Abstract
A novel mechanism aimed at controlling urease expression in Helicobacter pylori in the presence of ample nickel is described. Higher urease activities were observed in an hp0868 mutant (than in the wild type) in cells supplemented with nickel, suggesting that the HP0868 protein (herein named Mua for modulator of urease activity) represses urease activity when nickel concentrations are ample. The increase in urease activity in the Δmua mutant was linked to an increase in urease transcription and synthesis, as shown by quantitative real-time PCR, SDS-PAGE, and immunoblotting against UreAB. Increased urease synthesis was also detected in a Δmua ΔnikR double mutant strain. The Δmua mutant was more sensitive to nickel toxicity but more resistant to acid challenge than was the wild-type strain. Pure Mua protein binds 2 moles of Ni2+ per mole of dimer. Electrophoretic mobility shift assays did not reveal any binding of Mua to the ureA promoter or other selected promoters (nikR, arsRS, 5′ ureB-sRNAp). Previous yeast two-hybrid studies indicated that Mua and RpoD may interact; however, only a weak interaction was detected via cross-linking with pure components and this could not be verified by another approach. There was no significant difference in the intracellular nickel level between wild-type and mua mutant cells. Taken together, our results suggest the HP0868 gene product represses urease transcription when nickel levels are high through an as-yet-uncharacterized mechanism, thus counterbalancing the well-described NikR-mediated activation.
IMPORTANCE
Urease is a nickel-containing enzyme that buffers both the cytoplasm and the periplasm of Helicobacter pylori by converting urea into ammonia and carbon dioxide. The enzyme is the most abundant protein in H. pylori, accounting for an estimated 10% of the total protein content of the cell, and it is essential for early colonization and virulence. Numerous studies have focused on the transcription of the structural ureAB genes and its control by the regulatory proteins NikR and ArsR. Here we propose that urease transcription is under the control of another Ni-binding protein besides NikR, the Mua (HP0868) protein. Our results suggest that the Mua protein represses urease transcription when nickel levels are high. This mechanism would counterbalance the NikR-mediated activation of urease and ensure that, in the presence of a high nickel concentration, urease activation is limited and does not lead to massive production of detrimental ammonia.
INTRODUCTION
Helicobacter pylori is a spiral, Gram-negative, microaerophilic bacterium that colonizes the gastric epithelium in about 50% of the world’s population (1). H. pylori has been shown to be the etiological agent of peptic ulcer disease or chronic atrophic gastritis (2), which can subsequently develop into gastric cancers (3). Despite being a neutralophile, H. pylori successfully colonizes the extremely acidic environment of the human stomach by relying on a key enzyme, urease (UreAB). This enzyme is a nickel-containing dodecameric complex that converts urea into ammonia and carbon dioxide, thereby buffering the cytoplasm and the periplasm of the bacteria (4). It is one of the most abundant proteins synthesized by this microorganism (5), and it has been shown to be essential for colonization and persistence in different animal models (6, 7). Therefore, relentless efforts have focused on better understanding the transcription of the structural ureAB genes, as well as that of other genes involved in the Ni maturation of urease, in response to different stimuli such as pH or nickel.
The urease gene cluster in H. pylori is organized into two transcriptional units, ureAB and ureIEFGH, and produces primarily three transcripts, ureAB, ureIEFGH, and ureABIEFGH (8). The ureI gene encodes an inner membrane proton-gated channel involved in importing the urea substrate into the bacterial cell (9), and the ureEFGH accessory genes are required for the assembly of Ni into the urease apoenzyme (for reviews, see references 10 and 11). Based on promoter sequence comparisons, as well as transcriptional fusion studies of sequential deletions coupled with lacZ, it is assumed that the ureAB promoter belongs to the housekeeping σ70-dependent class of promoters (8, 12). Transcription of ureAB or ureIFGH has been shown to be acid induced in several independent microarrays (13–15); the acid-induced activation of both the PureA and PureI promoters is mediated directly by the phosphorylated ArsRS (acid-responsive signaling) two-component system (16). In addition, there is now evidence that the unphosphorylated ArsRS system represses the transcription of ureB through the expression of a cis-encoded antisense small RNA under neutral pH (17). Besides, transcription of the ureAB genes is also increased when Ni is added to the medium (18). This effect is mediated mostly by the Ni2+-responsive regulator NikR, which induces urease transcription in the presence of Ni2+ (19, 20).
In H. pylori, the hydrogenase accessory proteins HypA and HypB have also been shown to be required for maturation of urease (21). The hypA and hypB genes are located at two different loci; while hypB is part of a five-gene hypBCDEF operon, the hypA (hp0869) gene is located between the flagellar hook gene flgE (hp0870) and a gene with unknown function, the hp0868 gene (Fig. 1) (22). A few years ago, a yeast two-hybrid study aimed at discovering protein-protein interactions in H. pylori suggested that HypA and HP0868 could interact with one another (Hybrigenics PIMRider database) (23). This prompted researchers in our lab to (i) construct an H. pylori hp0868 mutant to see whether urease and hydrogenase activities would be affected and (ii) express and purify HP0868 and HypA to determine if the two proteins could actually form a complex. Since an hp0868::aphA3 mutant constructed in H. pylori strain 43504 had wild-type (WT) levels of urease and hydrogenase activities and since no interaction between HypA and HP0868 could be shown, we concluded that the HP0868 protein was not involved in either hydrogenase or urease maturation (24). Despite these findings, additional attempts (described herein) were made to discover a phenotype for this mutant and to characterize the role of this gene. A Δhp0868::aphA3 deletion mutant strain was generated in several H. pylori strains and grown in increasing Ni concentrations. We have found that urease activity, synthesis, and transcription were significantly higher in the Δhp0868 mutant background than in the WT background when cells were grown in medium supplemented with 5 or 50 µM NiCl2. In addition, our results indicate that the Mua protein can coordinate Ni2+ (2 mol per dimer). Therefore, we propose that the HP0868 (Mua) protein represses urease activity when supplies of nickel are high, thus providing a counterbalance mechanism to NikR activation.
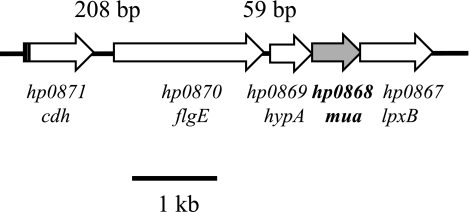
FIG 1 .
Physical map of the hypA-mua region in H. pylori strain 26695.
RESULTS AND DISCUSSION
Chromosomal location of hp0868 and distribution among bacterial species.
