of multiple diseases, further emphasizing the central role of NLRP3.187,188
C. NLRP1
Generalized vitiligo is a polygenic, multifactorial, non-contagious disorder that causes autoimmune loss of melanocytes and results in progressive loss of pigmentation from skin, overlying hair, and oral mucosa. Polymorphisms in NLRP1 predispose to generalized vitiligo and associated autoimmune diseases autoimmune thyroid disease, latent autoimmune diabetes in adults, rheumatoid arthritis, psoriasis, pernicious anemia, systemic lupus erythematosus, and Addison's disease (Fig. 2).189–192 Polymorphisms are located within the promoter region and could affect transcription of NLRP1 or cause one amino acid substitution (L155H), but whether these mutations affect inflammasome activity warrants further investigation.
D. NLRP2
As discussed above, little is known about NLRP2-mediated inflammasome activation. Beckwith-Wiedemann syndrome (BWS), a genetically complex congenital overgrowth syndrome, is a genomic imprinting disorder caused by mutation or altered expression of genes located in the imprinted 11p15.5 chromosomal region that are implicated in cell cycle and growth control, such as IGF2 (insulin-like growth factor-2), KCNQ1oT1 (potassium voltage-gated channel KQT-like subfamily member-1 [KCNQ1] overlapping transcript-1), and CDKN1C (cyclin-dependent kinase inhibitor 1C).193 A positional-candidate gene approach identified a homozygous frameshift mutation in exon 6 of NLRP2 in the mother of a BWS patient (Fig. 2).194 The frameshift c.1479delAG causes p.Arg493SerfsX32, resulting in a truncated protein lacking 539 amino acids encoding the LRR. The disturbed imprinting of BWS patients suggests a role of NLRP2 in establishing or maintaining genomic imprinting/methylation.194 This scenario is intriguing, because methylation might have evolved as a transposon defense mechanism. However, BWS patients do not show any inflammation.194,195
E. NLRP7
NLRP7 has been implicated in the regulation of caspase-1 activity based on one overexpression study. Therefore, information on its contribution, to inflammasome activation is also very limited.196 Familial recurrent hydatidiform mole (FRHM) is an autosomal-recessive condition that results in abnormal human pregnancy with no embryo and hydropic degeneration of chorionic villi.197 Like BWS, FRHM is a genomic imprinting disorder, thus linking a second NLR to imprinting defects. There have been 145 NLRP7 mutations described, and at least 23 hereditary mutations in NLRP7 have been identified as a causative for FRHM. Women homozygous or compound heterozygous for NLRP7 mutations were shown to be at risk for reproductive wastage (Fig. 2).166–168 Several NLRP7 mutations linked to FRHM have been identified, and the majority of the mutations affect the LRRs or the NACHT of NLRP7.198–205 There is speculation that FRHM might result from an immune-related defect during oogenesis or early embryonic development, but there is no information about whether NLRP7 mutations could cause gain-of-function, similar to cryopyrinopathies, or loss-of-function, as shown for Nod2 mutations in Crohn's disease. NLRP7 mutations might not be linked to inflammasomes after all, because NLRP7 has a role in cell proliferation, is up-regulated in testicular seminoma cells, and gene silencing of NLRP7 suppresses cell growth.206 Expression levels of NLRP7 are also correlated with the myometrical invasion in endometrial cancer.207
F. NLRP12
Little is known about any inflammasome-activating role of NLRP12, but using a candidate gene approach, nonsense and splice site mutations were identified in the NLRP12 gene in two families with a hereditary periodic fever syndromes referred to as Guadeloupe variant fever syndrome (FCAS2) (Fig. 2).208 FCAS2 causes clinical symptoms similar to those of FCAS and MWS, including cold-induced fever episodes, arthralgia, and myalgia. At least 25 mutations have been identified, and two mutations, c.2072+3insT, which causes a deletion of the LRRs, and R284X, which truncates the protein within the NACHT, are linked to FCAS2.208 NLRP12 (R284X) is significantly impaired in NF-κB inhibition, but NLRP12 (c.2072+3insT) is also less potent in NF-κB inhibition than the wild-type protein.208 Whether these mutations also affect inflammasome activity is not known.
G. NLRB1
Spinal muscular atropy (SMA) is an autosomal-recessive neuromuscular disease characterized by the degeneration of motor neurons in the spinal cord, which results in progressive weakness and wasting of voluntary muscles; IL-1β contributes to SMA.209 Deletions and mutation of NLRB1 have been shown to cause SMA (Fig. 2).210
H. NOD1 and NOD2
Autosomal dominant mutations in Nod2, mostly within the NACHT domain, are linked to Blau syndrome, which is characterized by granulomatous dermatitis, arthritis, and uveitis (Fig. 2).211 Blau syndrome-associated Nod2 mutations are gain-of-function mutations characterized by elevated NF-κB activation, and are mostly clustered in the central NACHT domain.166–168 In addition, one of the inflammatory bowel disease risk alleles is Nod2, and Nod2 mutations are linked to the susceptibility to Crohn's disease, which is characterized by diarrhea and abdominal pain.212,213 More than 77 mutations are linked to Crohn's disease and 24 to ulcerative colitis, which partially overlap with Crohn's disease; the most common ones are a frameshift mutation (c.L1007fsinsC) and substitutions within the LRR, such as R702W and G908R.213 In contrast to inflammatory bowel disease, the Crohn's disease-associated mutations reflect a loss-of-function phenotype.214 The Crohn's disease phenotype could result from reduced epithelial cell barrier function and α-defensin expression, and thus impaired growth restriction and clearance of invasive bacteria such as S. typhimurium.215–217 Accordingly, Crohn's disease-linked mutations show impaired cytokine release, including IL-1β.218,219 However, macrophages engineered for Nod2 Crohn's disease mutations show elevated IL-1β secretion, suggesting that mutations could rather reflect a gain-of-function mutation.220 Nod1 polymorphisms are also linked to susceptibility to Crohn's disease (Fig. 2).221,222 Seven gain-of-function Nod2 mutations are linked to early-onset sarcoidosis.223,224 Furthermore, the c.802C>T polymorphism is associated with chronic gastritis and predisposition to cancer in H. pylori-infected patients.225,226
VI. INFLAMMASOMES: A TARGET FOR THERAPEUTIC INTERVENTION
Direct IL-1β inhibitors, such as recombinant IL-1Ra (anakinra), IL-1TRAP (rilonacept), or humanized IL-1β antibody (canakinumab), are effective in several IL-1β autoinflammatory conditions, most prominently the cryopyrinopathies, but are also effective in rheumatic diseases and psoriasis.227–229 Based on the interaction of IL-1Ra with the IL-1RI, the small peptide antagonist peptide 101.1 was developed, and is effective in vivo in inflammatory bowel disease and phorbol 12-myristate 13-acetate-induced dermatitis mouse models.230 Caspase-1 has also been directly targeted (pralnacasan and VX765), which reduced several inflammatory conditions in vivo despite some toxicity.87,231,234 An adverse effect, as for any targeted biological anti-cytokine therapy, is that global cytokine blocking is linked to a slight increase in emerging opportunistic infections.235,236 Thus, a more targeted approach could be superior, such as specifically inhibiting a particular NLR. Colchicine, a microtubule inhibitor, is a standard treatment for FMF and gout, which not only impairs neutrophil migration and cytokine production, but also blocks phagocytosis.237 In addition, colchicine and nocodazole, another microtubule inhibitor, reduce the interaction of PSTPIP1 with pyrin to block pyrin-induced activation of caspase-1.148 The type 2 diabetes drug glyburide (glibenclamide), which inhibits ATP-sensitive K+ channels, blocks the activation of the NLRP3 inflammasome by a distinct mechanism downstream of the P2X7R, and is the first NLRP-specific drug.238 Potentially, the pannexin peptide could be used to similarly block NLRP3 activation, although no in vivo application has yet been reported.72 Increasing evidence points to an intrinsic ATPase activity of NLRs, which is required for receptor function, whether this is modulation of NF-κB in the case of NLRP12 or activation of caspase-1 by NLRP1 and NLRP3.36,43,44 Thus, one potentially could explore exploiting the ATP-binding pocket specific for any given NLR as an NLR-targeted therapy.
