Abstract
Conjugative plasmid transfer results in the spread of antibiotic resistance genes and virulence factors between bacterial cells. Plasmid transfer is dependent upon the DNA nicking activity of a plasmid-encoded relaxase enzyme. Tyrosine residues within the relaxase cleave the DNA plasmid nic site in a highly sequence-specific manner. The conjugative resistance plasmid pCU1 encodes a relaxase with four tyrosine residues surrounding its active site (Y18,19,26,27). We use activity assays to demonstrate that the pCU1 relaxase preferentially uses Y26 or a combination of Y18+19 to nick DNA at wild type levels, and that an adjacent aspartic acid deprotonates these tyrosines to activate them for attack. Our findings illustrate the unique modifications that the pCU1 relaxase has introduced into the traditional relaxase-mediated DNA nicking mechanism.
1. Introduction
Bacteria use conjugative plasmid transfer (CPT) to disseminate genetic material to neighboring cells. During CPT, a donor bacterium transfers one strand of a double-stranded DNA plasmid to a recipient [1]. Each conjugative plasmid encodes a complex of proteins necessary for its transfer. One of these proteins, the relaxase, initiates plasmid transfer by creating a single-stranded break at the nic site in the transferred strand (T-strand). A DNA helicase then separates the T-strand from the parent strand, beginning at the nic site. Finally, the relaxase acts a second time to terminate the process by resealing, or ligating, the nicked T-strand. The relaxase, therefore, is the key determinant of initiation and termination of plasmid transfer [2-4]. Despite the extensive number and variety of conjugative plasmids, the basic mechanism by which relaxases initiate and terminate transfer is conserved. During both steps, the relaxase uses a catalytic tyrosine to cleave its respective plasmid's T-strand in a sequence-specific manner. In particular, the catalytic tyrosine initiates a bimolecular nucleophilic substitution-type (SN2) attack on the scissile phosphate at the T-strand nic site, thus generating a free 3’ hydroxyl and forming a covalent phosphotyrosine bond [2,3].
Two conditions must be met for this nicking reaction to occur efficiently. First, the tyrosine must act as a nucleophile. Since the pKa of tyrosine is 10, the tyrosine hydroxyl is protonated at physiological pH. To deprotonate the hydroxyl, a neighboring residue must act as a base to extract the proton, or the local pH of the active site must be considerably basic. Second, the scissile phosphate must act as an electrophile. The negatively charged scissile phosphate is positioned within the relaxase active site by a divalent cation, which is itself bound by the conserved relaxase histidine-hydrophobic residue-histidine (HUH) motif. As a result of this arrangement, the negative charge on the phosphate is neutralized by the positively charged metal center [5-9].
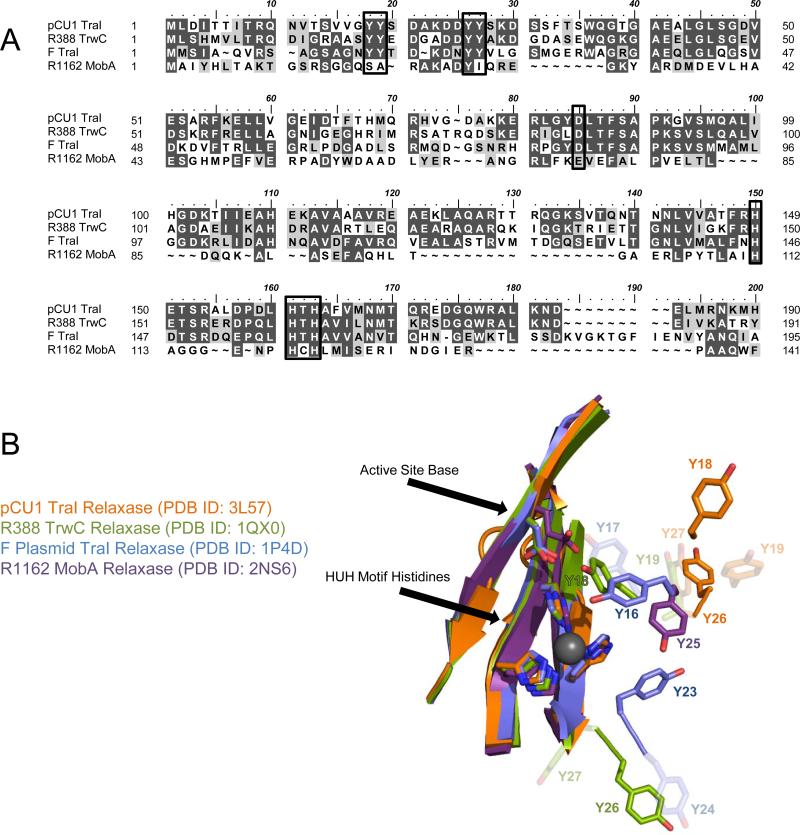
Structural data of the F plasmid and plasmid R388 relaxases reveal that, of the four tyrosines surrounding their active sites, the first in amino acid sequence is properly aligned for attack on the DNA substrate's scissile phosphate, and it is most likely activated for attack by an adjacent aspartic acid (Figures 1A,1B) [5,6,10-12]. In the case of the related R1162 relaxase, which contains only one tyrosine near the active site, structural data again reveal that this tyrosine is directed towards the HUH motif of the active site, though no neighboring base has been identified [13]. The role of these tyrosines has been verified by functional assays in which Y16 of the F relaxase, Y18 of the R388 relaxase, and Y25 of the R1162 relaxase were shown to be the primary DNA nicking residues of each enzyme [14-17]. In the case of R388, the third tyrosine in amino acid sequence, Y26, also nicked DNA efficiently if the relaxase was presented with substrates mimicking DNA transfer termination conditions [15,18].
