Abstract
Hydroxysteroid sulfotransferase (SULT2A) enzymes play important roles in hepatic steroid and xenobiotic metabolism. Unlike humans, which express one SULT2A, inspection of mouse genome information indicated the presence of seven SULT2A genes within a cluster on chromosome 7. The age- and sex-dependent expression of the seven murine SULT2A family members were characterized in the livers of C57BL/6 mice using real-time RT-PCR. The transcripts for three of the SULT2A forms (NCBI reference/model sequences XM_001471624, NM_009286 and NM_001111296) were abundant in pre-pubertal male and female mouse liver but were essentially silenced in the livers of adult male mice. The mRNAs of three other SULT2A forms (NM_001101534, XM_894052 and NM_001081325) were also expressed in pre-pubertal male and female mouse liver, but at markedly reduced levels relative to those of the abundant forms. The mRNA levels of these lower-abundance forms were further suppressed in adult animals. A seventh SULT2A mRNA (XM_983034) was expressed in adult male and female mouse liver, but was not detected in pre-pubertal mouse liver of either sex. Full-length amplifications with primers targeting untranslated regions confirmed that all SULT2A forms were expressed. However, while the XM_001471624, NM_001111296, NM_001101534, XM_894052 and NM_001081325 transcripts were detected at their predicted sizes, the NM_009286 and XM_983034 transcripts each lacked two predicted exons. These results demonstrate that seven murine SULT2As display different profiles of age- and sex-dependent hepatic expression.
Keywords: hydroxysteroid sulfotransferase, SULT2A gene family, mouse liver gene expression, transcriptional regulation, steroid and xenobiotic metabolism
1. Introduction
Hydroxysteroid sulfotransferase (SULT2A) enzymes catalyze the sulfonation of hydroxysteroids, xenobiotics and toxic hydrophobic bile acids, and therefore represent important determinants of detoxication, carcinogen bioactivation and cholesterol homeostasis [1;2]. Unlike humans which express a single SULT2A enzyme (SULT2A1) in liver [3], rats [4] and mice [5] express several hepatic SULT2A enzymes that are subject to differential age- and sex-dependent regulation. Since mouse models are becoming increasingly important paradigms for investigating metabolic networks, gaining a clearer understanding of the expression and regulation of murine hepatic SULT2A enzymes is essential.
Originally, Kong and Fei cloned two SULT2A cDNAs from a female B6CBA mouse liver cDNA library (mSTa1 and mSTa2) and classified these enzymes as orthologs of one of the female-predominant SULT2As expressed in rat liver [6]. Alnouti and Klaassen later found that the mRNA expression of these two murine SULT2As was abundant in the livers of 8 week old female mice but was not detected in the livers of adult male mice [5]. These findings confirmed earlier observations which noted that as male mice age past puberty, a marked reduction in hepatic SULT2A expression occurs, presumably to minimize SULT2A-catalyzed inactivation of the androgen precursor hormone dehydroepiandrosterone and to maximize the virilization of hepatic gene expression in the maturing male [7].
Using northern blot analysis, we previously demonstrated that murine hepatic SULT2A mRNA is more abundant in the livers of several strains of adult female relative to male mice [8]. In primary cultured hepatocytes prepared from adult female C57BL/6 mice, SULT2A mRNA expression was significantly induced in response to treatment with dexamethasone, both at a low concentration sufficient to activate the glucocorticoid receptor, as well as a higher concentration capable of additionally activating the pregnane X receptor (PXR)·retinoid X receptor (RXR) heterodimer [8]. However, neither basal nor dexamethasone-inducible SULT2A expression was detected in primary hepatocytes prepared from adult male mice [8]. Since then, further SULT2A transactivation studies in mouse models have ascribed a regulatory role to several RXR heterodimer nuclear receptors including PXR [9;10], constitutive androstane receptor [11;12], farnesoid X receptor [13;14], liver X receptor [15] and the vitamin D receptor [16;17]. Despite the extent of these prior investigations, the gene-specific regulation of murine hepatic SULT2A expression has yet to be clarified. An evaluation of current mouse genome information suggests the existence of a cluster containing up to seven distinct SULT2A genes on chromosome 7. Therefore, the present study was undertaken to characterize and quantify the age- and sex-dependent hepatic expression of these seven putative SULT2A genes.
2. Materials and Methods
2.1. Animals
Male and female C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in an AAALAC approved animal care facility under a 12 hr light-12 hr dark cycle and an ambient air temperature of 23°C. Mice were euthanized at either age 21 days or age 56 days, and livers were isolated for the preparation of genomic DNA and total RNA.