VII. CONCLUSIONS
The study of NLRs and related PRRs and the discovery of the inflammasome have offered tremendous insights into innate immunity and host defense over the past decade. They have also provided molecular mechanisms for the pathogenesis of inflammatory diseases and for adjuvant activity, and thus enabled the availability of novel treatment opportunities, such as anakinra and its derivatives and the prospect for novel therapies specifically targeting activation of inflammasomes for auto-inflammatory disease patients. However, there is still much room for further investigation, considering that we currently only have limited understanding of the nature of agonists initiating activation and of the physiological and potentially pathological consequences resulting from activation of the majority of NLRs.
ACKNOWLEDGEMENTS
Work in our laboratory is supported by National Institutes of Health grants GM071723, AI082406-02, and HL097183. S.K. is a recipient of a postdoctoral fellowship from the Arthritis Foundation (AF161715) and N.L. is a recipient of a predoctoral fellowship from the American Heart Association (10PRE2630172). We apologize to investigators whose contributions could not be cited due to space constraints.
ABBREVIATIONS
- AIM2
absent in melanoma-2
- ASC
apoptosis-associated speck-like protein containing a caspase recruitment domain
- bZIP
basic leucine zipper
- BWS
Beckwith-Wiedemann syndrome
- CAPS
cold-induced autoinflammatory syndrome-1 (CIAS1) gene-associated periodic syndrome
- CARD
caspase recruitment domain
- CC
coiled coil
- CD2BP1
cluster of differentiation-2 (CD2) binding protein-1
- CDKN1C
cyclin-dependent kinase inhibitor 1C
- CIAS1
cold-induced autoinflammatory syndrome-1
- CINCA
chronic infantile neurological cutaneous and articular syndrome
- DAMP
danger-associated molecular pattern
- dsDNA
double-stranded DNA
- HEK293
human embryonic kidney 293 cell line
- HSP90
heat-shock protein 90
- IκBα
inhibitor of nuclear factor of kappa light chain enhancer of activated B-cells (NF-κB) alpha
- IGF2
insulin-like growth factor-2
- IKK
IκB kinase
- IL
interleukin
- IL-18BP
interleukin-18 binding protein
- IL-1R
interleukin-1 receptor
- IL-1Ra
interleukin-1 receptor antagonist
- ERK
extra-cellular signal-regulated kinase
- FCAS
familial cold autoinflammatory syndrome
- FCAS2
Guadeloupe variant fever syndrome
- FCU
familial cold urticaria
- FIIND
function-to-find domain
- FMF
familial Mediterranean fever
- FRHM
familial recurrent hydatidiform mole
- KCNQ1oT1
potassium voltage-gated channel KQT-like subfamily member-1 (KCNQ1) overlapping transcript-1
- LeTx
lethal toxin
- LRR
leucine-rich repeat
- MEFV
Mediterranean fever gene
- MAPK
mitogen-activated protein kinase
- MARTX
multifunctional repeat-in toxins, MDP, muramyl dipeptide
- MWS
Muckle-Wells syndrome
- NAD
NACHT-associated domain
- NF-κB
nuclear factor kappa light-chain enhancer of activated B cells
- NLR
Nod-like receptor
- NLRB
NLR containing a baculovirus inhibitor of apoptosis domain
- NLRC
NLR containing a CARD
- NLRP
NLR containing a PYD
- Nod
nucleotide-binding oligomerization domain
- NOMID
neonatal-onset multisystem inflammatory disease
- P2X7R
ATP-activated purinergic (P2X) receptor-7
- PAI-2
plasminogen activator inhibitor-2
- PAMP
pathogen-associated molecular pattern
- PAPA
pyogenic arthritis, pyoderma gangrenosum and acne
- PI3K
phosphoinositide 3-kinase
- PRR
pattern recognition receptor
- PSTPIP1
proline serine threonine phosphatase-interacting protein-1
- PYD
pyrin N-terminal homology domain
- RIG1
retinoic acid-inducible gene-1
- RIP2
receptor-interacting protein-2
- RLR
retinoic acid inducible gene-1 like receptor
- ROS
reactive oxygen species
- SH3
Src homology-3
- SGT1
suppressor of G2 allele of SKP1
- SMA
spinal muscular atropy
- SREBP
sterol regulatory element-binding protein
- Syk
spleen tyrosine kinase
- TIR
Toll/interleukin-1 receptor
- TLR
Toll-like receptor
- TNFR1
tumor necrosis factor receptor-1
- TRIM
tripartite motif-containing protein
- TRX
thioredoxin
- TXNIP
thioredoxin interacting protein
REFERENCES
- 1.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–44. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. TLR signaling. Semin Immunol. 2007;19:24–32. doi: 10.1016/j.smim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–22. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Jha S, Ting JP. Inflammasome-associated nucleotide-binding domain, leucine-rich repeat proteins and inflammatory diseases. J Immunol. 2009;183:7623–9. doi: 10.4049/jimmunol.0902425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenbarth SC, Flavell RA. Innate instruction of adaptive immunity revisited: the inflammasome. EMBO Mol Med. 2009;1:92–8. doi: 10.1002/emmm.200900014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–65. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, Shaw MH, Kim YG, Nuñez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol. 2009;4:365–98. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 9.Alnemri ES. Sensing cytoplasmic danger signals by the inflammasome. J Clin Immunol. 2010 Jul;30(4):512–9. doi: 10.1007/s10875-010-9419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–50. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 11.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, Wong W, Kamen R, Tracey D, Allen H. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature. 1997;386:619–23. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 12.Li P, Allen H, Banerjee S, Franklin S, Herzog L, Johnston C, McDowell J, Paskind M, Rodman L, Salfeld J. Mice deficient in IL-1b-converting enzyme are defective in production of mature IL-1b and resistant to endotoxic shock. Cell. 1995;80:401–11. doi: 10.1016/0092-8674(95)90490-5. [DOI] [PubMed] [Google Scholar]
- 13.Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, Flavell RA. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–3. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- 14.Netea MG, Nold-Petry CA, Nold MF, Joosten LA, Opitz B, van der Meer JH, van de Veerdonk FL, Ferwerda G, Heinhuis B, Devesa I, Funk CJ, Mason RJ, Kullberg BJ, Rubartelli A, van der Meer JW, Dinarello CA. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood. 2009;113:2324–35. doi: 10.1182/blood-2008-03-146720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stehlik C. Multiple interleukin-1beta-converting enzymes contribute to inflammatory arthritis. Arthritis Rheum. 2009;60:3524–30. doi: 10.1002/art.24961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller M, Rüegg A, Werner S, Beer HD. Active caspase-1 is a regulator of unconventional protein secretion. Cell. 2008;132:818–31. doi: 10.1016/j.cell.2007.12.040. [DOI] [PubMed] [Google Scholar]
- 17.Gurcel L, Abrami L, Girardin S, Tschopp J, van der Goot FG. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 2006;126:1135–45. doi: 10.1016/j.cell.2006.07.033. [DOI] [PubMed] [Google Scholar]
- 18.Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–32. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Brennan MA, Cookson BT. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol. 2000;38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- 21.Cervantes J, Nagata T, Uchijima M, Shibata K, Koide Y. Intracytosolic Listeria monocytogenes induces cell death through caspase-1 activation in murine macrophages. Cell Microbiol. 2008;10:41–52. doi: 10.1111/j.1462-5822.2007.01012.x. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes-Alnemri T, Wu J, Yu JW, Datta P, Miller B, Jankowski W, Rosenberg S, Zhang J, Alnemri ES. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–16. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujisawa A, Kambe N, Saito M, Nishikomori R, Tanizaki H, Kanazawa N, Adachi S, Heike T, Sagara J, Suda T, Nakahata T, Miyachi Y. Disease-associated mutations in CIAS1 induce cathepsin B-dependent rapid cell death of human THP-1 monocytic cells. Blood. 2007;109:2903–11. doi: 10.1182/blood-2006-07-033597. [DOI] [PubMed] [Google Scholar]