Figure 1. Relaxase Sequence Alignment and Structural Comparison.
A) Protein Sequence Alignment.
The N-termini of the relaxase domains of pCU1 TraI, R388 TrwC, F TraI, and R1162 MobA were aligned. The active site tyrosines, proposed active site bases, and three histidines forming the active site are boxed. Identical residues are shaded black and similar residues are shaded gray.
B) Comparison of Relaxase Active Site Structure.
The crystal structures of the plasmid pCU1 TraI (orange, PDB ID: 3L57), plasmid R388 TrwC (green, PDB ID: 1QX0), F plasmid TraI (blue, PDB ID: 1P4D), and plasmid R1162 MobA (purple, PDB ID: 2NS6) relaxase active sites are compared. For each protein, the catalytic tyrosines, proposed active site base, and active site HUH motif histidines are shown as sticks; the first and third tyrosines are darkly shaded; all others are lightly shaded; a grey sphere represents the location of the bound Mg2+ as observed in PDB ID: 1P4D.
The plasmid pCU1 relaxase, originally isolated from Salmonella typhimurium [19,20], also contains four possible catalytic tyrosines, as determined by sequence alignment with homologous enzymes (Figure 1A). However, the tyrosines of the pCU1 relaxase are located on a loop at the periphery of the active site, in an orientation unique from that of other relaxases. For any one of these tyrosines to attack the scissile phosphate of a bound DNA substrate, the loop on which the residues are located must undergo a conformational change in order to direct the nicking tyrosine towards the scissile phosphate (Figure 1B). Therefore, it is unclear from the present structural data which tyrosine would then be oriented for attack. To determine the tyrosine(s) responsible for DNA cleavage, we performed a series of DNA nicking experiments with tyrosine mutants of the pCU1 relaxase. We then complemented these data with DNA nicking experiments involving aspartic acid mutants to demonstrate that interactions between the four tyrosines and this adjacent aspartic acid determine the nicking activity of the pCU1 relaxase. In summary, we present data characterizing the DNA nicking activities of the relaxase of the resistance plasmid pCU1 and highlight the unique aspects of the mechanism utilized by this relaxase relative to those previously studied.
2. Methods
2.1. Preparation of TraI Constructs
The pCU1 TraI relaxase domain is located at the N-terminus of the TraI protein, extending from residues 1 to 299 [21,22]. The wild type relaxase construct was cloned into the pTYB2 vector of the IMPACT system (New England Biolabs) as described previously to generate WT_299 [21]. Tyrosine and aspartic acid mutations were made within the wild type construct in the IMPACT system pTYB2 vector using Quick Change site directed mutagenesis (Stratagene). All cloning and mutagenesis was verified by sequencing at the UNC-CH Genome Analysis Facility.
Both wild type and mutant proteins were expressed and each protein was then purified on Chitin Resin (NEB) as described previously [21]. A final size exclusion chromatography purification step was then performed. Protein was loaded onto a HiLoad 16/60 Superdex 200 column (GE Healthcare) pre-equilibrated with Buffer S (500 mM NaCl, 20 mM Tris-HCl pH 7.5, 5% glycerol, 5 mM EDTA, 0.01% azide) on an ATKA Express FPLC (GE Healthcare). TraI was eluted from the column in Buffer S. A final dialysis into Buffer D (100 mM NaCl, 20 mM Tris-HCl pH 7.5, 5% glycerol, 0.01% azide) was performed to decrease the salt concentration and remove EDTA from the sample. The protein was immediately flash frozen in liquid nitrogen and stored in 60 μL aliquots at -80°C. All protein was visualized by SDS-PAGE and found to be >95% pure. All water was obtained from the laboratory Barnstead E-pure water filtration system, at > 17 megohm (ddH2O).