In H. pylori sequenced strain 26695 (22), the hp0868 gene is located downstream of the hypA (hp0869) gene (Fig. 1). hypA encodes a Ni-binding protein previously shown to be required for the maturation of both the urease and hydrogenase enzymes (21). There are homologs of HP0868 only in Helicobacter species, i.e., in all H. pylori strains sequenced to date (26695, J99, HPAG-1, G27, 98-10, and B128), as well as in Helicobacter hepaticus, Helicobacter mustelae, and Helicobacter acinonychis. In H. pylori species, the genetic organization is well conserved; hypA and hp0868 homologs are located downstream of flgE (hp0870 in strain 26695), a gene that encodes a protein known as the “flagellar hook” protein, and upstream of lpxB (hp0867), which encodes lipid A disaccharide synthetase (Fig. 1). The organization of the locus suggests that hypA, hp0868, and lpxB are cotranscribed and likely constitute a 3-gene operon, as confirmed by a recent transcriptomic study (25). In addition, there is a well-conserved 59-bp-long intergenic region between flgE and hypA, suggesting that hp0868 and hypA might not be cotranscribed along with flgE. The hp0868 gene is annotated as hypothetical (22). However, results of a yeast two-hybrid study suggested a possible role for HP0868 in urease or/and hydrogenase maturation; indeed, a significant score was reported for a putative interaction between HP0868 and HypA (23).
Construction and genetic characterization of Δhp0868 (Δmua) mutant.
A hp0868::aphA3 mutant constructed in H. pylori strain 43504 in our lab had WT levels of urease and hydrogenase activities, and no interaction between the purified HP0868 and HypA proteins could be detected, leading to the conclusion at the time that the hp0868 gene was involved in neither urease nor hydrogenase activity (24). Despite these original observations, additional attempts were made to identify the role of the hp0868 gene. To rule out the possibility of a truncated protein still being synthesized in the original strain, the hp0868::aphA3 insertional mutant, a Δhp0868 deletion mutant, was constructed in strain 26695 (see Table S1 in the supplemental material). In this mutant, 98% of the hp0868 open reading frame (ORF) is missing. The chromosomal deletion of the hp0868 gene following homologous recombination was confirmed by PCR (data not shown). The aphA3 cassette used to generate the Δhp0868 mutant was previously described as having no polar effect on the transcription of downstream genes (26). Nevertheless, the absence of a polar effect was confirmed by reverse transcriptase PCR (RT-PCR) using primers specific for lpxB (see Table S2 in the supplemental material). When total RNA from either the WT or the Δhp0868 mutant strain was used as the template, a PCR product with the expected size was obtained and there was no product in the control reaction mixture lacking RT (data not shown), confirming that lpxB was transcribed in the WT strain, as well as in the Δhp0868 mutant. Therefore, the transcription of lpxB is not affected by the construction of the Δhp0868 mutant and the phenotypes described herein are concluded to be due solely to disruption of the hp0868 gene (herein named mua for modulator of urease activity).
The Δmua mutant has higher urease activity than the WT at high Ni concentrations.
When WT strain 26695 and the Δmua mutant were grown on non-Ni-supplemented Brucella agar (BA) plates, there was no significant difference in urease activity between these two strains (Table 1), as previously reported for the mutant 43504 hp0868::aphA3 and its isogenic parental strain (24). Upon the addition of increasing NiCl2 concentrations to the growth medium, the urease activity concomitantly increased in the WT strain, as previously shown in several independent studies (for example, see reference 19). Interestingly, this increase was even more dramatic in the Δmua mutant (Table 1). Indeed, when grown on plates supplemented with identical Ni concentrations, there was 25% (for 5 µM supplemental Ni) to 90% (for 50 µM supplemental Ni) higher urease activity in the Δmua mutant than in the WT strain, suggesting that the mua gene product was acting as a repressor of urease activity, but only when ample Ni was available. The same trend was observed in other H. pylori strains into which the Δmua mutation was introduced; the 43504 Δmua and X47 Δmua mutant strains had significantly more urease activity in the presence of high Ni concentrations (10 to 50 µM) than did the isogenic parental strains (data not shown). Likewise, the original mutant constructed in our lab (43504 hp0868::aphA3) displayed the same phenotype; the urease activity of this mutant was significantly higher than that of the WT when cells were grown in the presence of 50 µM NiCl2 (data not shown).
TABLE 1 .
Urease activity in H. pylori parent and mutant strains as a function of nickel added to the growth mediuma
| Strain | Urease activity at supplemental nickel concn (µM) of: |
|||
|---|---|---|---|---|
| 0 | 0.5 | 5 | 50 | |
| WT (26695) | 5 ± 1 | 31 ± 6 | 158 ± 22 (a) | 190 ± 15 (c) |
| Δmua | 6 ± 2 | 34 ± 7 | 195 ± 20 (a) | 358 ± 54 (c) |
| ΔnikR | 6 ± 2 | 28 ± 4 | 46 ± 5 (b) | 46 ± 10 |
| Δmua ΔnikR | 5 ± 1 | 31 ± 9 | 57 ± 6 (b) | 46 ± 4 |
Urease activity is expressed as micromoles of NH3 produced per minute per milligram of total protein. Results shown are the mean ± standard deviation of three to seven independent experiments. Like letters after standard deviation values indicate a significant difference between the two mean values based on Student’s t test (P < 0.01).
Increased UreAB protein levels account for increased urease activity in the Δmua mutant.
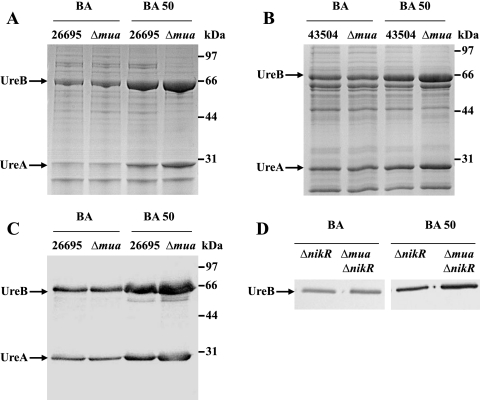
In H. pylori, it is known that the increase in urease activity under Ni-supplemented conditions is mediated mostly at the transcriptional level (18) through the action of a Ni-responsive regulator, NikR (19, 20). The marked increase in urease activity in the Δmua mutant when supplemented with Ni (compared to that in the WT) could be due to (i) increased transcription of the structural ureAB genes, possibly leading to increased UreAB protein levels, or (ii) increased Ni maturation of the urease enzyme (in this case, the amount of urease would remain the same as in the WT). To test the first hypothesis, the same amount of cell-free extracts from WT strain 26695 or the Δmua mutant (grown in the absence or presence of 50 µM Ni) was subjected to SDS-PAGE for direct comparison of the UreAB protein levels (Fig. 2A). The amounts of UreA (26.5 kDa) and UreB (61.7 kDa) synthesized in each strain grown without supplemental Ni were similar (Fig. 2A, lanes 1 and 2). The amounts of UreA and UreB increased in WT cells grown on medium with 50 µM supplemental Ni (Fig. 2A, lane 3), as previously reported by van Vliet and coworkers (19); however, under these conditions (50 µM supplemental Ni), both urease subunits were unequivocally more abundant in the Δmua mutant (Fig. 2A, lane 4) than in the WT (Fig. 2A, lane 3). The same phenotype was observed in parental strain 43504 and the isogenic Δmua mutant (Fig. 2B, compare lane 4 to lane 3). To confirm that the proteins whose synthesis was affected by the Δmua mutation were indeed UreA and UreB, an immunoblot assay using both anti-UreA and anti-UreB antibodies was performed (Fig. 2C). The immunoblot assay confirmed that the amounts of UreA and UreB were the same in the WT (strain 26695) and the Δmua mutant (lanes 1 and 2); there was more urease synthesized in the Δmua mutant than in the WT strain for cells grown with 50 µM supplemental Ni (Fig. 2C, compare lane 4 to lane 3). Taken together, these results indicated that the marked increase in urease activity observed in Δmua mutants under Ni-supplemented conditions was linked to higher urease expression, suggesting a role for Mua as a Ni-dependent transcriptional repressor of urease in H. pylori.