2.2. Evaluation of mouse genomic information and preparation of reagents for RT-PCR analysis
Because National Center for Biotechnology Information (NCBI) mouse genome information suggested the existence of multiple SULT2A genes, our initial goal was to develop quantitative RT-PCR assays that would permit quantification of expression of each of these genes. Initially, the available information suggested the existence of five candidate SULT2A genes within a region spanning from nt 12633712 to 13393106 of chromosome 7 (NC_000073.4; note assembly 4, May 2, 2006), corresponding to mRNA reference or model sequences NM_009286, XM_485863 (subsequently renamed NM_001101534), XM_894052, XM_894065 (subsequently renamed NM_001081325) and XM_983034. The coding regions of these reference sequences were identified and globally aligned using Clustal X [18] (Supplemental Fig. 1). The probable exon-intron structure of each gene was identified by aligning the transcript reference sequence against the corresponding chromosome 7 location, using Spidey (NCBI). The global alignment suggested that exon 2 contained sufficient differences among sequences to permit the design of PCR primer pairs that could specifically amplify each transcript (Table 1, Supplemental Fig. 1). Each primer pair was designed using Oligo Primer Analysis Software (version 6.71, Molecular Biology Insights, Cascade, CO) such that the 3’-end of at least one of the two primers contained a mismatch from all other SULT2A sequences at the terminal nucleotide. These PCR primers (Integrated DNA Technologies, Coralville, IA) were used in 20 μl PCR reactions containing 50 ng mouse genomic DNA (prepared using the RNeasy Blood and Tissue Kit, Qiagen Inc., Valencia, CA) as template, 1X PCR reaction buffer, 1.5 mM magnesium chloride, 200 μM dNTPs, 0.1 μM forward and reverse primer, and 0.5 U Taq DNA polymerase (Invitrogen Corporation, Carlsbad, CA). Cycling parameters were 30-40 cycles of 94°C for 15 sec, 56°C (or 50°C for XM_894052) for 15 sec, and 72°C for 45 sec. Following cycling, aliquots of reactions were resolved on 1% agarose gels, and ethidium bromide-stained DNA fragments were recovered using the QIAquick Gel Extraction Kit (Qiagen), ligated into the pGEM-T Easy plasmid (Promega Corporation, Madison, WI) and sequenced, using the services of the Applied Genomics Technology Center at Wayne State University (Detroit, MI). The sequence of each PCR product was identical to that of the corresponding reference sequence, suggesting that the various primer pairs were specific to the targeted SULT2A sequences. To prepare an exon 2 standard for each sequence, new sets of primers were designed, in which sufficient nucleotides were added to the 5’-ends of the original primers to yield a PCR product spanning from the beginning to the end of exon 2 (Table 1). Also, a Hind III restriction site was added to the 5' end of each forward primer, and an Xba I site was added to the 5' end of each reverse primer. These primers were used in PCR reactions using the plasmids containing the original cloned SULT2A amplicons as templates, and the products were ligated into a pGEM-4Z-based poly(A) tract-containing plasmid, as previously described [19]. After preparing these initial reagents, re-evaluation of the NCBI mouse genome information indicated that a new assembly of mouse chromosome 7 had been generated (NC_000073.5, July 10, 2007), which included an additional SULT2A gene, corresponding to mRNA reference sequence XM_001471625 (subsequently renamed NM_001111296). Moreover, additional BLAST (NCBI) analysis suggested the existence of yet another SULT2A gene on an alternate (Celera) assembly of chromosome 7, corresponding to mRNA reference sequence XM_001471624. We therefore included these sequences in the global alignment and Spidey analyses, and determined, using the terminal mismatch criterion described above, that all of the originally designed assays should remain specific for detection of their targets. We then designed new assays for detection of NM_001111296 and XM_001471624, using the approach described above.
Table 1. Sequences of primers used in this study.
Primers for preparation of standards begin with a GCG clamp, followed by either a Hind III site (for forward primers) or an Xba I site (for reverse primers). Sizes of amplicons are indicated; for UTR primers, predicted and (actual) sizes are presented. F, forward primer; R, reverse primer.
| RefSeq | Purpose | Primers | Size (nt) |
|---|---|---|---|
| XM_001471624 | qRT-PCR | F:AGACCAAGGGAGATCCG | 117 |
| R:ATATGAGTCGTGGTCCCTCT | |||
| Exon 2 standard | F:GCGAAGCTTGAACAAACTGGTTGGTTGAGATTGTCTGCTTGATTCAGACCAAGGGAGATCCG | 209 | |
| R:GCGTCTAGACTTGGCCTTGGAACTGAAGAAAGACTTGGAGAAGAGATGAATGGGAAGATGGGAGGATATGAGTCGTGGTCCCTCT | |||
| UTR | F: CCTGGACTGGAATC | 982 (982) | |
| R: TATGACCATATTCAGGTA | |||
| NM_009286 | qRT-PCR | F:AACTGGCTGAATGAGATTGTA | 138 |
| R:GTGGTCCTTCCTTATTGATTAT | |||
| Exon 2 standard | F:GCGAAGCTTGAACGAACTGGCTGAATGAGATTGTA | 209 | |
| R:GCGTCTAGACTTGGCCTTGGAACTGAAGAAAGACTTGGAGAAGAGATGGATGGGAAGATGGGAGGTTATGAGTCGTGGTCCTTCCTTATTGATTAT | |||
| UTR | F: GGCTGGAATCCTAAG | 978 (705) | |
| R: TATGACCATATTCAGGTA | |||
| NM_001111296 | qRT-PCR | F:AACTGTGCCCATTTGGA | 80 |
| R:GTGGTCCTTCCTTATTGATTAA | |||
| Exon 2 standard | F:GCGAAGCTTGAACGAACTGGCTGATTGAGATTGTATGCTTGATTCAGACCAAGGGAGATCCGAAGTGGATCCAAACTGTGCCCATTTGGA | 209 | |
| R:GCGTCTAGACTTGGCCTTGGAACTGAAGAAAGACTTGGAGAAGAGATGGATGGGAAGATGGGAGGTTATGAGTCGTGGTCCTTCCTTATTGATTAA | |||
| UTR | F: GGCTGGAATCCTAAGA | 972 (972) | |
| R: CCACATCTAGGTATTCAAGA | |||
| NM_001101534 | qRT-PCR | F:ATCCAAACTGTGTCCATTCA | 109 |
| R:TGGGAAGATGGGAGGTTA | |||
| Exon 2 standard | F:GCGAAGCTTGAACAAACTGGTTGATTGAGATTGTCTGCTTGATTCAGACCAAGGGAGATTCAAAGTGGATCCAAACTGTGTCCATTCA | 209 | |
| R:GCGTCTAGACTTGGCCTTAGAACTGAAGAAAGACTTGGAGAAGAGATGGATGGGAAGATGGGAGGTTA | |||
| UTR | F: CTGGGCTGGAATTCT | 988 (988) | |
| R: GGATTTATTCTATGACCACA | |||
| XM_894052 | qRT-PCR | F:CTTGATTCAGACCAGG | 155 |
| R:TGAAGAAGAGATGGACA | |||
| Exon 2 standard | F:GCGAAGCTTGAACACACTGGCTGATTGAGATTGTCTGCTTGATTCAGACCAGG | 209 | |
| R:GCGTCTAGACTTGGCCTTGGAACTGAAAAAAGACTTGAAGAAGAGATGGACA | |||
| UTR | F: GTCTTCGGCTGGAATT | 971 (971) | |
| R: CAGGTGTTCAAGATACAGAA | |||
| NM_001081325 | qRT-PCR | F: AGATTGTCTGCTTGATTCAG | 125 |
| R: GTGGTCCTTCCTTACTGG | |||
| Exon 2 standard | F:GCGAAGCTTGAACAAACTGGCTGATTGAGATTGTCTGCTTGATTCAG | 209 | |
| R:GCGTCTAGACTTAGCTTTGGAACTGAAGAAAGACTTGGAGAAAAGATGGATGGGAAGATGGGAGGTCATGAGTCGTGGTCCTTCCTTACTGG | |||
| UTR | F: GGTCAGGGCTGGAAC | 981 (981) | |
| R: TATTCTATGACCGCATTCAA | |||
| XM_983034 | qRT-PCR | F:TGATTCAGAAGAAGGGAGATAC | 170 |
| R:TGGAACTAAAGAGAGACTTGTG | |||
| Exon 2 standard | F:GCGAAGCTTGAATGACCTGGCTTGTTGAGATTGTCTGCTTGATTCAGAAGAAGGGAGATAC | 209 | |
| R:GCGTCTAGACTTGGTCTTGGAACTAAAGAGAGACTTGTG | |||
| UTR | F: CATCAGACTACGTTTGGT | 1003 (738) | |
| R: TTGTACAAGAACTGGAAAAT | |||
2.3. Real-time RT-PCR analysis of SULT2A expression
RNA was isolated from mouse liver samples using the RNeasy Mini Kit (Qiagen), with on-column DNase treatment, according to the manufacturer's instructions. Samples of total RNA (2.5 μg) were reverse transcribed in a 20 μl reaction volume using oligo(dT) and Superscript II (Invitrogen). As negative controls, equivalent amounts of total RNA were “mock-reversed transcribed” by performing the reactions in the absence of the reverse transcriptase enzyme.