2.2. DNA Nicking Assays
The 5’ fluorescein-labeled (“FAM”) DNA substrates, FAM-35/7oriT_hairpin and FAM-20/7oriT_half-hairpin, were commercially synthesized and HPLC purified (Integrated DNA Technologies). Upon arrival, the substrates were resuspended in Buffer R (50 mM NaCl, 10 mM Tris-HCl pH 7.5, 0.05 mM EDTA, 0.01% azide), heated to 95°C for 10 minutes, and then allowed to cool passively to room temperature. Their nucleic acid sequences are listed below:
FAM-35/7oriT_hairpin: TGTGATAGCGTGATTTATCGCGCTGCGTTAGGTGT^ATAGCAG
FAM-20/7oriT_half-hairpin: TATCGCGCTGCGTTAGGTGT^ATAGCAG
The nic site is indicated by a caret and bases forming the inverted repeat are underlined. DNA secondary structure predictions were performed using the M-fold server (http://mfold.bioinfo.rpi.edu/cgi-bin/dna-form1.cgi).
In general, each assay sample (10 μL total) contained 5 μM purified TraI and 1 μM 5’ fluorescently labeled DNA substrate in 50 mM NaCl, 18 mM Tris-HCl pH 7.5, 4.5% glycerol, and 4 mM MnCl2. The reaction was initiated upon addition of the enzyme. Each reaction was incubated at 37°C for 1 hour, quenched by 10 μL 2X quenching solution (0.01% xylene cyanol, 85% formamide, 20 mM EDTA, 2X TAE (80 mM Tris base, 80 mM acetic acid, 2 mM EDTA), 0.2% SDS), and then separated over a 16% denaturing acrylamide gel. Gels were visualized using a VersaDoc Imaging System, 4400 MP (BioRad) and the accompanying Quantity One software (BioRad). Band intensities were quantified using ImageJ 1.42 (Rasband, W.S., NIH 2008). Prior to quantification, standard background subtraction was performed on all gels.
DNA nicking activity was reported as “Percent of Substrate Nicked” and represented the intensity of the product band divided by the sum of product and substrates band intensities, all multiplied by 100%. Data points represent the average of three or more experiments (each which were performed in triplicate) and error bars represent the standard deviation of these data points. All data processing was performed in Excel 2007, and all plots were generated in Graphpad PRISM v5.03 (Graphpad, 2010).
3. Results
3.1. DNA Nicking by pCU1 Relaxase Tyrosine Mutants
The DNA nicking activity of the wild type pCU1 relaxase domain (WT_299) was compared to that of Y→F relaxase mutants of the four potential DNA nicking tyrosines (Y18, 19, 26, and 27). All mutants and mutations discussed in this manuscript refer to those made within the relaxase domain of pCU1 TraI (residues 1-299). DNA nicking was measured against the substrates FAM-35/7oriT-hairpin (Figure 2A) and FAM-20/7oriT-half_hairpin (Figure 2B). FAM-35/7oriT-hairpin is a pCU1 oriT-encoding DNA substrate that includes the intact hairpin upstream of the nic site, while FAM-20/7oriT-half_hairpin incorporates only the proximal arm of the hairpin sequence. It has been shown that WT_299 nicks a greater percentage of FAM-20/7oriT-half_hairpin, as compared to FAM-35/7oriT-hairpin [21]. Therefore, these two substrates were chosen for this current work in order to determine if individual tyrosine residues or combinations of tyrosine residues within the relaxase domain would also demonstrate a difference in relative nicking activity in the presence of these two substrates.
Figure 2. DNA Nicking Activity of pCU1 TraI Relaxase Tyrosine Mutants.
The DNA nicking activity of each TraI construct is represented as the percent DNA substrate nicked and error bars represent the standard deviation of three experiments, each performed in triplicate. Each construct is labeled according to whether a tyrosine (Y) or phenylalanine (F) is present at residues 18, 19, 26, and 27. ** indicates nicking activity is inhibited relative to WT_299 (Y – Y – Y – Y) with 95% confidence. Solid and dashed lines represent the average nicking activity of WT_299 and one standard deviation above and below this value, respectively.
A) The substrate tested, FAM-35/7oriT-hairpin, spans the pCU1 nic site and forms a hairpin upstream of the nic site.
B) The substrate tested, FAM-20/7oriT-half_hairpin, spans the pCU1 nic site and incorporates only one arm of the hairpin.