FIG 2 .
Effect of Ni on urease synthesis in WT or Δmua mutant bacteria. Shown is SDS-12.5% PAGE of 5 µg of cell-free extracts of WT strain 26695 or the 26695 Δmua mutant (A) or WT strain 43504 and the 43504 Δmua mutant (B). Cells were grown on medium without supplemental Ni (BA) or on medium with 50 µM supplemental Ni (BA 50). UreA and UreB are indicated by arrows, and the positions of molecular mass standards are indicated on the right. (C) Five micrograms of cell-free extracts of the WT strain (26695) or the 26695 Δmua mutant strain grown on medium without supplemental Ni (BA) or medium with 50 µM supplemental Ni (BA 50) was subjected to SDS-PAGE, transferred to nitrocellulose, and blotted with two antisera directed to subunits UreA and UreB. (D) Cell-free extracts (1 μ each) of the ΔnikR or Δmua ΔnikR mutant strain grown on medium without supplemented Ni (BA, left) or on medium with 50 µM supplemental Ni (BA 50, right) were separated on SDS-PAGE, transferred to nitrocellulose, and blotted with antiserum directed to the urease large subunit, UreB.
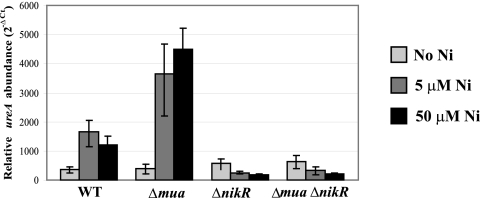
This role for Mua was further confirmed by monitoring the level of ureA gene expression using quantitative real-time PCR (qPCR). The WT and mutant strains were grown on medium not supplemented with NiCl2 or supplemented with 5 or 50 µM NiCl2, and total RNA was extracted and used as the template to synthesize cDNA. This cDNA was used in the qPCR to estimate the amount of ureA transcripts in each strain and for each Ni condition, relative to the internal control, gyrA (Fig. 3). Compared to the non-Ni-supplemented condition, there were, on average, 3.0- and 2.8-fold more ureA transcripts in the WT cells grown with 5 µM and 50 µM NiCl2-supplemented medium, respectively (Fig. 3), thus confirming the increased transcription of the ureAB genes in the presence of Ni in WT H. pylori. This result was in agreement with the immunoblotting results showing increased urease synthesis in the presence of Ni, as well as previously published results obtained with beta-galactosidase transcriptional fusions (19). In contrast, we found this ratio to be 5.4 with 5 µM Ni and 6.3 with 50 µM Ni for the Δmua mutant (Fig. 3). When the two strains were compared, this resulted in 2.1-fold (for 5 µM Ni) and 2.6-fold (for 50 µM Ni) more ureA transcripts synthesized in the Δmua mutant than in the WT strain. There was no significant difference in urease transcription between the two strains grown on non-Ni-supplemented medium. In summary, these results indicate a role for Mua as a Ni-dependent transcriptional repressor of urease.
FIG 3 .
Effects of nickel supplementation on the transcription of ureA in WT H. pylori 26695 or Δmua, ΔnikR, or Δmua ΔnikR mutants. The levels of ureA transcripts were quantified after 24 h of growth on BA medium supplemented with 0, 5, or 50 µM NiCl2. The relative abundance of each transcript was calculated with the 2−ΔCT formula and using the gyrA gene (hp0701) as a housekeeping gene control. Results are the mean and standard deviation of assays performed in triplicate. The error bars shown represent 2−ΔCT – 2−[ΔCT + SD] and 2−[ΔCT − SD] – 2−ΔCT.
H. pylori Mua represses PureA-xylE fusions in Escherichia coli.
To better understand the role of Mua, the ureA promoter was cloned upstream of the promotorless reporter gene xylE, and the resulting PureA-xylE fusion was inserted either into the cloning vector pBS-KS or into plasmid pBS-KS into which the mua ORF had been cloned (in the opposite direction and under the control of PlacZ). Each plasmid was introduced into E. coli, and cells were grown with or without 1 mM supplemental Ni before XylE activity was measured (Table 2). E. coli cells provide a good background for these assays because they possess no homolog of the genes (ureAB, mua, and xylE) studied herein. As controls, we used the hydrogenase promoter (PhydA) fused to xylE, as well as promoterless xylE. The presence or absence of mua on plasmids harboring either xylE only or PhydA-xylE fusions had no effect on the respective XylE activity. In contrast, the presence of mua on plasmids harboring PureA-xylE fusions led to 2.8- and 3.8-fold less XylE activity in Luria-Bertani liquid medium (LB) only and in LB with 1 mM Ni added, respectively. This suggested that mua was able to repress the transcription of PureA-xylE in E. coli, thus confirming the putative role of mua as a transcriptional repressor of urease. Although we saw an effect of Ni on the Mua-dependent repression of PureA-xylE, the difference does not appear to be statistically significant, suggesting that the mode of action of Mua on PureA might be different in this heterologous system.
TABLE 2 .
Expression of xylE transcriptional fusions in E. coli as a function of Nia
| Plasmid | xylE fusion | Presence of PlacZ-mua | XylE activity (units/108 cells) |
|
|---|---|---|---|---|
| No Ni added | 1 mM Ni added | |||
| pSB306 | xylE only | No | 0.12 ± 0.01 | 0.07 ± 0.01 |
| pSB328 | xylE only | Yes | 0.22 ± 0.02 | 0.17 ± 0.03 |
| pSB308 | PhydA-xylE | No | 10.47 ± 1.10 | 8.80 ± 0.29 |
| pSB330 | PhydA-xylE | Yes | 9.50 ± 0.59 | 9.08 ± 1.26 |
| pSB309 | PhydA-xylE | No | 4.93 ± 0.31 | 5.45 ± 0.25 |
| pSB331 | PhydA-xylE | Yes | 1.78 ± 0.37 | 1.43 ± 0.28 |
XylE activity is expressed as nanomoles of catechol oxidized per minute. Results shown are the mean ± standard deviation of two or three independent experiments.
Link between nikR and mua.
In H. pylori, the Ni2+-responsive regulator NikR induces urease transcription in the presence of Ni2+ (19, 20). In order to investigate the respective roles of Mua and NikR and to determine whether Mua could still repress urease transcription in the absence of NikR under high-Ni conditions, first a single ΔnikR::cat mutant was generated in the same parental strain (26695); the ΔnikR mutation was also introduced into the Δmua::aphA3 mutant to generate a Δmua ΔnikR double mutant. Both mutants were grown on non-Ni-supplemented BA or on BA supplemented with 0.5, 5, or 50 µM NiCl2 and subsequently characterized for urease activity (Table 1), urease protein synthesis (Fig. 2D), and urease transcript levels (Fig. 3). As previously described by several groups (19, 20), the deletion of the nikR gene in strain 26695 resulted in diminished Ni responsiveness of the urease activity (Table 1). In agreement with these results, immunoblotting using anti-UreB antiserum confirmed that urease synthesis was not Ni induced anymore in the ΔnikR mutant (Fig. 2D). Besides, qPCR analysis using ureA-specific primers revealed a 2- to 3-fold lesser amount of ureA transcripts in ΔnikR mutant cells grown in the presence of 5 or 50 µM NiCl2 than under the condition with no added Ni, respectively (Fig. 3). These results confirmed the central role of NikR as a primary Ni-responsive activator of urease in H. pylori.