SULT2A RNA standards were prepared as described previously [19]. The above-described plasmids containing the SULT2A exon 2 fragments were linearized with EcoR I, and transcripts were generated in vitro using the MEGAScript T7 Kit (Ambion, An Applied Biosystems Business, Austin, TX) according to the manufacturer's instructions. The qualities of the synthetic RNAs were evaluated using an RNA 6000 Nano LabChip kit and 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE) according to the manufacturer's instructions. RNA concentrations were calculated from UV absorbencies and concentrations converted to molarities based on known transcript sizes. For each standard, a range of RNA amounts was reverse transcribed in parallel with the liver RNAs.
Real-time RT-PCR was performed as previously described [19]. All PCR amplifications were performed using an ABI Prism 7000 Sequence Detection System in a final volume of 25 μl SYBER Green PCR Master Mix (Applied Biosystems, Foster City, CA). The PCR thermocycling parameters were as follows: 52°C for 2 min, 94°C for 10 min, and 40 cycles at 95°C for 15 sec and 60°C for 1 min. Cycle threshold (Ct) values were determined using SDS software (Applied Biosystems). For each SULT2A form, a standard curve of Ct value versus pmol input synthetic RNA was prepared. Amounts of the corresponding hepatic SULT2A mRNA were then expressed as attomol SULT2A per μg total RNA. Since the RT-PCR assays were performed using primer pairs targeting a single exon, control reactions containing equivalent amounts of the mock-reverse transcribed samples were also performed to rule out the possibility that any fluorescent signals were derived from contaminating genomic DNA. Real-time RT-PCR data were analyzed using one-way analysis of variance followed by the Newman-Keuls multiple comparison test, with p<0.05 considered as significantly different.
2.4. Standard RT-PCR and sequence analysis
Primers corresponding to sequences within the SULT2A mRNA untranslated regions (UTRs) were designed to permit amplification of the entire SULT2A coding regions. Since UTR sequences were not published for all of the SULT2A mRNA sequences, the NM_001111296 sequence, which contained the longest reported 5’- and 3’-UTRs (54 and 103 nt, respectively), was used as the basis for the assumption that the 5’- and 3’-UTRs of all SULT2A transcripts were at least 50 nt and 100 nt, respectively. Sequences were then retrieved from NC_000073.5 (NC_000073.4 for NM_009286) as necessary to produce SULT2A mRNA sequences with 50 nt 5’-UTR and ~100 nt 3’-UTR (Supplemental Fig. 1). Primers were then designed using Oligo Primer Analysis Software (Table 1, Supplemental Fig. 1). Total RNA from the liver of a pre-pubertal female mouse or an adult male mouse was reverse transcribed, as described above, and used as template for PCR reactions (the female template was used for all SULT2A PCR reactions except for XM_983034; the male template was used for XM_983034). PCR reactions (50 μl) contained 2 μl template, 1X Herculase II Reaction Buffer (Stratagene, La Jolla, CA), 250 μM dNTPs, 0.25 μM forward and reverse primer, and 0.5 U Herculase II Fusion DNA Polymerase (Stratagene). Cycling parameters were 95°C for 2 min followed by 35 cycles of 95°C for 20 sec, annealing temperature (50°C for XM_001471624 and NM_009286; 55°C for NM_001111296, NM_001101534 and XM_983034; and 58° for XM_894052 and NM_001081325) for 20 sec and 72°C for 2 min, followed by a final extension of 72°C for 3 min. After cycling, entire reactions were resolved on 0.8% agarose gels, and ethidium bromide-stained DNA fragments were visualized and recovered using the QIAEX II Gel Extraction Kit (Qiagen). The fragments were eluted with water and A-tailed by the addition of 1X PCR Reaction Buffer, 1.5 mM magnesium chloride, 200 μM dATP and 5 U Taq DNA Polymerase. Following incubation for 15 min at 70°C, the samples were extracted with phenol-chloroform followed by chloroform according to standard procedures. Glycogen (5 μg, Ambion) was then added and the samples were ethanol precipitated overnight at -80°C. After centrifugation and aspiration of supernatants, PCR products were ligated into the pGEM-T Easy plasmid by addition 3 μl water, 1X Rapid Ligation Buffer (Promega), 50 ng pGEM-T Easy and 3 U T4 ligase directly to the pellets. Following the application of standard molecular cloning procedures, plasmid DNA was isolated from individual clones using the QIAprep Spin Miniprep Kit (Qiagen), digested with EcoR I, visualized on 0.8% agarose gels, and sequenced.