As outlined in Figure 2, DNA nicking was inhibited by mutation of all four tyrosines to phenylalanines. Triple mutants that preserved either Y19 or Y27 were also incapable of nicking both substrates. Further, single mutants Y19F_299 and Y27F_299 nicked DNA at wild type levels. Unexpectedly, only limited DNA nicking occurred in the presence of Y18 alone, while, in contrast, nicking occurred at nearly wild type levels in the presence of Y26 alone. DNA nicking by single mutants Y18F_299 and Y26F_299 were both inhibited relative to wild type. These data indicate that as single residues, neither Y19 nor Y27 is sufficient, or required, for DNA nicking by pCU1 relaxase, while both Y18 and Y26 are capable of nicking DNA. Therefore, Y19 and Y27 will be termed “partner tyrosines”, and Y18 and Y26 will be termed “primary tyrosines”.
After establishing the identity of the primary and partner tyrosines, we first considered the impact the two partner tyrosines might have on the two primary tyrosines during DNA nicking. First, we examined the potential for interaction between pairs of adjacent primary and partner tyrosines (Y18+19 and Y26+27). As seen in Figure 2, the activity of Y18 was enhanced to that of wild type by the presence of its partner tyrosine Y19, but the presence of the partner tyrosine Y27 had no statistically significant impact on DNA nicking by the primary tyrosine Y26. Therefore, efficient DNA nicking by the pCU1 relaxase was achieved by either the primary tyrosine Y26 alone or by the combination of primary and partner tyrosines Y18+19 (Table 1). Second, we examined the potential for interaction between pairs of non-adjacent primary and partner tyrosines (Y18+27 and Y26+19), where Y27 and Y19 will be termed the “alternate partners” of Y18 and Y26, respectively. Both primary tyrosines (Y18 and Y26), when present in combination with their respective partner tyrosines, actually suffered a reduction in nicking activity when their alternate partner was also present; for example, the activity of Y18+19+27 was reduced relative to Y18+19; similarly, Y19+26+27 activity was reduced relative to Y26+27 (Figure 2, Table 1). Therefore, alternate partner tyrosines had a dominant negative effect on the pCU1 relaxase catalytic function.
Table 1. Chart of Tyrosine Activity of Each Relaxase Mutant.
A summary of the activity of each tyrosine mutant is provided, as well as the proposed role of each of the four tyrosines and the active site aspartic acid during DNA nicking.
| Relaxase Construct | % Activity FAM-35/7 | % Activity FAM-20/7 | Role of Individual Residues during DNA Nicking |
|---|---|---|---|
| Y – Y – Y – Y (Wild type relaxase) | 100 | 100 | Y18 attacks scissile PO4 after D84 indirectly deprotonates Y18 via Y19 OR Y26 attacks scissile PO4 after D84 directly deprotonates Y26 |
| Y – F – F – F | 30** | 20** | Y18 is too distant from D84 to be deprotonated, is limited to inefficient attack on scissile PO4 |
| Y – Y – F – F | 125 | 113 | Y18 now benefits from indirect deprotonation by D84 via Y19, is capable of efficient attack on scissile PO4 |
| Y – Y – F – Y | 45** | 55** | D84 deprotonates Y19. Y19 forms bifurcated interaction with Y18 (capable of efficient attack on scissile PO4) and Y27 (limited ability to attack scissile PO4) |
| F – F – Y – F | 80 | 90 | D84 deprotonates Y26, which in turn is capable of efficient attack on scissile PO4 |
| F – F – Y – Y | 100 | 110 | No change in DNA nicking as compared to F – F – Y – F; Y27 is not deprotonated by either D84 or Y26 |
| F – Y – Y – Y | 35** | 70** | D84 deprotonates both Y26 (capable of efficient attack on scissile PO4) and Y19 (does not attack scissile PO4) |
| F – Y – F – F | 0** | 0** | Y19 cannot not attack scissile PO4 |
| F – F – F – Y | 0** | 0** | Y27 is not deprotonated by D84 |
| F –Y – F – Y | 25** | 70** | Y19 forms bridge between D84 and Y27, which has a limited ability to attack scissile PO4 |
| Y – F – Y – F | 95 | 60** | Activity of this mutant primarily reflects that of Y26 |
| Y – Y – Y – F | 125 | 90 | Activity of this mutant reflects that of both Y26 and Y18+Y19 |
| Y – F – Y – Y | 115 | 95 | Activity of this mutant primarily reflects that of Y26 |
| D84A Mutants | D84 mediated deprotonation of tyrosines is eliminated AND Net charge on active site increases |
||
| D84A – Y – Y – Y – Y | 65‡ | 60‡ | Nicking by WT tyrosines (Y26 and Y18+19) is less efficient due to elimination of tyrosine deprotonation by D84 |
| D84A – Y – F – F – F | 60† | 70‡ | Nicking by Y18 is more efficient due to increase in net active site charge (note: Y – F – F – F is not deprotonated by D84) |
| D84A – Y – Y – F – F | 35‡ | 45‡ | Nicking by Y18+19 is less efficient due to elimination of tyrosine deprotonation by D84 |
| D84A – F – F – Y – F | 25‡ | 35‡ | Nicking by Y26 is less efficient due to elimination of tyrosine deprotonation by D84 |
| D84A – F – F – Y – Y | 40‡ | 40‡ | Nicking by Y26 is less efficient due to elimination of tyrosine deprotonation by D84 |
indicates nicking activity is inhibited relative to WT with 95% confidence.
indicates nicking activity of the D84A, Y→F mutant is inhibited relative to its D84, Y→F counterpart with 95% confidence.
indicates nicking activity of the D84A, Y→F mutant is enhanced relative to its D84, Y→F counterpart with 90% confidence.