Compared to the ΔnikR mutant, the Δmua ΔnikR double mutant showed similar or slightly higher urease activity under the Ni-supplemented condition (Table 1); however, while there were similar amounts of UreB enzyme produced in both strains grown without added Ni (Fig. 2D, left panel) or in the presence of 5 µM NiCl2 (data not shown), we found significantly higher UreB synthesis in the double mutant than in the ΔnikR mutant when cells were grown on medium supplemented with 50 µM Ni (Fig. 2D, right panel). Analysis of ureA transcripts in the double mutant by qPCR revealed a decrease in urease transcription with Ni supplementation compared to that under the condition with no added Ni, similar to the decrease observed in the ΔnikR mutant (Fig. 3). A comparison of the two strains (Δmua ΔnikR and ΔnikR mutants) revealed similar amounts of ureA transcripts for cells grown with no supplemental NiCl2 or 5 µM supplemental Ni and about 1.5-fold more ureA transcripts in the double mutant when cells were grown with 50 µM supplemental NiCl2 (Fig. 3).
Although these results suggest that Mua might repress ureAB in the absence of NikR at high Ni concentrations, they also clearly show that NikR remains the master regulator; indeed, all our experiments (urease assays, immunoblottings, and qPCRs) revealed that the Ni-dependent induction of urease seen in the WT strain was severely affected in the double mutant, as well as in the ΔnikR mutant.
Inactivation of the mua gene renders H. pylori nickel sensitive.
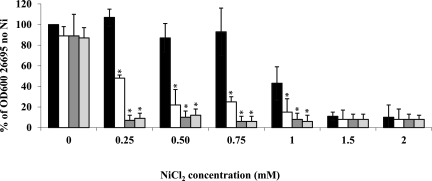
The effect of the mua deletion in the parental and ΔnikR mutant backgrounds was tested by comparing the growth of the WT and that of the three mutants (Δmua, ΔnikR, and Δmua ΔnikR) in media with increasing NiCl2 concentrations (Fig. 4). While the growth of each of the three mutants in unsupplemented medium was not significantly different from that of the WT strain, the growth of the Δmua mutant was clearly affected when concentrations equal to or higher than 0.25 mM NiCl2 were added to the medium (Fig. 4), highlighting the importance of the mua gene in nickel homeostasis in H. pylori. The ΔnikR mutant strain was also severely affected by 0.25 mM or higher supplemental Ni concentrations, as previously reported in an independent study using similar growth conditions (19). There was no significant difference in sensitivity between the ΔnikR mutant and the Δmua ΔnikR double mutant. In addition, a control experiment done with ZnCl2 revealed no sensitivity difference between the WT and Δmua mutant strains (data not shown). The increased Ni sensitivity of the Δmua mutant strain might be related to higher levels of toxic ammonia generated by urease in the presence of high Ni concentrations. Alternatively, other proteins involved in Ni efflux, Ni import, or Ni binding might be affected in the Δmua mutant, although measurements of intracellular Ni pools (see below) do not support this hypothesis.
FIG 4 .
Insertional inactivation of the mua and/or nikR gene(s) renders H. pylori Ni sensitive. WT H. pylori strain 26695 and its Δmua, ΔnikR, and Δmua ΔnikR isogenic mutants were grown in BHI with the indicated supplemental NiCl2 concentrations. Growth yield was monitored by measuring the OD600 24 h after inoculation and is expressed as a percentage of the OD600 of the WT strain in unsupplemented medium (set at 100%; no error bar). Black bars represent WT H. pylori 26695, white bars the Δmua mutant, dark grey bars the ΔnikR mutant, and light grey bars the Δmua ΔnikR double mutant strain. Results shown are the averages and standard deviations of three independent growth experiments. Asterisks indicate a significant difference in growth yield between each mutant and the WT (P < 0.01 for 0.25, 0.5, and 0.75 mM NiCl2; P < 0.02 for 1 mM NiCl2 [determined by Student’s t test]).
The Δmua mutant is more resistant to acid exposure than the WT is.
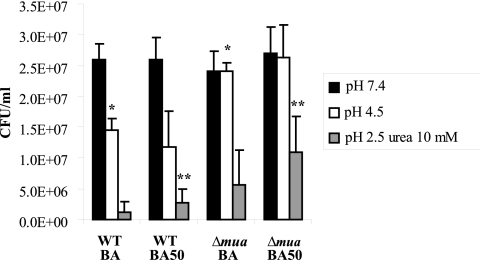
Given the higher levels of urease activity found in the Δmua mutant (than in the WT) in the presence of Ni and because urease is the primary tool H. pylori cells use to combat acidity, we hypothesized that the mutant would be better than the WT at surviving acid challenges, especially if grown with supplemental Ni. Therefore, the WT and Δmua mutant strains were grown on medium left unsupplemented or supplemented with 50 µM NiCl2 and resuspended in pH 7.4, 4.5, or 2.5 brain heart infusion broth (BHI) with 10 mM urea for 60 min. After serial dilution in BHI at pH 7.4, cells were spread on plates and colony-forming units were counted after 3 to 5 days (Fig. 5). The level of survival at pH 4.5 or pH 2.5 with 10 mM urea was higher in the mutant than in the WT when cells were grown on unsupplemented BA medium. When grown on BA supplemented with 50 µM NiCl2, the WT cells exhibited higher resistance to acid challenge (than did WT cells grown on BA); this phenotype can probably be linked to the previously described increase in urease synthesis. Interestingly, the Δmua mutant grown in the presence of Ni was more resistant to acid challenge (pH 4.5 or pH 2.5) than the WT grown under the same condition. Therefore, these results indicate that Mua, nickel, and the ability of H. pylori to survive acid challenges are linked; they also suggest that the presence of Mua is actually detrimental to H. pylori cells whenever they are confronted with high Ni concentrations coupled with acid conditions.
FIG 5 .
The Δmua mutant is more resistant to acid challenge than the WT is. WT H. pylori strain 26695 and its Δmua isogenic mutant were grown on unsupplemented BA medium or BA supplemented with 50 µM NiCl2. Cells were harvested, resuspended in BHI at pH 7.4, pH 4.5, or pH 2.5 with 10 mM urea, and left for 60 min before being serially diluted, spread on plates, and counted a few days later. Results shown are the averages and standard deviations of three independent growth and acid challenge experiments. Asterisks indicate a significant difference between the Δmua mutant and the WT strain (*, P < 0.005; **, P < 0.05 [determined by Student’s t test]).
Mua dimers can bind 2 moles of nickel.