3. Results
Information published to date has supported the existence at least two SULT2A family members that are expressed in mouse liver [5;6]. However, inspection of the NCBI mouse genome information suggested the presence of a SULT2A cluster containing at least five genes within a ~800,000 nt region of chromosome 7 (NC_000073.4), corresponding to mRNA reference/model sequences NM_009286, NM_001101534, XM_894052, NM_001081325 and XM_983034. The genes generally contained comparable intron-exon structures, with NM_009286, NM_001101534, XM_894052 and NM_001081325 containing 6 predicted exons, and XM_983034 containing 7 predicted exons (Supplemental Fig. 1). All of the transcripts contained open reading frames encoding predicted proteins with 284 or 285 amino acids. Of these sequences, NM_009286 was assigned based on the mSTa2 sequence described by Kong and Fei [6] (GenBank Accession no. L27121). However, NM_009286 exhibited 19 mismatches when compared to the assigned locus on chromosome 7 (12633719-12673171 of NC_00073.4). None of the SULT2A genes predicted to exist in NC_000073.4 appeared to correspond to the mSTa1 sequence (L02335) that was previously described by Kong and Fei [6]. To evaluate the expression of these multiple NCBI-predicted SULT2A genes, we developed quantitative RT-PCR assays, according to the approaches described in Methods. However, after developing these initial assays, re-inspection of the NCBI mouse genome information indicated that a new assembly of chromosome 7 had been generated (NC_00073.5) and evaluation of that information indicated several differences between the latter and former assemblies. NM_001101534, XM_894052, NM_001081325 and XM_983034 were present in both assemblies. By contrast, NM_009286 was not identified as existing on NC_000073.5, while four other sequences, XM_001479804, XM_001479810, XM_001479814 and NM_001111296 were identified as being present.
XM_001479804, XM_001479810 and XM_001479814 are identified as transcript variants (TVs) from a single gene (LOC100043194, located at 14318854-14363985 of NC_000073.5), with XM_001479804 (TV1) and XM_001479814 (TV3) being very closely-related sequences, each predicted to encode a protein containing 275 amino acids. However, the predicted intron-exon structures of TV1 and TV3 differ substantially from those of all of the other SULT2A sequences, raising doubt as to the actual existence of these transcripts. By comparison, XM_001479810 (TV2) represents a shortened transcript, containing only predicted exons 1, 2, 3 and 6. Although NM_009286 is not identified as existing on NC_000073.5, it is grouped together with XM_001479804, XM_001479810 and XM_001479814 in one UniGene cluster (Mm.457986). The predicted exon 1, 2, 3 and 6 regions of NM_009286 align closely with the corresponding regions of XM_001479804, XM_001479810 and XM_001479814, while the predicted exon 4 and 5 regions of NM_009286 do not show significant alignment to the corresponding regions of XM_001479804 and XM_001479814.
The sequence of NM_001111296 was identical to the mSTa1 sequence of Kong and Fei [6] (L02335), except for a 3 nt insertion in L02335 near the start codon. Therefore, NM_001111296 appears to be mSTa1. Additional BLAST analysis identified another sequence, termed XM_001471624, which is not identified as existing on NC_000073.5, but rather is indicated as being present on an alternate (Celera) assembly of chromosome 7. Alignment of XM_001471624 against the SULT2A cluster region of NC_000073.5 identified a region (14209376-14256150) of perfect alignment, with conservation of all of the splice sites seen for the other SULT2A genes. Therefore, the basis for non-assignment of XM_001471624 to this location of NC_000073.5 is not clear.
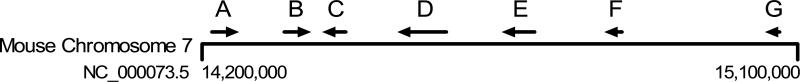
Based on the above-described information, we deduced the existence of seven SULT2A genes within the region of chromosome 7 extending from nt 14209376 to 15078275 (NC_000073.5). These genes are predicted to generate transcripts corresponding to XM_001471624 (A), NM_009286 (and/or one or more of the three above-described TVs, B), NM_001111296 (C), NM_001101534 (D), XM_894052 (E), NM_001081325 (F) and XM_983034 (G). The predicted locations and orientations of these genes on chromosome 7 are indicated in Fig. 1, and additional information, including UniGene clusters (if any), gene symbols and locations on chromosome 7 (NC_000073.5), and protein reference sequences and sizes, is contained in Table 2. Global alignments of all sequences, predicted exon demarcations, and locations of primer pairs used for RT-PCR are shown in Supplemental Fig. 1.
Fig. 1.
Representation of positions and orientations of seven mouse SULT2A genes on chromosome 7, assembly NC_000073.5. Additional information about these genes is provided in Table 2.
Table 2.
NCBI information about mouse SULT2A formsa
| SULT2A | RefSeq | UniGene | Gene Symbol | NC_000073.5 | Protein | #Amino acidse |
|---|---|---|---|---|---|---|
| A | XM_001471624 | None | EG434264 | 14209376 | XP_001471674 | 285 |
| 14256150b | ||||||
| B | XM_001479804 | Mm.457986 | LOC100043194c | 14318854 | XP_001479854 | 275 |
| XM_001479810 | 14364985 | XP_001479860 | 193 | |||
| XM_001479814 | XP_001479864 | 275 | ||||
| NM_009286 | C730007P19Rik | NP_033312 | 285 (194) | |||
| C | NM_001111296 | Mm.260026 | Sult2a1 | 14381602 | NP_001104766 | 285 |
| 14422724Cd | ||||||
| D | NM_001101534 | Mm.260026 | EG434121 | 14495025 | NP_001095004 | 284 |
| 14574940C | ||||||
| E | XM_894052 | None | EG629203 | 14652803 | XP_899145 | 284 |
| 14708342C | ||||||
| F | NM_001081325 | Mm.439627 | EG629219 | 14807826 | NP_001074794 | 285 |
| 14840182C | ||||||
| G | XM_983034 | None | EG638251 | 15050508 | XP_988128 | 298 (184) |
| 15078275C | ||||||
The information is updated frequently. All information is current as of July 14, 2008.
The gene corresponding to XM_001471624 is not identified as existing on the NC_000073.5 assembly. The indicated coordinates were derived from BLAST and Spidey analysis.
Gene Symbol LOC100043194 is given to the gene corresponding to mRNA sequences XM_001479804, XM_001479810 and XM_001479814 on assembly NC_000073.5. Gene symbol C730007P19Rik is given to the gene corresponding to mRNA sequence NM_009286 on assembly NC_000073.4.
C, complementary orientation.
Numbers of amino acids are those stated by NCBI for the indicated reference proteins. Numbers in parentheses are predicted numbers of amino acids based on in silico translation of RT-PCR product sequences.