Finally we considered the role of substrate length on the relative activities of each relaxase construct. The DNA nicking reaction of the relaxase exists in equilibrium with ligation of the nicked products, which regenerates the original DNA substrate [18,23]. Both wild type and mutant constructs nicked a greater overall percentage of FAM-20/7oriT-half_hairpin as compared to FAM-35/7oriT-hairpin (Figure 2) [21], where the relative DNA nicking activity of the majority of the tyrosine mutants as compared to wild type was similar between substrates (Figure 2, Table 1). Two mutants did not follow this trend, however. First, Y19+27 nicked the substrate FAM-20/7oriT-half_hairpin more effectively than the substrate FAM-35/7oriT-hairpin, though in both cases the mutant activity was significantly reduced relative to wild type. Second, Y18+26 nicked the substrate FAM-35/7oriT-hairpin at wild type levels, but its activity fell to 60% of wild type when nicking FAM-20/7oriT-half_hairpin (Figure 2). In conclusion, though, because the wild type pCU1 relaxase nicked a greater percentage of the shorter DNA substrate that did not form a hairpin, it appears that a longer substrate including a complete hairpin shifts the nicking/ligation equilibrium of the pCU1 relaxase in favor of ligation.
Taken together, these data support four conclusions concerning the DNA nicking activity of the pCU1 relaxase that are unique relative to the multi-tyrosine relaxases characterized to date [14,15,17,18]. First, Y26 alone nicks DNA at wild type levels. Second, Y18 requires the presence of its partner Y19 to nick a statistically significant percent of the two DNA substrates investigated. Third, DNA nicking by both Y18 and Y26 is affected by the presence of their alternate partner, Y27 or Y19, respectively. Fourth, the combination of Y19+27 exhibits limited DNA nicking in a substrate-dependent manner. As discussed below, these conclusions contrast those regarding other conjugative relaxases characterized previously [14,15,17,18].
3.2. DNA Nicking by pCU1 Relaxase Aspartic Acid Mutants
Multi-tyrosine relaxases homologous to the pCU1 relaxase primarily utilize the first tyrosine in the primary sequence to nick DNA [14,15,17,18]. Therefore, we sought to understand the reason for the enhanced activity of Y26 alone relative to Y18 alone, as well as the role of the partner tyrosines, Y19 and Y27, during DNA nicking by the pCU1 relaxase (Figure 2). We hypothesized that a neighboring base capable of activating a subset of the four tyrosines could mediate the observed pattern in DNA nicking activity.
Aspartic acid residues near the active site of the F plasmid relaxase (D81) and the plasmid R388 relaxase (D85) are known to affect the efficiency of DNA nicking and plasmid transfer for these two systems. The role of these aspartic acids is to both improve metal binding by the HUH motif, as well as to activate the primary nicking tyrosine for attack [5,6,9]. The pCU1 relaxase contains an aspartic acid at residue 84 that is predicted by structural alignment to interact with H162 of the pCU1 relaxase HUH motif. D84 is also the primary candidate in the pCU1 relaxase to act as a general base to deprotonate the nicking tyrosine's ring hydroxyl and activate it for catalytic attack (Figure 1).
Therefore, to investigate the role of D84 during DNA nicking by the pCU1 relaxase, D84A mutants were created in the background of the wild type (WT) relaxase, as well as Y19,26,27F, Y26,27F, Y18,19,27F, and Y18,19F mutants of the relaxase, and the DNA nicking activity of each protein variant was examined using two DNA substrates (Figure 3). By mutating D84 in the background of each of these tyrosine mutants, the impact of D84 on specific tyrosine residues could be specified. The DNA nicking activities of WT_299, Y26,27F_299, Y18,19F_299, and Y18,19,27F_299 were all reduced upon mutation of D84 (Figure 3). Interestingly, though, Y19,26,27F_299 exhibited an increase in DNA nicking activity in the presence of both substrates upon D84A mutation (Figure 3). Therefore, mutation of D84 revealed yet another difference in nicking activity between Y18 and Y26. As discussed below, we propose that these data indicate D84 activates Y18 for catalysis indirectly through Y19, but activates Y26 directly (Figure 4). Taken together, these data again illustrate that the pCU1 relaxase incorporates variations into its catalytic mechanism that are distinct from other conjugative relaxases characterized previously [14,15,17,18].