- 26.Willingham SB, Bergstralh DT, O'Connor W, Morrison AC, Taxman DJ, Duncan JA, Barnoy S, Venkatesan MM, Flavell RA, Deshmukh M, Hoffman HM, Ting JP. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2:147–59. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-1b. Mol. Cell. 2002;10:417–26. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 28.Nadiri A, Wolinski MK, Saleh M. The inflammatory caspases: key players in the host response to pathogenic invasion and sepsis. J Immunol. 2006;177:4239–45. doi: 10.4049/jimmunol.177.7.4239. [DOI] [PubMed] [Google Scholar]
- 29.Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol Cell. 2003;11:529–41. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 30.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–8. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto M, Yaginuma K, Tsutsui H, Sagara J, Guan X, Seki E, Yasuda K, Yamamoto M, Akira S, Nakanishi K, Noda T, Taniguchi S. ASC is essential for LPS-induced activation of procaspase-1 independently of TLR-associated signal adaptor molecules. Genes Cells. 2004;9:1055–67. doi: 10.1111/j.1365-2443.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–22. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- 33.Stehlik C, Lee SH, Dorfleutner A, Stassinopoulos A, Sagara J, Reed JC. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. J Immunol. 2003;171:6154–63. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- 34.Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, Bertin J. PYPAF7: a novel PRYIN-containing Apaf1-like protien that regulates activation of NF-kB and Caspase-1-dependent cytokine processing. J Biol Chem. 2002;277:29874–80. doi: 10.1074/jbc.M203915200. [DOI] [PubMed] [Google Scholar]
- 35.Hsu LC, Ali SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, Eckmann L, Powell JJ, Nizet V, Dixit VM, Karin M. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A. 2008;105:7803–8. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faustin B, Lartigue L, Bruey JM, Luciano F, Sergienko E, Bailly-Maitre B, Volkmann N, Hanein D, Rouiller I, Reed JC. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol Cell. 2007;25:713–24. doi: 10.1016/j.molcel.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 37.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–25. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 38.Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozören N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Núñez G. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–82. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 39.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7:569–75. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 40.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galán JE, Askenase PW, Flavell RA. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–27. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Taxman DJ, Zhang J, Champagne C, Bergstralh DT, Iocca HA, Lich JD, Ting JP. Cutting edge: ASC mediates the induction of multiple cytokines by Porphyromonas gingivalis via caspase-1-dependent and -independent pathways. J Immunol. 2006;177:4252–6. doi: 10.4049/jimmunol.177.7.4252. [DOI] [PubMed] [Google Scholar]
- 42.Dowds TA, Masumoto J, Zhu L, Inohara N, Núñez G. Cryopyrin-induced interleukin 1beta secretion in monocytic cells: enhanced activity of disease-associated mutants and requirement for ASC. J Biol Chem. 2004;279:21924–8. doi: 10.1074/jbc.M401178200. [DOI] [PubMed] [Google Scholar]
- 43.Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, Zimmermann AG, Ting JP. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci U S A. 2007;104:8041–6. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye Z, Lich JD, Moore CB, Duncan JA, Williams KL, Ting JP. ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2008;28:1841–50. doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arthur JC, Lich JD, Aziz RK, Kotb M, Ting JP. Heat shock protein 90 associates with monarch-1 and regulates its ability to promote degradation of NF-kappaB-inducing kinase. J Immunol. 2007;179:6291–6. doi: 10.4049/jimmunol.179.9.6291. [DOI] [PubMed] [Google Scholar]
- 46.Mayor A, Martinon F, De Smedt T, Pétrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- 47.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP, Inohara N, Mackenzie A, Maltais LJ, Nunez G, Ogura Y, Otten LA, Philpott D, Reed JC, Reith W, Schreiber S, Steimle V, Ward PA. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–7. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–32. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci U S A. 1998 May 26;95(11):5857–64. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan Q, Mathison J, Fearns C, Kravchenko VV, Da Silva Correia J, Hoffman HM, Kobayashi KS, Bertin J, Grant EP, Coyle AJ, Sutterwala FS, Ogura Y, Flavell RA, Ulevitch RJ. MDP-induced interleukin-1beta processing requires Nod2 and CIAS1/NALP3. J Leukoc Biol. 2007;82:177–83. doi: 10.1189/jlb.1006627. [DOI] [PubMed] [Google Scholar]
- 51.Sarkar A, Duncan M, Hart J, Hertlein E, Guttridge DC, Wewers MD. ASC directs NF-kappaB activation by regulating receptor interacting protein-2 (RIP2) caspase-1 interactions. J Immunol. 2006;176:4979–86. doi: 10.4049/jimmunol.176.8.4979. [DOI] [PubMed] [Google Scholar]
- 52.Thome M, Hofmann K, Burns K, Martinon F, Bodmer JL, Mattmann C, Tschopp J. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol. 1998 Jul 16;8(15):885–8. doi: 10.1016/s0960-9822(07)00352-1. [DOI] [PubMed] [Google Scholar]
- 53.Humke EW, Shriver SK, Starovasnik MA, Fairbrother WJ, Dixit VM. ICEBERG: a novel inhibitor of interleukin-1beta generation. Cell. 2000 Sep 29;103(1):99–111. doi: 10.1016/s0092-8674(00)00108-2. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi K, Inohara N, Hernandez LD, Galán JE, Núñez G, Janeway CA, Medzhitov R, Flavell RA. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–9. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 55.Damiano JS, Oliveira V, Welsh K, Reed JC. Heterotypic interactions among NACHT domains: implications for regulation of innate immune responses. Biochem J. 2004;381:213–9. doi: 10.1042/BJ20031506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinon F, Agostini L, Meylan E, Tschopp J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr Biol. 2004;14:1929–34. doi: 10.1016/j.cub.2004.10.027. [DOI] [PubMed] [Google Scholar]
- 57.Boyden ED, Dietrich WF. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat Genet. 2006;38:240–4. doi: 10.1038/ng1724. [DOI] [PubMed] [Google Scholar]
- 58.Newman ZL, Leppla SH, Moayeri M. CA-074Me protection against anthrax lethal toxin. Infect Immun. 2009;77:4327–36. doi: 10.1128/IAI.00730-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park JM, Greten FR, Li ZW, Karin M. Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science. 2002;297:2048–51. doi: 10.1126/science.1073163. [DOI] [PubMed] [Google Scholar]
- 60.Park JM, Greten FR, Wong A, Westrick RJ, Arthur JS, Otsu K, Hoffmann A, Montminy M, Karin M. Signaling pathways and genes that inhibit pathogen-induced macrophage apoptosis--CREB and NF-kappaB as key regulators. Immunity. 2005;23:319–29. doi: 10.1016/j.immuni.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 61.Greten FR, Arkan MC, Bollrath J, Hsu LC, Goode J, Miething C, Göktuna SI, Neuenhahn M, Fierer J, Paxian S, Van Rooijen N, Xu Y, O'Cain T, Jaffee BB, Busch DH, Duyster J, Schmid RM, Eckmann L, Karin M. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell. 2007;130:918–31. doi: 10.1016/j.cell.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bruey JM, Bruey-Sedano N, Newman R, Chandler S, Stehlik C, Reed JC. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J Biol Chem. 2004;279:51897–907. doi: 10.1074/jbc.M406741200. [DOI] [PubMed] [Google Scholar]