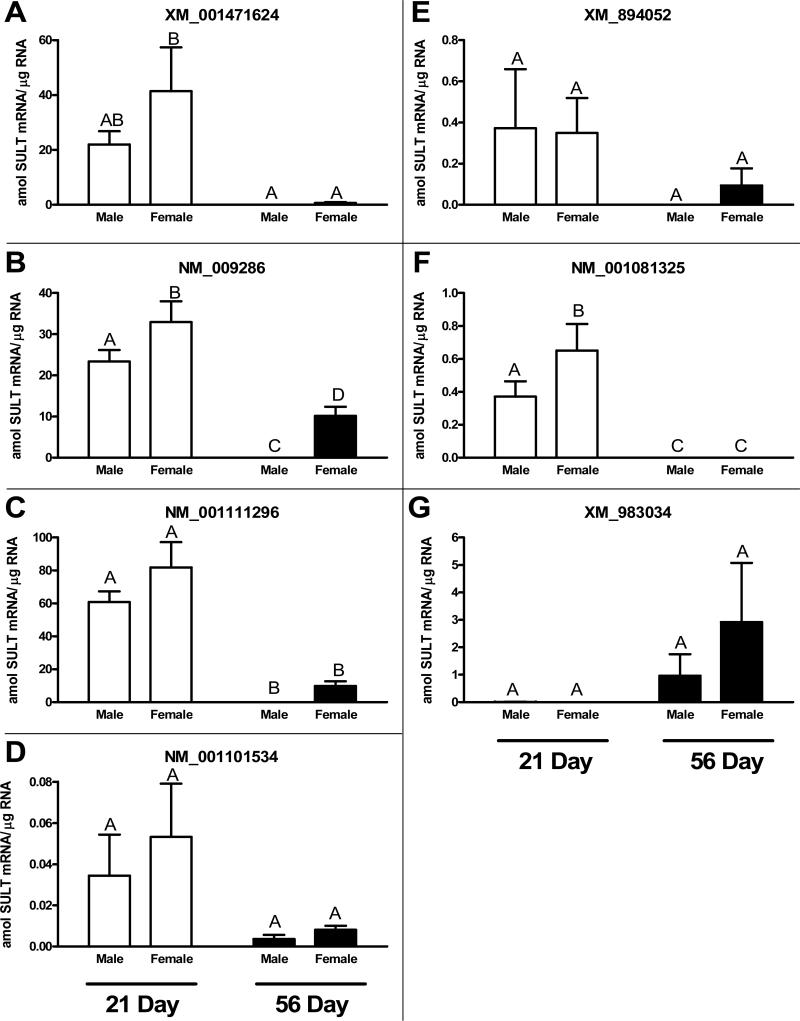
We evaluated the hepatic expression of the seven candidate SULT2A transcripts in prepubertal (21 day) and adult (56 day) male and female mice (Fig. 2). Qualitatively, the expression pattern was comparable for most of the transcripts, with mRNA levels being relatively higher in the young mice of both sexes than they were in the adult mice, and being higher in the adult female mice than they were in the adult male mice. XM_001471624, NM_009286 and NM_001111296 were the most abundant transcripts in young mice (~20 to 80 amol SULT2A RNA per μg cellular RNA). All of these transcripts were present at negligible levels in adult male mice (<0.01 amol SULT2A RNA per μg RNA). While NM_009286 and NM_001111296 were expressed at substantial levels (~10 amol SULT2A RNA per μg RNA) in the livers of adult female mice, XM_001471624 was expressed at a lower level (~0.7 amol SULT2A RNA per μg RNA). NM_001101534, XM_894052 and NM_001081325 also displayed expression patterns that were characterized by higher expression in young mice than in adult mice. However, the absolute expression levels of these three transcripts were much lower than were the levels measured for XM_001471624, NM_009286 and NM_001111296. The expression pattern for XM_983034 was different from that of the other transcripts, with negligible expression detectable in the livers of young mice of either sex, and clear expression detectable in the livers of adult mice. The average transcript level for XM_983034 in adult male mouse liver (~1 amol SULT2A RNA per μg RNA) was more than 100-fold higher than that of any of the other SULT2A transcripts.
Fig. 2.
Quantitative expression of seven SULT2A mRNAs in pre-pubertal (age 21 days) and adult (age 56 days) male and female mouse liver. Real-time RT-PCR was applied to RNA samples isolated from the livers of male and female C57BL/6 mice euthanized at the indicated ages, as described in Methods. For each transcript, data are expressed as attomol (amol) SULT2A mRNA/μg total RNA, based on interpolations from a standard curve prepared by the parallel analysis of a range of amounts of a synthetic SULT2A RNA fragment. Each bar represents the mean ± SEM of measurements in 4 (young) or 6 (adult) mice. Groups not sharing a letter are significantly different from each other, p<0.05.
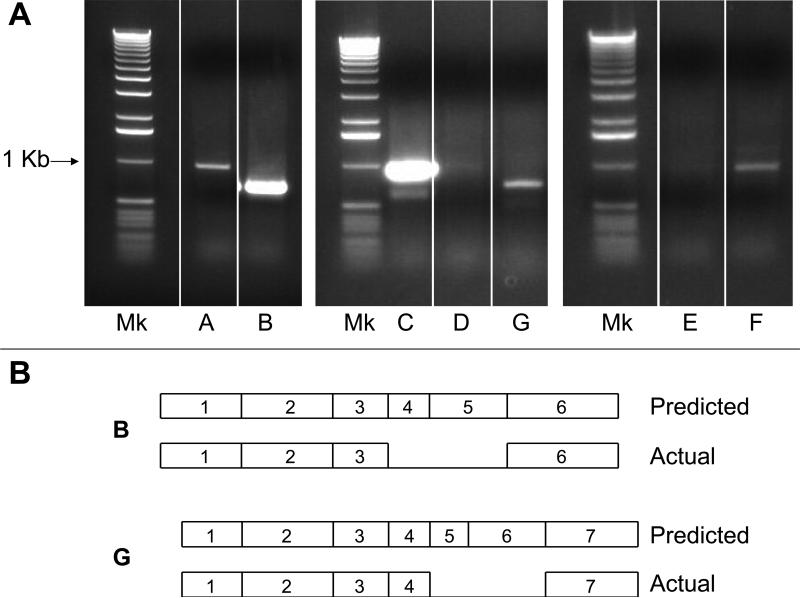
To determine whether the seven SULT2A genes were expressed as full-length transcripts (in accordance with the sequences shown in Supplemental Fig. 1), standard RT-PCR reactions were performed using primer pairs targeting the UTRs of the sequences (Fig. 3). Although these findings cannot be considered to be quantitative, the results were in general agreement with the real-time RT-PCR data described above. Thus, NM_009286 and XM_001111296 were amplified in abundance, while a clear PCR product was also obtained for XM_001471624, using a total RNA sample from a pre-pubertal female mouse liver (Fig. 3A, lanes B, C and A). By comparison, small amounts of NM_001101534, XM_894052 and NM_001081325 were amplified from the pre-pubertal female mouse liver RNA sample (Fig. 3A, lanes D, E and F). In particular, the NM_001101534 and XM_894052 amplicons were barely visible on ethidium bromide-stained gels, although the amounts were nevertheless sufficient for successful cloning. Also consistent with the real-time RT-PCR results, a clear PCR product for XM_983034 was amplified from a sample of adult male liver RNA (Fig. 3A, lane G). One clone of each amplicon was sequenced, and, with the exceptions described below, the sequences were found to be identical to the published sequences. Thus, XM_001471624, NM_001111296, NM_001101534, XM_894052 and NM_001081325 were detected as their full-length transcripts. The NM_009286 PCR product was shorter than predicted, and the sequence revealed the absence of exons 4 and 5 (represented in Fig. 3B). The sequence of the exons that were present was identical to the corresponding published sequence of NM_009286 except for two mismatches. Moreover, the amplicon sequence was identical to the published sequence of XM_001479810 (i.e., TV2 described above). The sequence of the XM_983034 PCR product revealed the absence of the predicted extra exon for this form (exon 5, see Supplemental Fig. 1) as well as exon 6 (Fig. 3B). The sequence of the exons that were present was identical to the published XM_983034 sequence.