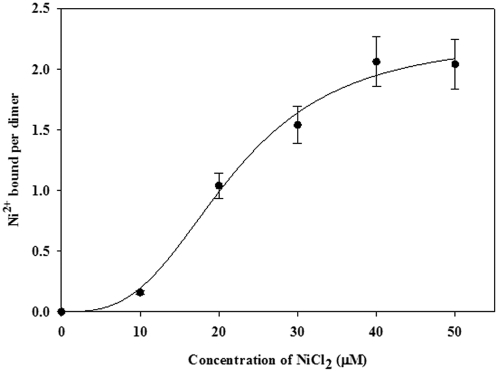
H. pylori Mua was expressed in E. coli and purified to near homogeneity by using an anion-exchange step followed by size exclusion chromatography (data not shown) as previously described (24). Cross-linking and bacterial adenylate cyclase two-hybrid (BACTH) experiments indicated that Mua can form dimers in solutions (see below), and preliminary results obtained with a BIAcore Ni-nitrilotriacetic acid (NTA) chip and with Ni-NTA agarose suggested that the Mua protein can bind Ni (data not shown); however, they did not provide quantitative data. Therefore, the purified Mua protein was dialyzed against increasing concentrations of ultrapure NiCl2 solutions and atomic absorption spectrophotometry was used to determine the amount of Ni in the dialysis baths and within the dialysis bags. Mua was able to bind up to 2 moles of Ni2+ per mole of dimer (Fig. 6), with half saturation at around a 20 µM Ni2+ ion concentration. Statistical analysis indicated positive cooperativity in nickel binding, with a Hill coefficient of approximately 3. Analysis of the amino acid sequence of Mua (22) did not reveal any significant homology with proteins of known function. The 153-amino-acid Mua protein has an unusually high content of charged amino acids, with about 22% positively (Arg or Lys) charged residues and 18% negatively (Asp or Glu) charged residues, for a total of 40% charged amino acids. Ni binding could involve two histidyl residues (His119 and His138), which are conserved among Mua homologs in all Helicobacter species. Site-directed mutagenesis of either residue will help determine their role in the Ni-binding ability of the purified protein and ultimately their involvement in the Ni-mediated repression of urease transcription.
FIG 6 .
Nickel ion binding by Mua protein based on equilibrium dialysis. Purified Mua protein (3 to 4 µM dimer concentration) in 50 mM NaCl (pH 8.25) was equilibrated with the indicated concentrations of NiCl2, and the number of nickel ions bound per dimer was determined by graphite furnace atomic absorption spectrophotometry. Standard deviations were ≤10% for each set of measurements. SigmaPlot, version 11, was used to generate the graph, as well as to calculate the KD for Ni (50% saturation).
Mua does not regulate ureAB genes through direct binding to the PureA promoter.
While it appears from the above results (higher ureA transcription in the H. pylori Δmua mutant and lower PureA-xylE activity in E. coli expressing Mua) that the Mua protein modulates urease activity through control of ureA transcription, different mechanisms could be involved. To address whether this control would be achieved by direct binding of Mua to the ureAB promoter (PureA), electrophoretic mobility shift assays (EMSA) were carried out with 32P-labeled PureA and purified Mua. Since the binding of H. pylori NikR (HpNikR) to PureA has been extensively documented (27–30), T7-tagged HpNikR was expressed in E. coli and purified to homogeneity (data not shown); it was used as a positive control in our study. Various concentrations of purified Mua or purified T7-NikR (from 0 to 4 µM in 0.05-µM increments) were incubated with 100 to 200 pM of a 208-bp-long DNA sequence containing the PureA promoter. NiSO4 (0.8 mM) or EDTA (1 mM) was added to the mixture, the gel, and the running buffer. While the labeled PureA DNA was retarded upon incubation with T7-NikR (50 nM and higher) only in the presence of Ni2+, the addition of Mua did not result in retardation of labeled PureA and it did not disturb the NikR-PureA complex either (data not shown). Therefore, in contrast to the NikR regulator, Mua does not appear to regulate the ureAB genes by direct binding to the ureA promoter DNA sequence.
Alternatively, since the transcription of ureAB genes is under the control of NikR, as well as under the antagonistic control of ArsRS (acid induction of ureA by phosphorylated ArsRS, repression of ureB and possibly ureA by unphosphorylated ArsRS) (16, 17), we hypothesized that Mua could exert its control of ureA indirectly, via either NikR or ArsRS. However, EMSA carried out with Mua and the PnikR, the ParsRS (Php0166), or the antisense small RNA ureB promoter did not reveal any binding under the conditions tested (excess EDTA or Ni), suggesting that Mua exerts its control of urease transcription through an independent mechanism.
Interactions between Mua and other H. pylori proteins.
We investigated the possibility of Mua regulating gene transcription through binding to the housekeeping sigma factor (σ70 or RpoD). Indeed, yeast two-hybrid studies aimed at discovering protein-protein interactions in H. pylori suggested such an interaction; the highest possible score was reported for an interaction between HP0088 (RpoD) and HP0868 (Mua) (Hybrigenics PIMRider database) (23). Therefore, Mua and RpoD were expressed separately in E. coli, purified to near homogeneity (data not shown), and incubated alone or mixed together in the presence of the homobifunctional cross-linker dimethyl suberimidate, with or without supplemental Ni in the mixture. When Mua alone was incubated with the cross-linker for only 1 h, the protein (theoretical molecular mass of 18,411 Da) ran with an estimated mass of approximately 42 to 44 kDa, whether Ni was added to the mixture or not, indicating that Mua forms dimers in solution (data not shown). When Mua and RpoD were mixed together, a protein adduct whose apparent mass was consistent with the formation of a Mua-RpoD complex was observed in the presence of Ni, but only when proteins were incubated for an extended period (16 h) with the cross-linker (data not shown), suggesting a weak interaction, if any. When purified NikR and Mua were mixed together in the presence of the cross-linker, we did not see any protein adduct, suggesting that the two proteins do not interact in vitro.
To further investigate the putative RpoD-Mua interaction, as well as to test other interactions (NikR-Mua, HypA-Mua) which could have accounted for the mutants’ phenotypes, we used the BACTH system based on the interaction-mediated reconstitution of the adenylate cyclase activity in E. coli (31). Cyclic AMP (cAMP) produced by the reconstituted chimeric adenylate cyclase binds to the catabolic activator protein (CAP), and the cAMP/CAP complex controls the transcription of several genes in E. coli, including lacZ. Each gene (mua, nikR, hypA, or rpoD) was genetically fused to the C terminus or the N terminus of the plasmid-encoded T25 fragment or the T18 fragment of the adenylate cyclase, thus generating 14 different fusions (see Table S1 in the supplemental material). Next, E. coli cya mutants were cotransformed with each set of compatible plasmids and screened on solid medium (LB with isopropyl-β-d-thiogalactopyranoside [IPTG], 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [X-Gal], and no added Ni or 100 µM added Ni) before being assayed for beta-galactosidase activity. Among all the possible plasmid combinations, only cells harboring two plasmids with the mua gene (pUT18-Mua and pKT25-Mua) had significant beta-galactosidase activity, ranging from 1,925 to 2,975 Miller units per mg of bacterial dry weight (bdw), regardless of the Ni added to the medium (770 U/mg of bdw and 10,800 U/mg of bdw for the negative and positive controls provided with the kit, respectively). All other 2-plasmid combinations, involving one plasmid with mua and one plasmid with rpoD, nikR, or hypA, showed a plate phenotype and/or beta-galactosidase activity similar to those of the vector-only negative control. These results indicated that interactions between Mua and RpoD, NikR, or HypA seem unlikely or too transient to be captured under these conditions. They nevertheless confirmed that Mua can make dimers in vivo, in agreement with the result obtained in vitro with purified Mua and the cross-linker.