- 63.Razmara M, Srinivasula SM, Wang L, Poyet JL, Geddes BJ, DiStefano PS, Bertin J, Alnemri ES. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277:13952–8. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- 64.Yu JW, Wu J, Zhang Z, Datta P, Ibrahimi I, Taniguchi S, Sagara J, Fernandes-Alnemri T, Alnemri ES. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13:236–49. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 65.Kanneganti TD, Ozören N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, Bertin J, Coyle A, Grant EP, Akira S, Núñez G. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006 Mar 9;440(7081):233–6. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- 66.Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 67.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Núñez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–8. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 68.Muruve DA, Pétrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–7. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- 69.Franchi L, Kanneganti TD, Dubyak GR, Núñez G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J Biol Chem. 2007;282:18810–8. doi: 10.1074/jbc.M610762200. [DOI] [PubMed] [Google Scholar]
- 70.Pétrilli V, Papin S, Dostert C, Mayor A, Martinon F, Tschopp J. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14:1583–9. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 71.Iyer SS, Pulskens WP, Sadler JJ, Butter LM, Teske GJ, Ulland TK, Eisenbarth SC, Florquin S, Flavell RA, Leemans JC, Sutterwala FS. Necrotic cells trigger a sterile inflammatory response through the Nlrp3 inflammasome. Proc Natl Acad Sci U S A. 2009;106:20388–93. doi: 10.1073/pnas.0908698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanneganti TD, Lamkanfi M, Kim YG, Chen G, Park JH, Franchi L, Vandenabeele P, Núñez G. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity. 2007;26:433–43. doi: 10.1016/j.immuni.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Cruz CM, Rinna A, Forman HJ, Ventura AL, Persechini PM, Ojcius DM. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J Biol Chem. 2007;282:2871–9. doi: 10.1074/jbc.M608083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Piccini A, Carta S, Tassi S, Lasiglié D, Fossati G, Rubartelli A. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1beta and IL-18 secretion in an autocrine way. Proc Natl Acad Sci U S A. 2008;105:8067–72. doi: 10.1073/pnas.0709684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gross O, Poeck H, Bscheider M, Dostert C, Hannesschläger N, Endres S, Hartmann G, Tardivel A, Schweighoffer E, Tybulewicz V, Mocsai A, Tschopp J, Ruland J. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature. 2009;459:433–6. doi: 10.1038/nature07965. [DOI] [PubMed] [Google Scholar]
- 76.Dostert C, Guarda G, Romero JF, Menu P, Gross O, Tardivel A, Suva ML, Stehle JC, Kopf M, Stamenkovic I, Corradin G, Tschopp J. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS One. 2009;4:e6510. doi: 10.1371/journal.pone.0006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shimada T, Park BG, Wolf AJ, Brikos C, Goodridge HS, Becker CA, Reyes CN, Miao EA, Aderem A, Götz F, Liu GY, Underhill DM. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe. 2010;7:38–49. doi: 10.1016/j.chom.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Franchi L, Eigenbrod T, Núñez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–6. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li H, Willingham SB, Ting JP, Re F. Cutting edge: inflammasome activation by alum and alum's adjuvant effect are mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Franchi L, Núñez G. The Nlrp3 inflammasome is critical for aluminium hydroxide-mediated IL-1beta secretion but dispensable for adjuvant activity. Eur J Immunol. 2008;38:2085–9. doi: 10.1002/eji.200838549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–56. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eisenbarth SC, Colegio OR, O'Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–6. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol. 2008;9:857–65. doi: 10.1038/ni.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nuñez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V, Latz E. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 2010;464:1357–61. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, Carter AB, Rothman PB, Flavell RA, Sutterwala FS. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci U S A. 2008;105:9035–40. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dostert C, Pétrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008 May 2;320(5876):674–7. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bauer C, Duewell P, Mayer C, Lehr HA, Fitzgerald KA, Dauer M, Tschopp J, Endres S, Latz E, Schnurr M. Colitis induced in mice with dextran sulfate sodium (DSS) is mediated by the NLRP3 inflammasome. Gut. 2010 Sep;59(9):1192–9. doi: 10.1136/gut.2009.197822. [DOI] [PubMed] [Google Scholar]
- 88.Terada K, Yamada J, Hayashi Y, Wu Z, Uchiyama Y, Peters C, Nakanishi H. Involvement of cathepsin B in the processing and secretion of interleukin-1beta in chromogranin A-stimulated microglia. Glia. 2010;58:114–24. doi: 10.1002/glia.20906. [DOI] [PubMed] [Google Scholar]
- 89.Hentze H, Lin XY, Choi MS, Porter AG. Critical role for cathepsin B in mediating caspase-1-dependent interleukin-18 maturation and caspase-1-independent necrosis triggered by the microbial toxin nigericin. Cell Death Differ. 2003;10:956–68. doi: 10.1038/sj.cdd.4401264. [DOI] [PubMed] [Google Scholar]
- 90.Toma C, Higa N, Koizumi Y, Nakasone N, Ogura Y, McCoy AJ, Franchi L, Uematsu S, Sagara J, Taniguchi S, Tsutsui H, Akira S, Tschopp J, Núñez G, Suzuki T. Pathogenic Vibrio activate NLRP3 inflammasome via cytotoxins and TLR/nucleotide-binding oligomerization domain-mediated NF-kappaB signaling. J Immunol. 2010;184:5287–97. doi: 10.4049/jimmunol.0903536. [DOI] [PubMed] [Google Scholar]
- 91.Ozören N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, Ertürk I, Jagirdar R, Zhu L, Inohara N, Bertin J, Coyle A, Grant EP, Núñez G. Distinct Roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–42. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- 92.Craven RR, Gao X, Allen IC, Gris D, Bubeck Wardenburg J, McElvania-Tekippe E, Ting JP, Duncan JA. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS One. 2009;4:e7446. doi: 10.1371/journal.pone.0007446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He X, Mekasha S, Mavrogiorgos N, Fitzgerald KA, Lien E, Ingalls RR. Inflammation and fibrosis during Chlamydia pneumoniae infection is regulated by IL-1 and the NLRP3/ ASC inflammasome. J Immunol. 2010;184:5743–54. doi: 10.4049/jimmunol.0903937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abdul-Sater AA, Saïd-Sadier N, Padilla EV, Ojcius DM. Chlamydial infection of monocytes stimulates IL-1beta secretion through activation of the NLRP3 inflammasome. Microbes Infect. 2010 Aug;12(8–9):652–61. doi: 10.1016/j.micinf.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas PG, Dash P, Aldridge JR, Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, Doherty PC, Kanneganti TD. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30:566–75. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med. 2009;206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30:556–65. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ichinohe T, Pang IK, Iwasaki A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol. 2010;11:404–10. doi: 10.1038/ni.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschläger N, Schlee M, Rothenfusser S, Barchet W, Kato H, Akira S, Inoue S, Endres S, Peschel C, Hartmann G, Hornung V, Ruland J. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–9. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 100.Shio MT, Eisenbarth SC, Savaria M, Vinet AF, Bellemare MJ, Harder KW, Sutterwala FS, Bohle DS, Descoteaux A, Flavell RA, Olivier M. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 2009;5:e1000559. doi: 10.1371/journal.ppat.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Saïd-Sadier N, Padilla E, Langsley G, Ojcius DM. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the Syk tyrosine kinase. PLoS One. 2010;5:e10008. doi: 10.1371/journal.pone.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106:2543–50. doi: 10.1182/blood-2005-03-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–40. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 104.Geddes BJ, Wang L, Huang WJ, Lavellee M, Manji GA, Brown M, Jurman M, Cao J, Morgenstern J, Merriam S, Glucksmann MA, DiStefano PS, Bertin J. Human CARD12 is a novel CED4/Apaf-1 family member that induces apoptosis. Biochem Biophys Res Comm. 2001;284:77–82. doi: 10.1006/bbrc.2001.4928. [DOI] [PubMed] [Google Scholar]