Fig. 3.
Amplification of full-length mouse SULT2A cDNAs using UTR primers. A: Total RNA isolated from the liver of a pre-pubertal female mouse (lanes A, B, C, D, E and F) or an adult male mouse (lane G) was subjected to RT-PCR using primers targeting the 5’- and 3’-UTRs of the SULT2A sequences, as described in Methods. Following PCR, ethidium bromide-stained products were visualized on 0.8% agarose gels. Mk=1 Kb ladder; the location of the ~1 Kb band is indicated. B: Sequencing of the shorter than predicted B and G PCR fragments revealed the absence of the indicated exons (see Supplemental Fig. 1). A through G are as defined in Table 2.
4. Discussion
We and others have described the robust SULT2A expression in adult female mouse liver, and the markedly repressed expression in adult male mouse liver [7;8]. Two female-predominant murine hepatic SULT2A family members, termed mSTa1 and mSTa2, were previously reported [5;6]. However, our evaluation of available mouse genome information suggested the presence of seven SULT2A genes within a cluster on chromosome 7. In the present study, the seven murine SULT2As are shown to exhibit different hepatic expression patterns, in terms of their quantitative abundance and ageand sex-dependent regulation. Our results suggest that XM_001471624, NM_009286 and NM_001111296 are the predominant SULT2A transcripts expressed in young mice of both sexes. However, because hepatic XM_001471624 expression is markedly suppressed in adult female mice, NM_009286 and NM_001111296 are the predominant transcripts expressed in adult female mice. It is noteworthy that these two transcripts likely correspond to the mSTa1 and mSTa2 cDNA sequences that were originally cloned by Kong and Fei from a female mouse liver cDNA library [6]. By comparison, NM_001101534, XM_894052 and NM_001081325 appear to be low-abundance SULT2A forms. In general, these latter SULT2As displayed age- and sex-dependent hepatic expression profiles that were qualitatively comparable to those of the major forms. By contrast, XM_983034 displayed an expression pattern that was different from the other SULT2As, and XM_983034 appears to be the predominant SULT2A transcript that is expressed in adult male mouse liver. It should also be noted that although some of the SULT2As have been termed minor forms, it is conceivable that these forms are inducibly expressed to higher levels under certain physiological or treatment conditions.
In general, our results reconcile with published information and with the current assembly of mouse chromosome 7 (NC_000073.5). However, one issue that remains unclear involves the imperfect alignment of NM_009286 with any of the SULT2A genes. As indicated above, NM_009286 was assigned based on the mSTa2 sequence identified by Kong and Fei [6]. In the previous assembly of mouse chromosome 7 (NC_000073.4), NM_009286 was assigned to Gene Symbol C7300007P19Rik, which occurs at positions 12633719-12673171. However, NM_009286 exhibits 19 mismatches when compared to the exons predicted from this locus. In the current assembly, Gene Symbol LOC100043194 is given to positions 14318854-14364985. Despite the fact that LOC100043194 corresponds to C7300007P19Rik from NC_000073.4 (based on perfect alignments of 10 Kb of the 5’-flanking regions through exon 3), NM_009286 does not match any of the mRNAs that are predicted to be expressed from LOC100043194. Rather, three TVs, termed XM_001479804 (TV1), XM_001479810 (TV2) and XM_001479814 (TV3), are the predicted gene products. As noted above, the predicted exon-intron structures of TV1 and TV3 differ from those of all of the other SULT2A genes, while TV2 is a short sequence, which lacks predicted exons 4 and 5. There is therefore uncertainty about the nature of NM_009286, as well as the actual transcripts that are expressed from LOC100043194. The sequence differences between NM_009286 and the mouse genome-derived information cannot be attributed to simple sequencing errors of the original cDNA since two identical clones (BC125365, cDNA clone MGC:159068 IMAGE:40129880 and BC125367, cDNA clone MGC:159070 IMAGE:40129882) with sequences nearly identical to that of NM_009286 (one mismatch) were also identified under the National Institutes of Health Mammalian Gene Collection Program [20]. Our analysis, using primers targeting the 5’- and 3’-UTRs of NM_009286, revealed that only a single PCR product was amplified from pre-pubertal female mouse liver RNA, and that the sequence of this product was identical to the short XM_001479810 (TV2) sequence. Thus, we have confirmed the expression of the TV2 sequence, but have found no evidence for the expression of the TV1 or TV3 sequences. Moreover, our data do not confirm the expression of NM_009286 as a full-length transcript in C57BL/6 mouse liver, and the origin of this transcript remains unclear.
In addition to the shortened version of NM_009286, the XM_983034 PCR product that was generated using UTR primers and adult male mouse liver RNA lacked two of its predicted exons. The expression of SULT2A transcripts lacking exons prompts the question of functional significance. Whereas full-length SULT2A transcripts (e.g., NM_001111296) encode proteins predicted to contain 284 or 285 amino acids, the short NM_009286 and XM_983034 sequences are predicted to encode proteins of 194 and 184 amino acids, respectively (Table 2). It seems unlikely that either of these proteins would retain sulfotransferase activity, since amino acids essential for co-factor binding would be missing [21].