Finally, a comparative analysis of two-dimensional protein gels loaded with membrane-free protein extracts of WT and Δmua mutant cells grown in BA with or without 50 µM supplemental Ni was carried out. Given that σ70 is the major sigma factor in H. pylori, one might expect a broader effect of the mua deletion on H. pylori proteins (besides urease). However, there was no significant difference between the WT and the mutant (data not shown). Therefore, this result suggests that RpoD is not involved in the Mua-mediated regulation of urease.
Ni levels in WT and Δmua mutant cells.
It is possible that increased levels of intracellular Ni in Δmua mutants could account for increased urease activity (and transcription). To test this hypothesis, WT strains 26695 and 43504, along with their isogenic Δmua mutants, were grown in BHI (with or without 100 µM Ni added to the medium). Intracellular levels of Ni were determined by atomic absorption spectrophotometry. There was no significant difference in the amount of intracellular Ni between each WT and its isogenic mutant, at 223 ± 15 and 218 ± 28 ng Ni per mg of total protein for 26695 and 26695 Δmua grown in the presence of supplemental Ni, respectively, and 361 ± 25 and 305 ± 12 ng Ni per mg of total protein for 43504 and 43054 Δmua grown with supplemental Ni, respectively. Therefore, the various phenotypes observed in Δmua mutants grown in the presence of Ni (urease activity and transcription, Ni toxicity) cannot be connected to larger intracellular Ni pools.
The mode of action of Mua is still not clear and requires more detailed investigation. In H. pylori, a complete set of proteins is designed to import Ni into the cell (encoded by nixA, fecA3, frpB4, and exbBD-tonB), to sequester it and release it when needed (Hpn, Hpn-like), and to export the metal from the cell (CznABC) to prevent the buildup of harmful Ni concentrations (for a review, see reference 11). Even with this battery of proteins, H. pylori might encounter an occasional surge in intracellular Ni. This would lead to increased apourease synthesis (mediated via NikR), a problem for H. pylori considering that it already produces huge amounts of urease (5). In addition, it is generally estimated that only a subset of the urease enzyme pool is active (i.e., Ni activated) (32); therefore, a massive and quick influx of Ni would also result in a significant portion of urease being activated at once, resulting in a massive production of ammonia that could be detrimental to the cell. This probably explains why, in the absence of Mua, increased urease activity (in the presence of a high Ni concentration) correlated with better acid resistance but also with higher Ni sensitivity. By putting a “cap” on urease activity when Ni concentrations are ample in the cell, the Mua protein would provide a safeguard mechanism. Homologs of the Mua protein can be found in all sequenced Helicobacter species (H. pylori, H. hepaticus, H. mustelae, and H. acinonychis). Therefore, this dual Ni-dependent fine-tuning (by NikR and Mua) of urease seems to constitute yet another hallmark of the Helicobacter genus.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The E. coli and H. pylori strains and the plasmids used in this study are listed in Table S1 in the supplemental material. The primers used in this study are listed in Table S2. All plasmids and PCR products used to generate H. pylori mutants, to overexpress the Mua, NikR, or RpoD protein in E. coli, or to express xylE transcriptional fusions in E. coli were sequenced at the Sequencing and Synthesis Facility of the University of Georgia, Athens, GA, and compared to DNA sequences from strain 26695 (22) to ensure that no error had been introduced following PCR amplification.
Growth conditions.
E. coli cells were grown aerobically in LB (liquid or) plates at 37°C. H. pylori was routinely grown on brucella agar plates supplemented with 10% defibrinated sheep blood (BA plates) at 37°C under microaerophilic conditions with 5% O2, 5% CO2, and 90% N2. For liquid growth, BHI with 0.4% β-cyclodextrin was used and cultures were grown in 10 ml of medium in 165-ml sealed glass bottles containing 5% CO2, 10% H2, 75% N2, and 10% O2. When needed, media were supplemented with ampicillin, kanamycin, or chloramphenicol to a final concentration of 100 µg/ml, 30 µg/ml, or 25 µg/ml, respectively. For nickel toxicity assays, H. pylori grown in BA was resuspended in BHI and inoculated to a final optical density at 600 nm (OD600) of 0.05 in 10 ml of BHI supplemented with 0.4% β-cyclodextrin and either no NiCl2 or 250, 500, 750, 1,000, 1,500, or 2,000 µM NiCl2. Cultures were shaken at 200 rpm for 24 h at 37°C, and growth (resistance to Ni toxicity) was determined by measuring the OD600. Results are the average of three to five independent experiments and are given as percentages of the average OD600 of the WT cells grown for 24 h without Ni. For acid challenge experiments, the protocol described by Wen et al. (33) was followed, with slight modification. Briefly, H. pylori cells grown in BA plates not supplemented with NiCl2 or supplemented with 50 µM NiCl2 were resuspended to a final OD600 of 0.1 in BHI at pH 7.4, 4.5, or 2.5, the latter supplemented with 10 mM urea. After 60 min, cells were serially diluted for CFU counting in BHI at pH 7.4, spread onto BA plates, and incubated for 3 to 5 days in a microaerobic atmosphere at 37°C. The average number of colony-forming units per milliliter in the original suspension was calculated. Results are the average of three independent growth (followed by acid challenge) experiments.
Construction of mutants. (i) Construction of Δmua mutant.
An overlapping PCR method was used to construct the Δmua mutant. Genomic DNA from strain 26695 and primers Δmua1 and Δmua2-kan (see Table S2 in the supplemental material) were used to amplify a 406-bp-long DNA sequence that contained part of the hp0867 gene, the last three codons of hp0868, and part of the aphA3 (Kanr) gene; primers Δmua3-kan and Δmua4 (see Table S2) were used to amplify a 379-bp-long DNA sequence that contained the hp0869 gene and part of the aphA3 gene. Each PCR product was gel purified and mixed together along with a purified 1.3-kbp-long aphA3 cassette. The final elongation step with primers Δmua1 and Δmua4 yielded a 2.1-kbp PCR product that was cloned into the pGEM-T vector to generate plasmid pGEM-Δmua::aphA3. This plasmid was introduced into various H. pylori strains by natural transformation to generate Δmua mutants. H. pylori cells were transferred after 16 h onto BA plates supplemented with 30 µg/ml kanamycin; clones appeared after 3 to 5 days of incubation. The disruption of the mua gene by homologous recombination was confirmed by PCR using genomic DNA from each mutant as the template and primers Δmua1 and Δmua4; a 2.1-kbp PCR product was visualized on a 0.7% Tris-acetate-EDTA (TAE) agarose gel.
(ii) Construction of ΔnikR single mutant and Δmua ΔnikR double mutant.