- 105.Damiano JS, Stehlik C, Pio F, Godzik A, Reed JC. CLAN, a novel human CED-4-like gene. Genomics. 2001;75:77–83. doi: 10.1006/geno.2001.6579. [DOI] [PubMed] [Google Scholar]
- 106.Poyet JL, Srinivasula SM, Tnani M, Razmara M, Fernandes-Alnemri T, Alnemri ES. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J Biol Chem. 2001 Jul 27;276(30):28309–13. doi: 10.1074/jbc.C100250200. [DOI] [PubMed] [Google Scholar]
- 107.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozören N, Brady G, Meshinchi S, Jagirdar R, Gewirtz A, Akira S, Núñez G. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem. 2006;281:35217–23. doi: 10.1074/jbc.M604933200. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, Mimuro H, Inohara N, Sasakawa C, Nuñez G. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sutterwala FS, Mijares LA, Li L, Ogura Y, Kazmierczak BI, Flavell RA. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J Exp Med. 2007;204:3235–45. doi: 10.1084/jem.20071239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lightfield KL, Persson J, Brubaker SW, Witte CE, von Moltke J, Dunipace EA, Henry T, Sun YH, Cado D, Dietrich WF, Monack DM, Tsolis RM, Vance RE. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008 Oct;9(10):1171–8. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hersh D, Monack DM, Smith MR, Ghori N, Falkow S, Zychlinsky A. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc Natl Acad Sci U S A. 1999;96:2396–401. doi: 10.1073/pnas.96.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen Y, Smith MR, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 1996;15:3853–60. [PMC free article] [PubMed] [Google Scholar]
- 113.Hilbi H, Moss JE, Hersh D, Chen Y, Arondel J, Banerjee S, Flavell RA, Yuan J, Sansonetti PJ, Zychlinsky A. Shigella-induced apoptosis is dependent on caspase-1 which binds to IpaB. J Biol Chem. 1998;273:32895–900. doi: 10.1074/jbc.273.49.32895. [DOI] [PubMed] [Google Scholar]
- 114.Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Aderem, Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107:3076–80. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Monack D, Falkow S. Apoptosis as a common bacterial virulence strategy. Int J Med Microbiol. 2000;290:7–13. doi: 10.1016/S1438-4221(00)80096-X. [DOI] [PubMed] [Google Scholar]
- 116.Navarre WW, Zychlinsky A. Pathogen-induced apoptosis of macrophages: a common end for different pathogenic strategies. Cell Microbiol. 2000;2:265–73. doi: 10.1046/j.1462-5822.2000.00056.x. [DOI] [PubMed] [Google Scholar]
- 117.Wright EK, Goodart SA, Growney JD, Hadinoto V, Endrizzi MG, Long EM, Sadigh K, Abney AL, Bernstein-Hanley I, Dietrich WF. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr Biol. 2003;13:27–36. doi: 10.1016/s0960-9822(02)01359-3. [DOI] [PubMed] [Google Scholar]
- 118.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE, Kuida K, Mariathasan S, Dixit VM, Flavell RA, Dietrich WF, Roy CR. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–25. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 119.Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K, Swanson MS. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Diez E, Yaraghi Z, MacKenzie A, Gros P. The neuronal apoptosis inhibitory protein (Naip) is expressed in macrophages and is modulated after phagocytosis and during intracellular infection with Legionella pneumophila. J Immunol. 2000;164:1470–7. doi: 10.4049/jimmunol.164.3.1470. [DOI] [PubMed] [Google Scholar]
- 121.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J Immunol. 2008;180:7558–64. doi: 10.4049/jimmunol.180.11.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu J, Fernandes-Alnemri T, Alnemri ES. Involvement of the AIM2, NLRC4, and NLRP3 Inflammasomes in Caspase-1 Activation by Listeria monocytogenes. J Clin Immunol. 2010 May 20; doi: 10.1007/s10875-010-9425-2. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Endrizzi MG, Hadinoto V, Growney JD, Miller W, Dietrich WF. Genomic sequence analysis of the mouse Naip gene array. Genome Res. 2000;10:1095–102. doi: 10.1101/gr.10.8.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GH, Glucksmann MA, DiStefano PS. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955–8. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 126.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Núñez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–7. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 127.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nuñez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003;4:702–7. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 128.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–7. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 129.Yoo NJ, Park WS, Kim SY, Reed JC, Son SG, Lee JY, Lee SH. Nod1, a CARD protein, enhances pro-interleukin-1beta processing through the interaction with pro-caspase-1. Biochem Biophys Res Commun. 2002;299:652–8. doi: 10.1016/s0006-291x(02)02714-6. [DOI] [PubMed] [Google Scholar]
- 130.Rosenzweig HL, Galster KT, Planck SR, Rosenbaum JT. NOD1 expression in the eye and functional contribution to IL-1beta-dependent ocular inflammation in mice. Invest Ophthalmol Vis Sci. 2009;50:1746–53. doi: 10.1167/iovs.08-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, Kurtz S, Coffield VM, Accavitti-Loper MA, Su L, Vogel SN, Braunstein M, Ting JP. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem. 2005;280:39914–24. doi: 10.1074/jbc.M502820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Williams KL, Taxman DJ, Linhoff MW, Reed W, Ting JP. Cutting Edge: Monarch-1: A Pyrin/Nucleotide-binding domain/Leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J Immunol. 2003;170:5354–8. doi: 10.4049/jimmunol.170.11.5354. [DOI] [PubMed] [Google Scholar]
- 133.Lich JD, Ting JP. Monarch-1/PYPAF7 and other CATERPILLER (CLR, NOD, NLR) proteins with negative regulatory functions. Microbes Infect. 2007;9:672–6. doi: 10.1016/j.micinf.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ludlow LE, Johnstone RW, Clarke CJ. The HIN-200 family: more than interferon-inducible genes? Exp Cell Res. 2005;308:1–17. doi: 10.1016/j.yexcr.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 135.Dawson MJ, Trapani JA. The interferon-inducible autoantigen, IFI 16: localization to the nucleolus and identification of a DNA-binding domain. Biochem Biophys Res Commun. 1995;214:152–62. doi: 10.1006/bbrc.1995.2269. [DOI] [PubMed] [Google Scholar]
- 136.Duhl DM, Gaczynski M, Olinski R, Briggs RC. Intra-nuclear distribution of the human myeloid cell nuclear differentiation antigen in HL-60 cells. J Cell Physiol. 1989;141:148–53. doi: 10.1002/jcp.1041410122. [DOI] [PubMed] [Google Scholar]
- 137.Choubey D, Lengyel P. Interferon action: cytoplasmic and nuclear localization of the interferon-inducible 52-kD protein that is encoded by the Ifi 200 gene from the gene 200 cluster. J Interferon Res. 1993;13:43–52. doi: 10.1089/jir.1993.13.43. [DOI] [PubMed] [Google Scholar]
- 138.Choubey D, Lengyel P. Interferon action: nucleolar and nucleoplasmic localization of the interferon-inducible 72-kD protein that is encoded by the Ifi 204 gene from the gene 200 cluster. J Cell Biol. 1992;116:1333–41. doi: 10.1083/jcb.116.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johnstone RW, Kerry JA, Trapani JA. The human interferon-inducible protein, IFI 16, is a repressor of transcription. J Biol Chem. 1998;273:17172–7. doi: 10.1074/jbc.273.27.17172. [DOI] [PubMed] [Google Scholar]