The hepatic contents of the SULT2A mRNAs are primarily determined by the transcription rates of the various genes, which are established by the functional dynamics of transcription factor-responsive motifs within the gene control regions. To provide a preliminary appreciation of the SULT2A regulatory sequences, the proximal promoter regions of the seven SULT2A genes have been aligned (Fig. 4). The alignment was limited to the region ranging from the approximate transcription start site through an IR0 motif, which has been identified as a functional response element that mediates ligand-mediated regulation by several nuclear receptors [9;10;16;17;22]. From the alignments, it is apparent that there is a high degree of sequence similarity among the proximal promoter regions of six of the SULT2A genes (XM_001471624, NM_009286, NM_001111296, NM_001101534, XM_894052 and NM_001081325), while the corresponding region of XM_983034 shows greater sequence divergence.
Fig. 4.
Computer-based identification of transcription factor binding sites in proximal promoter regions of mouse SULT2A genes. The regions of the seven mouse SULT2A genes spanning from the IR0 motif to the transcription start site (as estimated based on the 5’ position of NM_001111296) were aligned using Clustal X and were evaluated for the presence of transcription factor binding sites using MatInspector. A transcription factor binding site was considered to be a positive match if the matrix similarity of the sequence was greater than the optimized matrix threshold. The IR0 motif is not a computer-predicted site, but is included based on its demonstrated functional significance. The six SULT2A promoter sequences showing a high degree of similarity to each other are shown stacked, and * indicates positions of nucleotide identity among these six sequences. Positions and orientations of transcription factor binding sites for these six sequences are indicated with dashed arrows above the NM_009286 sequence, and sequences matching the binding sites are indicated with boldface type. The aligned promoter sequence of XM_983034 is shown below the other six sequences, and # indicates positions of nucleotide identity among all seven sequences. Positions and orientations of computer-predicted transcription factor binding sites for XM_983034 are indicated with dashed arrows above the XM_983034 sequence. GRE=glucocorticoid responsive and related elements (matrix V$ARE.01 or V$PRE.01); CEBP=CCAAT/enhancer binding protein (matrix V$CEBPB.01); HNF1=hepatic nuclear factor 1 (matrix V$HNF1.01, V$HNF1.02 or V$HNF1.03); NR2=nuclear receptor subfamily 2 factors [matrix V$PNR.01, V$TR2.01, V$TR4.01 or V$TR4.02; for some sequences this site was additionally identified with matrix V$RAR_RXR.01 or V$RAR_RXR.03 (RXR heterodimer binding sites) and/or with matrix V$PPAR_RXR.02 (peroxisome proliferator-activated receptor)]; HNF3β= fork head domain factors (V$HNF3B.01).
We previously demonstrated that glucocorticoid-inducible expression of rat hepatic SULT2A is mediated by both PXR-dependent and CCAAT/enhancer-binding protein-dependent mechanisms [23;24]. An IR0 motif with a sequence of GGGTCATGAACT was originally identified in the promoter of the rat SULT2A3 gene, and this element has been shown to mediate transcriptional regulation by several nuclear receptors, including PXR and constitutive androstane receptor [12;22]. An IR0 motif is located at approximately the same position in the promoters of several of the mouse SULT2A genes (Fig. 4). The IR0 sequence of XM_001471624, NM_009286 and NM_001111296 is the same as that found in the rat gene, and this motif has been shown to be a functional response element for PXR, vitamin D receptor and farnesoid X receptor [9;10;16;17;22]. The IR0 sequence of XM_894052 differs by one nt (GGGTAATGAACT) from the sequence indicated above, and the functionality of this motif as a liver X receptor response element has also been demonstrated [15]. The corresponding locations of NM_001101534 and NM_001081325 contain IR0 sequences with other nt changes in the upstream hexamer, and the functionality of these motifs has not been established. By contrast, an IR0 motif is not located at the corresponding location of XM_983034.
Computer-based analysis of the various SULT2A proximal promoter regions for transcription factor binding sites, using MatInspector [25], identifies a spectrum of candidate transcription factor binding sites. Some of these sites, including sites for liver-enriched transcription factors and nuclear receptors, are indicated in Fig. 4. This analysis suggests that the six most similar SULT2A promoters contain many of the same transcription factor binding sites, but that there are also differences among genes. For example, NM_009286 and NM_001111296, the two most highly expressed SULT2A transcripts in adult female mouse liver, contain the same nearly continuous series of predicted binding sites throughout their proximal promoter regions, while XM_894052 and NM_001081325, two of the low-abundance SULT2A forms, match fewer of these sites. However, XM_001471624, an abundant SULT2A form in pre-pubertal mice, and XM_894052, a low-abundance form, contain comparable patterns of predicted transcription factor binding sites.
The escalating application of transgenic mouse models to elucidate the pathways involved in xenobiotic detoxication, hepatic metabolism and diabetogenesis have implicated an important role for hepatic SULT2A in the metabolism of endogenous and toxic intermediates and as a modifier of intra-hepatic endocrine homeostasis. However, given the relatively large number of SULT2A genes that exist in the mouse, coupled with the knowledge that these genes are not regulated identically, it is essential that the specific gene under investigation in any given study be clearly identified. For example, chromatin immunoprecipitation (ChIP) analysis has been applied with increasing frequency to investigate interactions between regulatory transcription factors and endogenous promoters. Using in silico PCR (UCSC In-Silico PCR, http://genome.ucsc.edu/cgi-bin/hgPcr) as a tool, we determined that recently reported ChIP analyses focused on the IR0-containing region of murine SULT2A likely interrogated different genes. For example, the ChIP primers that were used in the study by Uppal et al. [15] are directed toward the IR0-containing region of XM_894052, while the primers used by Seo et al. [17] are predicted to amplify products from NM_009286, NM_001111296 and XM_894052. Achieving a comprehensive portrait of murine hepatic SULT2A form-specific expression would be particularly germane to investigations aiming to “knock-down” SULT2A expression in mouse liver. In light of the multiplicity of the SULT2A family in the mouse, future studies will require improved consideration of the evident age- and sex-dependent expression profiles exhibited by the individual SULT2As. Our analysis provides a solid foundation for such investigations.