Figure 3. DNA Nicking Activity of pCU1 TraI Relaxase Aspartic Acid Mutants.
The DNA nicking activity of each TraI construct is represented as the percent DNA substrate nicked and error bars represent the standard deviation of three experiments performed in triplicate. Each construct is labeled according to whether a tyrosine (Y) or phenylalanine (F) is present at residues 18, 19, 26, and 27. ** and * indicates nicking activity is altered relative to the corresponding D84 tyrosine mutant with 95% and 90% confidence, respectively.
A) The substrate tested, FAM-35/7oriT-hairpin, spans the pCU1 nic site and forms a hairpin upstream of the nic site.
B) The substrate tested, FAM-20/7oriT-half_hairpin, spans the pCU1 nic site and incorporates only one arm of the hairpin.
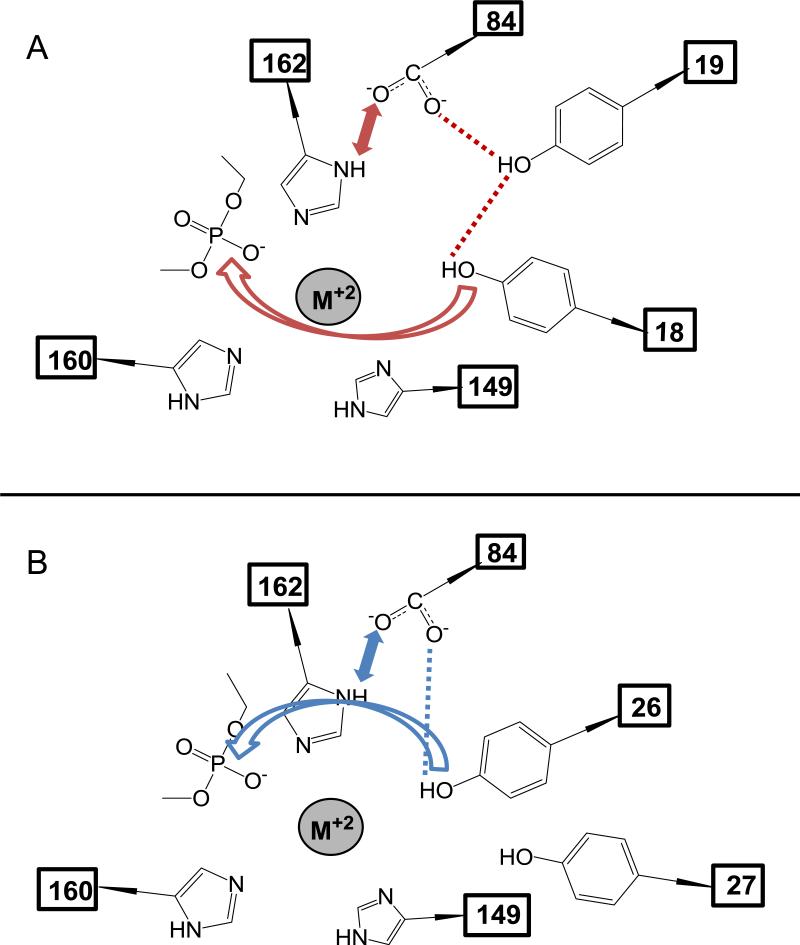
Figure 4. pCU1 TraI Relaxase Active Site Schematic.
A) The proposed orientation of tyrosines 18 and 19 relative to the active site base, aspartic acid 84, and the scissile phosphate of the DNA substrate, based upon DNA nicking activity data. Red dotted lines and bidirectional arrows indicate the presence of residue-residue interactions. The red curved arrow indicates the SN2 attack of Y18 on the scissile phosphate.
B) As in (A), the proposed orientation of tyrosines 26 and 27 relative to the active site base, aspartic acid 84, and the scissile phosphate of the DNA substrate, based upon DNA nicking activity data. Blue dotted lines and bidirectional arrows indicate the presence of residue-residue interactions. The blue curved arrow indicates the SN2 attack of Y26 on the scissile phosphate.
4. Discussion
The active site of conjugative relaxases consists of the HUH motif, a neighboring basic residue, up to four catalytic tyrosines, and one divalent cation that together accomplish cleavage of the scissile phosphate of a bound DNA substrate [5,7,9-12,21,24,25]. Here, we investigate the activities of the basic residue and the catalytic tyrosines in the context of the pCU1 relaxase catalytic mechanism. This work expands our understanding of the range of approaches a relaxase can employ to accomplish DNA cleavage.