- 140.Albrecht M, Choubey D, Lengauer T. The HIN domain of IFI-200 proteins consists of two OB folds. Biochem Biophys Res Commun. 2005;327:679–87. doi: 10.1016/j.bbrc.2004.12.056. [DOI] [PubMed] [Google Scholar]
- 141.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–8. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 144.Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323:1057–60. doi: 10.1126/science.1169841. [DOI] [PubMed] [Google Scholar]
- 145.Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, Eisenlohr L, Landel CP, Alnemri ES. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010 May;11(5):385–93. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, Kastner DL. Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci U S A. 2003;100:13501–6. doi: 10.1073/pnas.2135380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Seshadri S, Duncan MD, Hart JM, Gavrilin MA, Wewers MD. Pyrin levels in human monocytes and monocyte-derived macrophages regulate IL-1beta processing and release. J Immunol. 2007;179:1274–81. doi: 10.4049/jimmunol.179.2.1274. [DOI] [PubMed] [Google Scholar]
- 148.Yu JW, Fernandes-Alnemri T, Datta P, Wu J, Juliana C, Solorzano L, McCormick M, Zhang Z, Alnemri ES. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol Cell. 2007;28:214–27. doi: 10.1016/j.molcel.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Centola M, Wood G, Frucht DM, Galon J, Aringer M, Farrell C, Kingma DW, Horwitz ME, Mansfield E, Holland SM, O'Shea JJ, Rosenberg HF, Malech HL, Kastner DL. The gene for familial Mediterranean fever, MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood. 2000;95:3223–31. [PubMed] [Google Scholar]
- 150.Sebastian S, Luban J. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005;2:40. doi: 10.1186/1742-4690-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Barr SD, Smiley JR, Bushman FD. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 2008;4:e1000007. doi: 10.1371/journal.ppat.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–60. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*) Annu Rev Immunol. 2009;27:621–68. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP, Kastner DL. Targeted disruption of Pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol. Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 155.Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N, Núñez G. Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem. Biophys. Res. Commun. 2003;302:575–80. doi: 10.1016/s0006-291x(03)00221-3. [DOI] [PubMed] [Google Scholar]
- 156.Richards N, Schaner P, Diaz A, Stuckey J, Shelden E, Wadhwa A, Gumucio DL. Interaction between Pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J Biol Chem. 2001;276:39320–9. doi: 10.1074/jbc.M104730200. [DOI] [PubMed] [Google Scholar]
- 157.Papin S, Cuenin S, Agostini L, Martinon F, Werner S, Beer HD, Grütter C, Grütter M, Tschopp J. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1beta processing. Cell Death Differ. 2007;14:1457–66. doi: 10.1038/sj.cdd.4402142. [DOI] [PubMed] [Google Scholar]
- 158.Chae JJ, Wood G, Masters SL, Richard K, Park G, Smith BJ, Kastner DL. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc Natl Acad Sci U S A. 2006;103:9982–7. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chae JJ, Wood G, Richard K, Jaffe H, Colburn NT, Masters SL, Gumucio DL, Shoham NG, Kastner DL. The familial Mediterranean fever protein, pyrin, is cleaved by caspase-1 and activates NF-kappaB through its N-terminal fragment. Blood. 2008;112:1794–803. doi: 10.1182/blood-2008-01-134932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Barksby HE, Lea SR, Preshaw PM, Taylor JJ. The expanding family of interleukin-1 cytokines and their role in destructive inflammatory disorders. Clin Exp Immunol. 2007;149:217–25. doi: 10.1111/j.1365-2249.2007.03441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo AM, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Amos CI, Mulley J, Quane KA, Molloy MG, Ranki A, Powell RJ, Hitman GA, O'Shea JJ, Kastner DL. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97:133–44. doi: 10.1016/s0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 162.McGonagle D, McDermott MF. A proposed classification of the immunological diseases. PLoS Med. 2006;3:e297. doi: 10.1371/journal.pmed.0030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guz G, Kanbay M, Ozturk MA. Current perspectives on familial Mediterranean fever. Curr Opin Infect Dis. 2009;22:309–15. doi: 10.1097/QCO.0b013e328329d15e. [DOI] [PubMed] [Google Scholar]
- 164.French FMF Consortium A candidate gene for familial mediterranean fever. Nat Genet. 1997 Sep;17(1):25–31. doi: 10.1038/ng0997-25. [DOI] [PubMed] [Google Scholar]
- 165.Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium. Cell. 1997;90:797–807. doi: 10.1016/s0092-8674(00)80539-5. No Authors Listed. [DOI] [PubMed] [Google Scholar]
- 166.Sarrauste de Menthière C, Terrière S, Pugnère D, Ruiz M, Demaille J, Touitou I. INFEVERS: the Registry for FMF and hereditary inflammatory disorders mutations. Nucleic Acids Res. 2003 Jan 1;31(1):282–5. doi: 10.1093/nar/gkg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Milhavet F, Cuisset L, Hoffman HM, Slim R, El-Shanti H, Aksentijevich I, Lesage S, Waterham H, Wise C, Sarrauste de Menthiere C, Touitou I. The infevers autoinflammatory mutation online registry: update with new genes and functions. Hum Mutat. 2008;29:803–8. doi: 10.1002/humu.20720. [DOI] [PubMed] [Google Scholar]
- 168.Touitou I, Lesage S, McDermott M, Cuisset L, Hoffman H, Dode C, Shoham N, Aganna E, Hugot JP, Wise C, Waterham H, Pugnere D, Demaille J, Sarrauste de Menthiere C. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum Mutat. 2004;24:194–8. doi: 10.1002/humu.20080. [DOI] [PubMed] [Google Scholar]
- 169.Chae JJ, Aksentijevich I, Kastner DL. Advances in the understanding of familial Mediterranean fever and possibilities for targeted therapy. Br J Haematol. 2009;146:467–78. doi: 10.1111/j.1365-2141.2009.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Touitou I. Standardized testing for mutations in familial Mediterranean fever. Clin Chem. 2003;49:1781–2. doi: 10.1373/clinchem.2003.025791. [DOI] [PubMed] [Google Scholar]
- 171.Schaner P, Richards N, Wadhwa A, Aksentijevich I, Kastner D, Tucker P, Gumucio D. Episodic evolution of pyrin in primates: human mutations recapitulate ancestral amino acid states. Nat Genet. 2001;27:318–21. doi: 10.1038/85893. [DOI] [PubMed] [Google Scholar]
- 172.Chae JJ, Centola M, Aksentijevich I, Dutra A, Tran M, Wood G, Nagaraju K, Kingma DW, Liu PP, Kastner DL. Isolation, genomic organization, and expression analysis of the mouse and rat homologs of MEFV, the gene for familial mediterranean fever. Mamm Genome. 2000;11:428–35. doi: 10.1007/s003350010082. [DOI] [PubMed] [Google Scholar]
- 173.Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29:301–5. doi: 10.1038/ng756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71:198–203. doi: 10.1086/341357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, Stein L, Russo R, Goldsmith D, Dent P, Rosenberg HF, Austin F, Remmers EF, Balow JE, Jr, Rosenzweig S, Komarow H, Shoham NG, Wood G, Jones J, Mangra N, Carrero H, Adams BS, Moore TL, Schikler K, Hoffman H, Lovell DJ, Lipnick R, Barron K, O'Shea JJ, Kastner DL, Goldbach-Mansky R. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 2002;46:3340–8. doi: 10.1002/art.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tassi S, Carta S, Delfino L, Caorsi R, Martini A, Gattorno M, Rubartelli A. Altered redox state of monocytes from cryopyrin-associated periodic syndromes causes accelerated IL-1beta secretion. Proc Natl Acad Sci U S A. 2010;107:9789–94. doi: 10.1073/pnas.1000779107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Neven B, Prieur AM, Quartier dit Maire P, Medscape Cryopyrinopathies: update on pathogenesis and treatment. Nat Clin Pract Rheumatol. 2008;4:481–9. doi: 10.1038/ncprheum0874. [DOI] [PubMed] [Google Scholar]