Supplementary Material
Acknowledgements
This work was conducted with support from National Institutes of Health Grants ES05823 (to M.R.-M.) and HL50710 (to T.A.K.) and services provided by the Microarray and Bioinformatics Facility Core and Imaging and Cytometry Facility Core of EHS Center Grant ES06639.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 2.Okuda H, Nojima H, Watanabe N, Watabe T. Sulphotransferase-mediated activation of the carcinogen 5-hydroxymethyl-chrysene. Species and sex differences in tissue distribution of the enzyme activity and a possible participation of hydroxysteroid sulphotransferases. Biochem.Pharmacol. 1989;38:3003–3009. doi: 10.1016/0006-2952(89)90008-7. [DOI] [PubMed] [Google Scholar]
- 3.Comer KA, Falany JL, Falany CN. Cloning and expression of human liver dehydroepiandrosterone sulphotransferase. Biochem.J. 1993;289:233–240. doi: 10.1042/bj2890233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Klaassen CD. Ontogeny and hormonal basis of female-dominant rat hepatic sulfotransferases. J.Pharmacol.Exp.Ther. 1996;279:386–391. [PubMed] [Google Scholar]
- 5.Alnouti Y, Klaassen CD. Tissue distribution and ontogeny of sulfotransferase enzymes in mice. Toxicol.Sci. 2006;93:242–255. doi: 10.1093/toxsci/kfl050. [DOI] [PubMed] [Google Scholar]
- 6.Kong A-N, Fei P. Molecular cloning of three sulfotransferase cDNAs from mouse liver. Chemico-Biol.Interact. 1994;92:161–168. doi: 10.1016/0009-2797(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 7.Leiter EH, Chapman HD, Coleman DL. The influence of genetic background on the expression of mutations at the diabetes locus in the mouse. V. Interaction between the db gene and hepatic sex steroid sulfotransferases correlates with gender-dependent susceptibility to hyperglycemia. Endocrinology. 1989;124:912–922. doi: 10.1210/endo-124-2-912. [DOI] [PubMed] [Google Scholar]
- 8.Wu W, Kocarek TA, Runge-Morris M. Sex-dependent regulation by dexamethasone of murine hydroxysteroid sulfotransferase gene expression. Toxicol.Lett. 2001;119:235–246. doi: 10.1016/s0378-4274(01)00263-6. [DOI] [PubMed] [Google Scholar]
- 9.Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane X receptor (PXR). Proc.Natl.Acad.Sci.USA. 2002;99:13801–13806. doi: 10.1073/pnas.212494599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echchgadda I, SongOh T-S, Cho S-H, Rivera OJ, Chatterjee B. Gene regulation for the senescence marker protein DHEA-sulfotransferase by the xenobiotic-activated nuclear pregnane X receptor (PXR). Mech.Aging Devel. 2004;125:733–745. doi: 10.1016/j.mad.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, Zelcer N, Adachi M, Strom S, Evans RM, Moore DD, Borst P, Schuetz JD. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. J.Biol.Chem. 2004;279:22250–22257. doi: 10.1074/jbc.M314111200. [DOI] [PubMed] [Google Scholar]
- 12.Saini SPS, Sonoda J, Xu L, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. A novel constitutive androstane receptor-mediated and CYP3A-independent pathway of bile acid detoxification. Mol.Pharmacol. 2004;65:292–300. doi: 10.1124/mol.65.2.292. [DOI] [PubMed] [Google Scholar]
- 13.Miyata M, Matsuda Y, Tsuchiya H, Kitada H, Akase T, Shimada M, Nagata K, Gonzalez FJ, Yamazoe Y. Chenodeoxycholic acid-mediated activation of the farnesoid X receptor negatively regulates hydroxysteroid sulfotransferase. Drug Metab.Pharmacokinet. 2006;21:315–323. doi: 10.2133/dmpk.21.315. [DOI] [PubMed] [Google Scholar]
- 14.Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, Nagata K, Sinal CJ, Guo GL, Gonzalez FJ, Yamazoe Y. Protective role of hyroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J.Biol.Chem. 2003;278:17838–17844. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- 15.Uppal H, Saini SPS, Moschetta A, Zhou J, Gong H, Zhai Y, Ren S, Michalopoulos GK, Mangelsdorf DJ, Xie W. Activation of LXRs prevents bile acid toxicity and cholestasis in female mice. Hepatology. 2007;45:422–432. doi: 10.1002/hep.21494. [DOI] [PubMed] [Google Scholar]
- 16.Echchgadda I, Song CS, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin D receptor. Mol.Pharmacol. 2003;65:720–729. doi: 10.1124/mol.65.3.720. [DOI] [PubMed] [Google Scholar]
- 17.Seo Y-K, Chung Y-T, Kim S, Echchgadda I, Song CS, Chatterjee B. Xenobiotic- and vitamin D-responsive induction of the steroid/bile acid-sulfotransferase Sult2A1 in young and old mice: The role of a gene enhancer in the liver chromatin. Gene. 2007;386:218–223. doi: 10.1016/j.gene.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins DG, Sharp PM. CLUSTAL: A package for performing multiple sequence alignments on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 19.Thomas RD, Green MR, Wilson C, Weckle AL, Duanmu Z, Kocarek TA, Runge-Morris M. Cytochrome P450 expression and metabolic activation of cooked food mutagen 2-amino-1-methyl-6-phenylimidazo(4,5-b)pyridine (PhIP) in MCF10A breast epithelial cells. Chemico-Biol.Interact. 2006;160:204–216. doi: 10.1016/j.cbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 20.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc.Natl.Acad.Sci.USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS.Biol. 2007;5:1063–1078. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song CS, Echchgadda I, Baek B-S, Ahn SC, Oh T, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J.Biol.Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- 23.Runge-Morris M, Wu W, Kocarek TA. Regulation of rat hepatic hydroxysteroid sulfotransferase (SULT2-40/41) gene expression by glucocorticoids: Evidence for a dual mechanism of transcriptional control. Mol.Pharmacol. 1999;56:1198–1206. doi: 10.1124/mol.56.6.1198. [DOI] [PubMed] [Google Scholar]
- 24.Fang H-L, Abdolalipour M, Duanmu Z, Smigelski JR, Weckle A, Kocarek TA, Runge-Morris M. Regulation of glucocorticoid-inducible hydroxysteroid sulfotransferase (SULT2A-40/41) gene transcription in primary cultured rat hepatocytes: role of CCAAT/enhancer-binding protein liver-enriched transcription factors. Drug Metab.Dispos. 2005;33:147–156. doi: 10.1124/dmd.104.000281. [DOI] [PubMed] [Google Scholar]
- 25.Catharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. Matinspector and beyond: analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.