Using a series of DNA nicking experiments involving tyrosine and aspartic acid mutants of the pCU1 relaxase, we generated a model in which a series of interactions between D84 and the four tyrosines mediate cleavage of the scissile phosphate of the bound pCU1 relaxase substrate (Figure 4, Table 1). In this model, D84 first serves as a base to deprotonate the ring hydroxyl of Y19 or Y26, thus generating a nucleophile at this residue. Second, Y18 or Y26 can assume an orientation from which it performs an SN2 attack on the scissile phosphate of a bound DNA substrate. Therefore, even when alone, Y26 can nick DNA with an activated, nucleophilic ring hydroxyl. However, when Y18 is in position to nick DNA, it still requires the presence of its partner tyrosine Y19 to serve as a bridge between itself and the activating effect of D84. Finally, Y27 appears to be a poor candidate for both acid-base interaction with D84 and scissile phosphate attack, except in the extreme case of the mutant Y18,26F_299. Here, Y27 appears capable of limited attack on the scissile phosphate when Y19 is present to form a bridge between Y27 and D84 (Figure 2).
While this model illustrates how the partner tyrosine Y19 enhances the DNA nicking activity of the primary tyrosine Y18, the presence of an alternate partner tyrosine was inhibitory under certain circumstances. In particular, the presence of the alternate partner Y19 reduced nicking by primary tyrosine Y26, and the alternate partner Y27 reduced nicking by primary tyrosine Y18(+19) (Figure 2). We propose that the role of D84 to polarize the tyrosine hydroxyls for attack actually results in this observed alternate partner effect. For example, in the case of Y26 or Y26+27, the addition of Y19 forces D84 to interact with either Y26 as before, leading to a productive nicking reaction, or to interact with Y19, which would lead to a nonproductive interaction. In the case of Y18+19, the addition of Y27 forces Y19, which is interacting with D84, to either maintain its interaction with Y18, leading to a productive nicking reaction, or to interact with Y27, which results in limited DNA nicking (Table 1).
Finally, the nicking activity of WT_299, Y26,27F_299, Y18,19F_299, and Y18,19,27F_299 are all inhibited to ~50% of wild type upon mutation of D84 to alanine. In contrast, mutation of D84 increased the nicking activity of Y18 alone from ~10% of wild type to ~50% of wild type (Figure 3). Two conclusions are drawn from these data. First, since mutation of D84A did not eliminate the activity of the wild type relaxase, we conclude that the nicking tyrosine can be deprotonated for attack via a second, though less effective method. Two scenarios could produce this partially deprotonated tyrosine. If the local pH of the relaxase active site is higher than that of bulk solution, the tyrosine hydroxyl will exist in a protonateddeprotonated equilibrium. Alternately, a second basic residue could weakly activate the tyrosine. Analysis of the structure of the pCU1 relaxase active site fails to reveal an obvious candidate for a secondary base; however, the surface of the active site cavity exhibits an overall negative potential which could increase the local pH. Therefore, it is most likely the tyrosine is weakly deprotonated due to a high local pH within the active site. Second, since Y19,26,27F_299 was the only mutant to increase its DNA nicking activity in the presence of D84A, we conclude this increased activity is likely a result of a change in the net charge on the scissile phosphate in the relaxase active site upon mutation of D84. D84 is positioned to polarize one of the HUH motif histidines of pCU1 relaxase (H162) by shifting the side chain proton on its εN to its δN, thus generating a partial negative charge at the εN, which faces the bound cation in the pCU1 active site (Figure 4). As a result, this increases the net negative charge of the HUH motif + scissile phosphate. Therefore, mutation of D84 to alanine shifts this net charge positive. Since Y18, when present alone, does not benefit from activation by D84 (Figure 4), this increase in net charge on the scissile phosphate is expected to enhance Y18's ability to attack the scissile phosphate within the active site (Table 1).
In summary, these data describe the mechanism utilized by the pCU1 relaxase to accomplish DNA nicking. This analysis provides another distinct example of the variety of approaches conjugative relaxases employ to achieve plasmid transfer [12-15,18,26]. Previous data have shown that, during transfer of the conjugative F plasmid, its multi-tyrosine relaxase only uses its first tyrosine in primary sequence to nick DNA [14]; in contrast, the closely related TrwC multi-tyrosine relaxase uses the first tyrosine during initiation of plasmid transfer, but the third during termination [15,18]. The structurally homologous Mob A relaxase accomplishes plasmid transfer with only one functional DNA nicking tyrosine [13]. For pCU1, as we have outlined here, either the third tyrosine or a combination of the first and second tyrosines perform the critical first DNA cleavage step by this relaxase. It is possible that as additional plasmid systems and their respective relaxase enzymes are investigated, further mechanistic variations may be observed.