The ΔnikR mutant was generated using an overlapping PCR method. Genomic DNA from strain 26695 and primers ΔnikR1 and ΔnikR2-cat (see Table S2) were used to amplify a 373-bp-long DNA sequence containing the intergenic region between hp1338 (nikR) and hp1339, part of nikR, and the beginning of a cat cassette; in a separate PCR, primers ΔnikR3-cat and ΔnikR4 were used to amplify a 300-bp-long sequence complementary to the end of nikR, part of hp1337, and the end of the cat cassette. The final amplification step included each PCR product and an 800-bp-long cat cassette (with its own promoter and no transcription termination sequence), as well as primers ΔnikR1 and ΔnikR4. The resulting 1,470-bp-long PCR product was introduced into H. pylori 26695 to generate a ΔnikR::cat mutant. This PCR product was also introduced into the Δmua mutant to create a Δmua ΔnikR double mutant. Mutants were isolated on BA supplemented with chloramphenicol. Following homologous recombination, the disruption of the nikR gene was confirmed by PCR using genomic DNA from each mutant as the template and primers ΔnikR1 and ΔnikR4; a 1.47-kbp PCR product was visualized on a 0.7% TAE agarose gel.
Construction of transcriptional xylE fusions and XylE assays.
The promoterless Pseudomonas putida xylE reporter gene (34) was digested with PstI and XhoI and cloned into pBluescript-KS to generate plasmid pSB306. Primers ureAPstI and ureABamHI (see Table S2) were designed to amplify a DNA sequence that corresponded to positions −140 to +49 relative to the transcription start site of H. pylori ureAE, which was determined previously (8). Similarly, a region of DNA corresponding to positions −152 to −6 relative to the translation start site of H. pylori hydA was amplified using primers hydAPstI and hydABamHI (see Table S2). Each PCR product was digested with PstI and BamHI and cloned into plasmid pSB306 to generate a PureA-xylE or PhydA-xylE transcriptional fusion, respectively (plasmid pSB308 or pSB309). Finally, the hp0868 ORF was amplified using primers Mua-Kpn2 and Mua-SacI, digested with KpnI and SacI, and cloned into pSB306, pSB308, or pSB309 to generate plasmid pSB328, pSB330, or pSB331, respectively. In each plasmid, the hp0868 gene is placed under the control of the lacZ promoter (opposite direction from xylE fusions). XylE activity was measured as described previously (35). E. coli strains containing the xylE reporter plasmids were grown in LB with or without 1 mM supplemental NiCl2 for 24 h, harvested, and then resuspended in 50 mM phosphate buffer, pH 7.4, to a cell density (OD600) of 1 (7 × 108 CFU/ml). Whole cells (50 to 100 µl) were added to a mixture containing 10 mM catechol in 50 mM potassium phosphate, pH 7.4. Catechol oxidation to 2-hydroxymuconic semialdehyde was monitored continuously at 375 nm. A unit of XylE activity corresponds to 1 nmol of catechol oxidized per min, and values were expressed as units per 108 cells. The results are the mean ± standard deviation for two or three independent assays (three to five replicates per assay).
BACTH system.
The BACTH system was purchased from Euromedex. Primers were designed to introduce XbaI and KpnI restriction sites and to PCR amplify mua, rpoD, hypA, or nikR (see Table S2). Each PCR product was gel purified and digested with XbaI and KpnI before being ligated into plasmid pUT18, pUT18C, pKT25, or pKNT25, thus generating 12 different plasmids with mua, rpoD, hypA, or nikR fused in frame with either the T18 or the T25 peptide (N- or C-terminal fusion) of Bordetella pertussis adenylate cyclase. E. coli BTH101 cya mutant cells were cotransformed with each set of compatible plasmids and screened on LB plates supplemented with antibiotics, 0.1 mM IPTG, and 40 µg/ml of X-Gal, with or without 100 µM NiCl2. Beta-galactosidase assays were performed as described by Miller (36). One Miller unit equals 1 nmol of ortho-nitrophenol released per min.
RNA isolation.
Total RNA was isolated from H. pylori WT or mutant cells grown for 24 h on BA plates not supplemented with NiCl2 or supplemented with 5 or 50 µM NiCl2 by using the Aurum total isolation kit (Bio-Rad, Hercules, CA) and the Turbo DNA-free kit (Ambion, Austin, TX). Total RNA from each sample was analyzed on nondenaturing 1.2% agarose gel and quantified by spectrophotometry after dilution in diethyl pyrocarbonate-treated water.
cDNA synthesis.
cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) and 200 ng of DNA-free total RNA as the template. The protocol included a brief denaturation step at 95°C, followed by 5 min at 25°C (annealing with random primers), 30 min at 42°C (reverse transcription), and 5 min at 85°C (inactivation of RT). The cDNA generated was used as the template for the qPCR experiments.
qPCR.
Primers were designed to amplify 100- to 300-bp regions of the ureA or gyrA (internal control) gene (see Table S2). A reaction mixture containing the master mix with SYBR green (Bio-Rad) and each set of primers was added to a 96-well plate along with diluted cDNA samples for a final reaction volume of 20 µl per well. Negative controls contained the reaction mixture but no DNA. Samples were incubated in an iCycler (Bio-Rad) for 45 cycles (15 s at 95°C, 30 s at 58°C, and 30 s at 72°C). Gene expression was quantified by the comparative CT (cycle threshold) method and normalized to the gyrA housekeeping gene’s CT in each sample. The relative abundance of each transcript was calculated by using the 2−ΔCT formula. Error bars correspond to 2−ΔCT − 2−[ΔCT + SD] and 2−[ΔCT − SD] − 2−ΔCT, where SD = √[(SD1 CT ureA)2 + (SD2 CT gyrA)2]. Results shown are from assays done in triplicate for one growth experiment. The same patterns were seen for two additional (independent) growth experiments.
RT-PCR.
The expression of the hp0867 gene in the WT and the Δmua mutant was analyzed by RT-PCR using the one-step RT-PCR kit from Qiagen (Valencia, CA). Total RNA isolated from the WT strain (26695) or the Δmua mutant grown in BA was used as the template. Positive controls were performed with genomic DNA from either strain as the template, and negative controls were performed with RNA that had not been subjected to retrotranscription. Primers Hp0867F and Hp0867R were used to amplify an hp0867-specific 741-bp-long PCR product (see Table S2). After the final PCR amplification step, 10 µl of the reaction mixture was loaded onto an agarose gel.
Urease assays.
Urease activities were determined as previously described (24), by following the method of Weatherburn (37). The results are the mean ± standard deviation for three to seven independent assays (performed in duplicate).
Immunoblottings.
Immunoblottings were done as previously described (24), with either anti-UreA or anti-UreB antibody (1:5,000 dilution).
Equilibrium dialysis and nickel determination.