- 178.Brydges SD, Mueller JL, McGeough MD, Pena CA, Misaghi A, Gandhi C, Putnam CD, Boyle DL, Firestein GS, Horner AA, Soroosh P, Watford WT, O'Shea JJ, Kastner DL, Hoffman HM. Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity. 2009;30:875–87. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Meng G, Zhang F, Fuss I, Kitani A, Strober W. A mutation in the Nlrp3 gene causing inflammasome hyperactivation potentiates Th17 cell-dominant immune responses. Immunity. 2009;30:860–74. doi: 10.1016/j.immuni.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Gasse P, Mary C, Guenon I, Noulin N, Charron S, Schnyder-Candrian S, Schnyder B, Akira S, Quesniaux VF, Lagente V, Ryffel B, Couillin I. IL-1R1/MyD88 signaling and the inflammasome are essential in pulmonary inflammation and fibrosis in mice. J Clin Invest. 2007;117:3786–99. doi: 10.1172/JCI32285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pizzirani C, Falzoni S, Govoni M, La Corte R, Donadei S, Di Virgilio F, Trotta F, Lo Monaco A. Dysfunctional inflammasome in Schnitzler's syndrome. Rheumatology (Oxford) 2009;48:1304–8. doi: 10.1093/rheumatology/kep222. [DOI] [PubMed] [Google Scholar]
- 182.Loock J, Lamprecht P, Timmann C, Mrowietz U, Csernok E, Gross WL. Genetic predisposition (NLRP3 V198M mutation) for IL-1-mediated inflammation in a patient with Schnitzler syndrome. J Allergy Clin Immunol. 2010;125:500–2. doi: 10.1016/j.jaci.2009.10.066. [DOI] [PubMed] [Google Scholar]
- 183.Schoultz I, Verma D, Halfvarsson J, Törkvist L, Fredrikson M, Sjöqvist U, Lördal M, Tysk C, Lerm M, Söderkvist P, Söderholm JD. Combined polymorphisms in genes encoding the inflammasome components NALP3 and CARD8 confer susceptibility to Crohn's disease in Swedish men. Am J Gastroenterol. 2009;104:1180–8. doi: 10.1038/ajg.2009.29. [DOI] [PubMed] [Google Scholar]
- 184.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Fortin PR, Wither JE, Sarfati M, Rutgeerts P, Rioux JD, Vermeire S, Hudson TJ, Franchimont D. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–6. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kastbom A, Verma D, Eriksson P, Skogh T, Wingren G, Söderkvist P. Genetic variation in proteins of the cryopyrin inflammasome influences susceptibility and severity of rheumatoid arthritis (the Swedish TIRA project) Rheumatology (Oxford) 2008;47:415–7. doi: 10.1093/rheumatology/kem372. [DOI] [PubMed] [Google Scholar]
- 186.Kastbom A, Johansson M, Verma D, Söderkvist P, Rant-apää-Dahlqvist S. CARD8 p.C10X polymorphism is associated with inflammatory activity in early rheumatoid arthritis. Ann Rheum Dis. 2010;69:723–6. doi: 10.1136/ard.2008.106989. [DOI] [PubMed] [Google Scholar]
- 187.Hitomi Y, Ebisawa M, Tomikawa M, Imai T, Komata T, Hirota T, Harada M, Sakashita M, Suzuki Y, Shimojo N, Kohno Y, Fujita K, Miyatake A, Doi S, Enomoto T, Taniguchi M, Higashi N, Nakamura Y, Tamari M. Associations of functional NLRP3 polymorphisms with susceptibility to food-induced anaphylaxis and aspirin-induced asthma. J Allergy Clin Immunol. 2009;124:779–85. doi: 10.1016/j.jaci.2009.07.044. [DOI] [PubMed] [Google Scholar]
- 188.Pontillo A, Brandao L, Guimaraes R, Segat L, Araujo J, Crovella S. Two SNPs in NLRP3 gene are involved in the predisposition to type-1 diabetes and celiac disease in a pediatric population from northeast Brazil. Autoimmunity. 2010 Apr 7; doi: 10.3109/08916930903540432. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 189.Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, Bennett DC, Fain PR, Spritz RA. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356:1216–25. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 190.Jin Y, Birlea SA, Fain PR, Spritz RA. Genetic variations in NALP1 are associated with generalized vitiligo in a Romanian population. J Invest Dermatol. 2007;127:2558–62. doi: 10.1038/sj.jid.5700953. [DOI] [PubMed] [Google Scholar]
- 191.Magitta NF, Bøe Wolff AS, Johansson S, Skinningsrud B, Lie BA, Myhr KM, Undlien DE, Joner G, Njølstad PR, Kvien TK, Førre Ø , Knappskog PM, Husebye ES. A coding polymorphism in NALP1 confers risk for autoimmune Addison's disease and type 1 diabetes. Genes Immun. 2009;10:120–4. doi: 10.1038/gene.2008.85. [DOI] [PubMed] [Google Scholar]
- 192.Glinsky GV. Disease phenocode analysis identifies SNP-guided microRNA maps (MirMaps) associated with human “master” disease genes. Cell Cycle. 2008;7:3680–94. doi: 10.4161/cc.7.23.7153. [DOI] [PubMed] [Google Scholar]
- 193.Cooper WN, Luharia A, Evans GA, Raza H, Haire AC, Grundy R, Bowdin SC, Riccio A, Sebastio G, Bliek J, Schofield PN, Reik W, Macdonald F, Maher ER. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur J Hum Genet. 2005;13:1025–32. doi: 10.1038/sj.ejhg.5201463. [DOI] [PubMed] [Google Scholar]
- 194.Meyer E, Lim D, Pasha S, Tee LJ, Rahman F, Yates JR, Woods CG, Reik W, Maher ER. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome) PLoS Genet. 2009;5:e1000423. doi: 10.1371/journal.pgen.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bestor TH. Cytosine methylation mediates sexual conflict. Trends Genet. 2003;19:185–90. doi: 10.1016/S0168-9525(03)00049-0. [DOI] [PubMed] [Google Scholar]
- 196.Kinoshita T, Wang Y, Hasegawa M, Imamura R, Suda T. PYPAF3, a PYRIN-containing APAF-1-like protein, is a feedback regulator of caspase-1-dependent interleukin-1beta secretion. J Biol Chem. 2005;280:21720–5. doi: 10.1074/jbc.M410057200. [DOI] [PubMed] [Google Scholar]
- 197.Slim R, Mehio A. The genetics of hydatidiform moles: new lights on an ancient disease. Clin Genet. 2007;71:25–34. doi: 10.1111/j.1399-0004.2006.00697.x. [DOI] [PubMed] [Google Scholar]
- 198.Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, Hanash S, Rouleau GA, Slim R. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–2. doi: 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- 199.Qian J, Deveault C, Bagga R, Xie X, Slim R. Women heterozygous for NALP7/NLRP7 mutations are at risk for reproductive wastage: report of two novel mutations. Hum Mutat. 2007;28:741. doi: 10.1002/humu.9498. [DOI] [PubMed] [Google Scholar]
- 200.Deveault C, Qian JH, Chebaro W, Ao A, Gilbert L, Mehio A, Khan R, Tan SL, Wischmeijer A, Coullin P, Xie X, Slim R. NLRP7 mutations in women with diploid androgenetic and triploid moles: a proposed mechanism for mole formation. Hum Mol Genet. 2009;18:888–97. doi: 10.1093/hmg/ddn418. [DOI] [PubMed] [Google Scholar]
- 201.Kou YC, Shao L, Peng HH, Rosetta R, del Gaudio D, Wagner AF, Al-Hussaini TK, Van den Veyver IB. A recurrent intragenic genomic duplication, other novel mutations in NLRP7 and imprinting defects in recurrent biparental hydatidiform moles. Mol Hum Reprod. 2008;14:33–40. doi: 10.1093/molehr/gam079. [DOI] [PubMed] [Google Scholar]
- 202.Hayward BE, De Vos M, Talati N, Abdollahi MR, Taylor GR, Meyer E, Williams D, Maher ER, Setna F, Nazir K, Hussaini S, Jafri H, Rashid Y, Sheridan E, Bonthron DT. Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat. 2009;30:E629–39. doi: 10.1002/humu.20993. [DOI] [PubMed] [Google Scholar]
- 203.Slim R, Bagga R, Chebaro W, Srinivasan R, Agarwal N. A strong founder effect for two NLRP7 mutations in the Indian population: an intriguing observation. Clin Genet. 2009;76:292–5. doi: 10.1111/j.1399-0004.2009.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Puechberty J, Rittore C, Philibert L, Lefort G, Burlet G, Bénos P, Reyftmann L, Sarda P, Touitou I. Homozygous NLRP7 mutations in a Moroccan woman with recurrent reproductive failure. Clin Genet. 2009;75:298–300. doi: 10.1111/j.1399-0004.2008.01098.x. [DOI] [PubMed] [Google Scholar]
- 205.Wang CM, Dixon PH, Decordova S, Hodges MD, Sebire NJ, Ozalp S, Fallahian M, Sensi A, Ashrafi F, Repiska V, Zhao J, Xiang Y, Savage PM, Seckl MJ, Fisher RA. Identification of 13 novel NLRP7 mutations in 20 families with recurrent hydatidiform mole; missense mutations cluster in the leucine-rich region. J Med Genet. 2009;46:569–75. doi: 10.1136/jmg.2008.064196. [DOI] [PubMed] [Google Scholar]