5. Acknowledgements
This work was supported by the National Institutes of Health (AI078924, M.R.R., ES016488, R.P.N., T32GM008719)
We thank Suzanne Paterson for the generous gift of the TraI gene. We thank Yuan Cheng and other members of the Redinbo lab for assistance with manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- 1.Willetts N, Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiology Reviews. 1984;48:24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Molecular Microbiology. 1997;25:1011–22. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- 3.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiology Reviews. 2010;34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 4.Lanka E, Wilkins BM. DNA processing reactions in bacterial conjugation. Annual Review of Biochemistry. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 5.Boer R, Russi S, Guasch A, Lucas M, Blanco A, Perez-Luque R, Coll M, de la Cruz F. Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. Journal of Molecular Biology. 2006;358:857–869. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nature Structural Biology. 2003;10:1002–10. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 7.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Research. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, Ilyina TV. Computer-assisted dissection of rolling circle DNA replication. BioSystems. 1993;30:241–268. doi: 10.1016/0303-2647(93)90074-m. [DOI] [PubMed] [Google Scholar]
- 9.Larkin C, Haft RJ, Harley MJ, Traxler B, Schildbach JF. Roles of active site residues and the HUH motif of the F plasmid TraI relaxase. Journal of Biological Chemistry. 2007;282:33707–13. doi: 10.1074/jbc.M703210200. [DOI] [PubMed] [Google Scholar]
- 10.Datta S, Larkin C, Schildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13:1533–1544. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 12.Lujan SA, Guogas LM, Ragonese H, Matson SW, Redinbo MR. Disrupting antibiotic resistance propagation by inhibiting the conjugative DNA relaxase. Proceedings of the National Academy of Science USA. 2007;104:12282–12287. doi: 10.1073/pnas.0702760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monzingo AF, Ozburn A, Xia S, Meyer RJ, Robertus JD. The structure of the minimal relaxase domain of MobA at 2.1 Å resolution. Journal of Molecular Biology. 2007;366:165–78. doi: 10.1016/j.jmb.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dostal L, Shao S, Schildbach JF. Tracking F plasmid TraI relaxase processing reactions provides insight into F plasmid transfer. Nucleic Acids Research, Advance Access. 2011 doi: 10.1093/nar/gkq1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. Journal of Molecular Biology. 2000;295:1163–72. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- 16.Scherzinger E, Kruft V, Otto S. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleavage/DNA-joining activity. FEBS Journal. 1993;217:929–938. doi: 10.1111/j.1432-1033.1993.tb18323.x. [DOI] [PubMed] [Google Scholar]
- 17.Street LM. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochimica et Biophysica Acta. 2003;1646:86–99. doi: 10.1016/s1570-9639(02)00553-8. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Perez B, Lucas M, Cooke LA, Vyle JS, de la Cruz F, Moncalian G. Analysis of DNA processing reactions in bacterial conjugation by using suicide oligonucleotides. EMBO Journal. 2007;26:3847–57. doi: 10.1038/sj.emboj.7601806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khatoon H, Iyer RV, Iyer VN. A new filamentous bacteriophage with sex-factor specificity. Virology. 1972;48:145–155. doi: 10.1016/0042-6822(72)90122-5. [DOI] [PubMed] [Google Scholar]
- 20.Konarska-Kozlowska M, Iyer VN. Physical and genetic organization of the IncN-group plasmid pCU1. Gene. 1981;14:195–204. doi: 10.1016/0378-1119(81)90115-3. [DOI] [PubMed] [Google Scholar]
- 21.Nash RP, Habibi S, Cheng Y, Lujan SA, Redinbo MR. The mechanism and control of DNA transfer by the conjugative relaxase of resistance plasmid pCU1. Nucleic Acids Research. 2010;38:5929–5943. doi: 10.1093/nar/gkq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paterson E, More M, Pillay G, Cellini C, Woodgate R, Walker G, Iyer V, Winans S. Genetic analysis of the mobilization and leading regions of the IncN plasmids pKM101 and pCU1. Journal of Bacteriology. 1999;181:2572–83. doi: 10.1128/jb.181.8.2572-2583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams SL, Schildbach JF. Examination of an inverted repeat within the F factor origin of transfer: context dependence of F TraI relaxase DNA specificity. Nucleic Acids Research. 2006;34:426–35. doi: 10.1093/nar/gkj444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dyda F, Hickman AB. A Mob of Reps. Structure. 2003;11:1310–1311. doi: 10.1016/j.str.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 25.Xia S, Robertus J. Effect of divalent ions on the minimal relaxase domain of MobA. Archives of Biochemistry and Biophysics. 2009;488:42–47. doi: 10.1016/j.abb.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiology Reviews. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]