The Ni-binding ability of the purified Mua protein was determined by equilibrium dialysis, followed by graphite furnace atomic absorption spectrophotometry (Shimadzu AA-6701F). Briefly, the purified Mua protein (3 to 4 µM dimer) was left for 48 h at 4°C in 6 independent dialysis baths of 1 liter each composed of 50 mM ultra pure NaCl (pH 8.25) and increasing concentrations of NiCl2 (0, 10, 20, 30, 40, and 50 µM). After dialysis, the protein concentration in each dialysis bag was determined again and the nickel concentrations of the protein solution (bound plus free Ni2+) and the dialysis buffer (free Ni2+) were measured. The concentration of protein-bound nickel was estimated by subtracting the two values. Results are the average of three to five sets of measurements, with each measurement set representing three to seven injections; standard deviations were ≤10% for each set of measurements. A SigmaPlot, version 11, graphical program was used to generate the graph shown (Fig. 6). The program was also used to fit a curve, %S = f(log[Ni2+]), with the equation %S = [Ni2+]n/([EC50]n + [Ni2+]n), where %S is percent saturation, EC50 is the effective concentration of Ni2+ at 50% saturation, and n is the estimated Hill coefficient.
Measurements of Ni levels in cells.
H. pylori WT and Δmua mutant cells were grown for 18 h in Ni-unsupplemented BHI or BHI supplemented with 100 µM NiCl2, pH 6.5, in sealed bottles before being harvested, washed once, and resuspended in ice-cold deionized water and broken by sonication. The amount of Ni in the cell extracts was determined using graphite furnace atomic absorption spectrophotometry (Shimadzu AA-6701F), and the amount of total protein was determined using the bicinchoninic acid assay kit.
Two-dimensional electrophoresis.
Cell-free, membrane-free protein extracts from H. pylori (26695 and 26695 Δmua) cells grown in BA or in BA supplemented with 50 µM NiCl2 were sent to Kendrick Laboratories (Madison, WI) for processing and data analysis.
SUPPLEMENTAL MATERIAL
ACKNOWLEDGMENTS
This work was supported by the Georgia Research Foundation and by National Institutes of Health grant RO1DK60061.
Antisera against H. pylori ureAE or UreB were kindly provided by H. L. Mobley, University of Michigan Medical School, Ann Arbor, MI. We thank Gerardo Gutierrez-Sanchez and Carl Bergmann (Complex Carbohydrate Research Center, University of Georgia) for their help with the BIAcore study and Erica Miller for comments on the manuscript.
Footnotes
Citation Benoit SL, Maier RJ. 2011. Mua (HP0868) is a nickel-binding protein that modulates urease activity in Helicobacter pylori. mBio 2(2):e00039-11. doi:10.1128/mBio.00039-11.
REFERENCES
- 1. Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328–1333 [DOI] [PubMed] [Google Scholar]
- 2. Blaser MJ. 1995. The role of Helicobacter pylori in gastritis and its progression to peptic ulcer disease. Aliment. Pharmacol. Ther. 9(Suppl. 1):27–30 [DOI] [PubMed] [Google Scholar]
- 3. Sipponen P, Hyvärinen H, Seppälä K, Blaser MJ. 1998. Review article: pathogenesis of the transformation from gastritis to malignancy. Aliment. Pharmacol. Ther. 12(Suppl. 1):61–71 [DOI] [PubMed] [Google Scholar]
- 4. Scott DR, Marcus EA, Weeks DL, Sachs G. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187–195 [DOI] [PubMed] [Google Scholar]
- 5. Bauerfeind P, Garner R, Dunn BE, Mobley HL. 1997. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40:25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andrutis KA, et al. 1995. Inability of an isogenic urease-negative mutant strain of Helicobacter mustelae to colonize the ferret stomach. Infect. Immun. 63:3722–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsuda M, Karita M, Morshed MG, Okita K, Nakazawa T. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akada JK, Shirai M, Takeuchi H, Tsuda M, Nakazawa T. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071–1084 [DOI] [PubMed] [Google Scholar]
- 9. Weeks DL, Eskandari S, Scott DR, Sachs G. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482–485 [DOI] [PubMed] [Google Scholar]
- 10. Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. 2009. Interplay of metal ions and urease. Metallomics 1:207–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maier RJ, Benoit SL, Seshadri S. 2007. Nickel-binding and accessory proteins facilitating Ni-enzyme maturation in Helicobacter pylori. Biometals 20:655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davies BJ, de Vries N, Rijpkema SG, van Vliet AH, Penn CW. 2002. Transcriptional and mutational analysis of the Helicobacter pylori urease promoter. FEMS Microbiol. Lett. 213:27–32 [DOI] [PubMed] [Google Scholar]
- 13. Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. 2003. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect. Immun. 71:3529–3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bury-Moné S, et al. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623–638 [DOI] [PubMed] [Google Scholar]
- 15. Wen Y, et al. 2003. Acid-adaptive genes of Helicobacter pylori. Infect. Immun. 71:5921–5939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pflock M, Kennard S, Delany I, Scarlato V, Beier D. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. 2010. A cis-encoded antisense small RNA regulated by the HP0165-HP0166 two-component system controls expression of ureB in Helicobacter pylori. J. Bacteriol. 193:40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Vliet AH, et al. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Vliet AH, et al. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49:947–963 [DOI] [PubMed] [Google Scholar]
- 21. Olson JW, Mehta NS, Maier RJ. 2001. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol. Microbiol. 39:176–182 [DOI] [PubMed] [Google Scholar]
- 22. Tomb JF, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547 [DOI] [PubMed] [Google Scholar]
- 23. Rain JC, et al. 2001. The protein-protein interaction map of Helicobacter pylori. Nature 409:211–215 [DOI] [PubMed] [Google Scholar]
- 24. Mehta N, Olson JW, Maier RJ. 2003. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J. Bacteriol. 185:726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma CM, et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250–255 [DOI] [PubMed] [Google Scholar]
- 26. Benoit S, Mehta N, Wang G, Gatlin M, Maier RJ. 2004. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb. Pathog. 36:153–157 [DOI] [PubMed] [Google Scholar]
- 27. Zambelli B, et al. 2008. High-affinity Ni2+ binding selectively promotes binding of Helicobacter pylori NikR to its target urease promoter. J. Mol. Biol. 383:1129–1143 [DOI] [PubMed] [Google Scholar]
- 28. Benanti EL, Chivers PT. 2007. The N-terminal arm of the Helicobacter pylori Ni2+-dependent transcription factor NikR is required for specific DNA binding. J. Biol. Chem. 282:20365–20375 [DOI] [PubMed] [Google Scholar]
- 29. Abraham LO, Li Y, Zamble DB. 2006. The metal- and DNA-binding activities of Helicobacter pylori NikR. J. Inorg. Biochem. 100:1005–1014 [DOI] [PubMed] [Google Scholar]
- 30. Ernst FD, et al. 2005. The nickel-responsive regulator NikR controls activation and repression of gene transcription in Helicobacter pylori. Infect. Immun. 73:7252–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stingl K, De Reuse H. 2005. Staying alive overdosed: how does Helicobacter pylori control urease activity? Int. J. Med. Microbiol. 295:307–315 [DOI] [PubMed] [Google Scholar]
- 33. Wen Y, Feng J, Scott DR, Marcus EA, Sachs G. 2009. The pH-responsive regulon of HP0244 (FlgS), the cytoplasmic histidine kinase of Helicobacter pylori. J. Bacteriol. 191:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zukowski MM, et al. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc. Natl. Acad. Sci. U. S. A. 80:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karita M, Tummuru MK, Wirth HP, Blaser MJ. 1996. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect. Immun. 64:4501–4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Weatherburn MW. 1967. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 39:971–974 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.