- 206.Okada K, Hirota E, Mizutani Y, Fujioka T, Shuin T, Miki T, Nakamura Y, Katagiri T. Oncogenic role of NALP7 in testicular seminomas. Cancer Sci. 2004;95:949–54. doi: 10.1111/j.1349-7006.2004.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ohno S, Kinoshita T, Ohno Y, Minamoto T, Suzuki N, Inoue M, Suda T. Expression of NLRP7 (PYPAF3, NALP7) protein in endometrial cancer tissues. Anticancer Res. 2008;28:2493–7. [PubMed] [Google Scholar]
- 208.Jéru I, Duquesnoy P, Fernandes-Alnemri T, Cochet E, Yu JW, Lackmy-Port-Lis M, Grimprel E, Landman-Parker J, Hentgen V, Marlin S, McElreavey K, Sarkisian T, Grateau G, Alnemri ES, Amselem S. Mutations in NALP12 cause hereditary periodic fever syndromes. Proc Natl Acad Sci U S A. 2008;105:1614–9. doi: 10.1073/pnas.0708616105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Simi A, Tsakiri N, Wang P, Rothwell NJ. Interleukin-1 and inflammatory neurodegeneration. Biochem Soc Trans. 2007;35:1122–6. doi: 10.1042/BST0351122. [DOI] [PubMed] [Google Scholar]
- 210.Roy N, Mahadevan MS, McLean M, Shutler G, Yaraghi Z, Farahani R, Baird S, Besner-Johnston A, Lefebvre C, Kang X, Salih M, Aubry H, Tamai K, Guan K, Ioannou P, Crawford T, de Jong PJ, Surh L, Ikeda J-E, Korneluk RG, MacKenzie A. The gene for neuronal apoptosis inhibitory protein is partially deleted in individuals with spinal muscular atrophy. Cell. 1995;80:167–78. doi: 10.1016/0092-8674(95)90461-1. [DOI] [PubMed] [Google Scholar]
- 211.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Häfner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat. Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 212.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nuñez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 213.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 214.Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nuñez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–12. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 215.Hisamatsu T, Suzuki M, Reinecker HC, Nadeau WJ, Mc-Cormick BA, Podolsky DK. CARD15/NOD2 functions as an antibacterial factor in human intestinal epithelial cells. Gastroenterology. 2003;124:993–1000. doi: 10.1053/gast.2003.50153. [DOI] [PubMed] [Google Scholar]
- 216.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schäffeler E, Schlee M, Herrlinger KR, Stallmach A, Noack F, Fritz P, Schröder JM, Bevins CL, Fellermann K, Stange EF. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–64. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nuñez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–4. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 218.van Heel DA, Ghosh S, Butler M, Hunt KA, Lundberg AM, Ahmad T, McGovern DP, Onnie C, Negoro K, Goldthorpe S, Foxwell BM, Mathew CG, Forbes A, Jewell DP, Playford RJ. Muramyl dipeptide and toll-like receptor sensitivity in NOD2-associated Crohn's disease. Lancet. 2005;365:1794–6. doi: 10.1016/S0140-6736(05)66582-8. [DOI] [PubMed] [Google Scholar]
- 219.Abraham C, Cho JH. Functional consequences of NOD2 (CARD15) mutations. Inflamm Bowel Dis. 2006;12:641–50. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 220.Maeda S, Hsu LC, Liu H, Bankston LA, Iimura M, Kagnoff MF, Eckmann L, Karin M. Nod2 mutation in Crohn's disease potentiates NF-kappaB activity and IL-1beta processing. Science. 2005;307:734–8. doi: 10.1126/science.1103685. [DOI] [PubMed] [Google Scholar]
- 221.McGovern DP, Hysi P, Ahmad T, van Heel DA, Moffatt MF, Carey A, Cookson WO, Jewell DP. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum Mol Genet. 2005;14:1245–50. doi: 10.1093/hmg/ddi135. [DOI] [PubMed] [Google Scholar]
- 222.Verma R, Ahuja V, Paul J. Frequency of single nucleotide polymorphisms in NOD1 gene of ulcerative colitis patients: a case-control study in the Indian population. BMC Med Genet. 2009;10:82. doi: 10.1186/1471-2350-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S, Fuji A, Yuasa T, Manki A, Sakurai Y, Nakajima M, Kobayashi H, Fujiwara I, Tsutsumi H, Utani A, Nishigori C, Heike T, Nakahata T, Miyachi Y. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: common genetic etiology with Blau syndrome. Blood. 2005;105:1195–7. doi: 10.1182/blood-2004-07-2972. [DOI] [PubMed] [Google Scholar]
- 224.Schürmann M, Valentonyte R, Hampe J, Müller-Quernheim J, Schwinger E, Schreiber S. CARD15 gene mutations in sarcoidosis. Eur Respir J. 2003;22:748–54. doi: 10.1183/09031936.03.00040602. [DOI] [PubMed] [Google Scholar]
- 225.Hnatyszyn A, Szalata M, Stanczyk J, Cichy W, Slomski R. Association of c.802C>T polymorphism of NOD2/CARD15 gene with the chronic gastritis and predisposition to cancer in H. pylori infected patients. Exp Mol Pathol. 2010;88:388–93. doi: 10.1016/j.yexmp.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 226.Tian Y, Li Y, Hu Z, Wang D, Sun X, Ren C. Differential effects of NOD2 polymorphisms on colorectal cancer risk: a meta-analysis. Int J Colorectal Dis. 2010;25:161–8. doi: 10.1007/s00384-009-0809-9. [DOI] [PubMed] [Google Scholar]
- 227.Hoffman HM. Therapy of autoinflammatory syndromes. J Allergy Clin Immunol. 2009;124:1129–38. doi: 10.1016/j.jaci.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hawkins PN, Lachmann HJ, McDermott MF. Interleukin-1-receptor antagonist in the Muckle-Wells syndrome. N Engl J Med. 2003;348:2583–4. doi: 10.1056/NEJM200306193482523. [DOI] [PubMed] [Google Scholar]
- 229.Braddock M, Quinn A. Targeting IL-1 in inflammatory disease: new opportunities for therapeutic intervention. Nat Rev Drug Discov. 2004;3:330–9. doi: 10.1038/nrd1342. [DOI] [PubMed] [Google Scholar]
- 230.Quiniou C, Sapieha P, Lahaie I, Hou X, Brault S, Beauchamp M, Leduc M, Rihakova L, Joyal JS, Nadeau S, Heveker N, Lubell W, Sennlaub F, Miller G, Pshezhetsky AV, Chemtob S. Development of a novel noncompetitive antagonist of IL-1 receptor. J Immunol. 2008;180:6977–87. doi: 10.4049/jimmunol.180.10.6977. [DOI] [PubMed] [Google Scholar]
- 231.Rudolphi K, Gerwin N, Verzijl N, van der Kraan P, van den Berg W. Pralnacasan, an inhibitor of interleukin-1beta converting enzyme, reduces joint damage in two murine models of osteoarthritis. Osteoarthritis Cartilage. 2003;11:738–46. doi: 10.1016/s1063-4584(03)00153-5. [DOI] [PubMed] [Google Scholar]
- 232.Loher F, Bauer C, Landauer N, Schmall K, Siegmund B, Lehr HA, Dauer M, Schoenharting M, Endres S, Eigler A. The interleukin-1 beta-converting enzyme inhibitor pralnacasan reduces dextran sulfate sodium-induced murine colitis and T helper 1 T-cell activation. J Pharmacol Exp Ther. 2004;308:583–90. doi: 10.1124/jpet.103.057059. [DOI] [PubMed] [Google Scholar]
- 233.Ku G, Faust T, Lauffer LL, Livingston DJ, Harding MW. Harding, Interleukin-1 beta converting enzyme inhibition blocks progression of type II collagen-induced arthritis in mice. Cytokine. 1996;8:377–86. doi: 10.1006/cyto.1996.0052. [DOI] [PubMed] [Google Scholar]
- 234.Ross J, Brough D, Gibson RM, Loddick SA, Rothwell NJ. A selective, non-peptide caspase-1 inhibitor, VRT-018858, markedly reduces brain damage induced by transient ischemia in the rat. Neuropharmacology. 2007;53:638–42. doi: 10.1016/j.neuropharm.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 235.Fleischmann RM, Tesser J, Schiff MH, Schechtman J, Burmester GR, Bennett R, Modafferi D, Zhou L, Bell D, Appleton B. Safety of extended treatment with anakinra in patients with rheumatoid arthritis. Ann Rheum Dis. 2006;65:1006–12. doi: 10.1136/ard.2005.048371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Botsios C. Safety of tumour necrosis factor and interleukin-1 blocking agents in rheumatic diseases. Autoimmun Rev. 2005;4:162–70. doi: 10.1016/j.autrev.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 237.Molad Y. Update on colchicine and its mechanism of action. Curr Rheumatol Rep. 2002;4:252–6. doi: 10.1007/s11926-002-0073-2. [DOI] [PubMed] [Google Scholar]
- 238.Lamkanfi M, Mueller JL, Vitari AC, Misaghi S, Fedorova A, Deshayes K, Lee WP, Hoffman HM, Dixit VM. Glyburide inhibits the Cryopyrin/Nalp3 inflammasome. J Cell Biol. 2009;187:61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]