Abstract
We report a pseudo-outbreak caused by Clostridium sordellii, an uncommon human pathogen. The pseudo-outbreak involved six patients and was temporally-associated with a change in the protocol of handling anaerobic culture specimens by the clinical microbiology laboratory. All isolates were genetically indistinguishable from a laboratory reference strain used for quality control.
INTRODUCTION
Clostridium sordellii is a soil-dwelling, gram positive, anaerobic, spore-forming bacillus that rarely causes overt infections.1 The mortality of C. sordellii infections is high (>70%) and correlates with the presence of a stereotypical toxic shock syndrome (TSS) (reviewed in 1). The severity of clinical C. sordellii disease is attributable in part to the production of two large clostridial cytotoxins, lethal toxin (TcsL) and hemorrhagic toxin (TcsH).2-4
C. sordellii has been an uncommon cause of pseudo-outbreaks,5-6 We now report a pseudo-outbreak with C. sordellii involving seven positive cultures obtained from six different patients over a seven week period.
METHODS
Epidemiological Investigation
This study was reviewed and approved by the University of Michigan Institutional Review Board. The Department of Infection Control and Epidemiology (ICE) was notified by personnel in the microbiology laboratory of an unusual cluster of C. sordellii isolates from clinical specimens; an investigation was begun. The case definition was any patient with C. sordellii isolated from a bacterial culture between April 1 and May 30, 2009. Cases were ascertained through review of ICE and microbiological laboratory records for the past one year. Clinical and laboratory data were retrospectively accrued from the patients’ electronic medical records using standardized data collection forms. For purposes of this study, C. sordellii TSS was defined according to a modification of previously published criteria for obstetric/post-abortion C. sordellii infections.7-8
Microbiological Evaluation
C. sordellii were isolated from clinical specimens using standard microbiological methods. These strains were confirmed to be C. sordellii by PCR amplification of a specific portion of the 16S rRNA gene (not shown). C. sordellii strain ATCC9714 was from the American Type Culture Collection (Manassas, VA). The clinical strain DA-108 was isolated from the bloodstream of a patient with postpartum endometritis.4 Reinforced clostridial medium was from BD Biosciences (San Jose, CA).
Pulsed field gel electrophoresis
PFGE typing was performed by the Michigan Department of Community Health using the restriction enzymes SmaI and ApaI using a standard protocol for the PFGE of C. difficile.9-10
Polymerase chain reaction (PCR)
DNA was isolated from C. sordellii cultures using an Easy-DNA™ Kit (Invitrogen). PCR amplifications were performed using primer pairs for the tcsL gene as detailed elsewhere.4
Environmental Investigation
An investigation of laboratory procedures for handling specimens was performed with laboratory and infection control personnel. All sterile supplies and reagents were reviewed for lot numbers and for possible contamination. The procedure for handling anaerobic cultures was reviewed with laboratory personnel. Normal cleaning processes of the anaerobic chamber was reviewed. Evaluation of phenotypic characteristics of the C. sordelli isolates were compared with clinical isolates.
RESULTS
Time course of C. sordellii isolation from clinical specimens
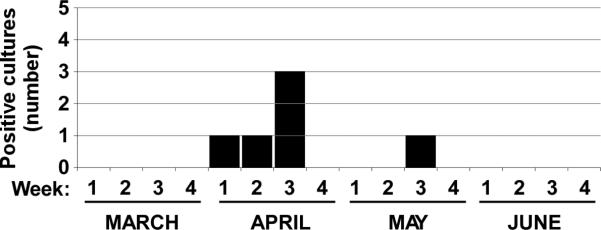
Seven isolates of C. sordellii were obtained from separate clinical specimens taken from six different patients and processed in the same hospital microbiology laboratory over the period of April 2 to May 18, 2009 (Fig. 1A). In the preceding 12 months no isolates of C. sordellii had been recovered in the same laboratory.
Figure 1.
(A) Chronological history of C. sordellii pseudo-outbreak. The number of positive cultures for C. sordellii is plotted against time. Months are broken into quarters along the x-axis. (B) Pulsed field gel electrophoresis (PFGE) analysis of C. sordellii isolates. PFGE using the restriction enzyme ApaI was performed on genomic DNA from C. sordellii isolates as detailed in the Materials and Methods section. Lane 1, clinical strain DA-108 not involved in this pseudo-outbreak; lane 2, reference strain ATCC9714; lanes 3-8, clinical C. sordellii isolates (1-6) involved in this pseudo-outbreak; lane 9, digest control.
Clinical and epidemiological characteristics of patients
Investigation into the patients whose samples were associated with C. sordellii did not reveal shared epidemiological risk factors for the acquisition of this pathogen. None of the patients had clinical features of C. sordellii infection, such as a leukemoid reaction or TSS. Patient 1 was a 59 year old male receiving chronic suppressive therapy with clindamycin for chronic sinusitis, whose sinus cultures grew Streptococcus milleri, Staphylococcus aureus, and C. sordellii. Clindamycin was extended 6 weeks in response to the C. sordellii isolate without adverse effect.. Patient 2 had two cultures positive for C. sordellii, including inguinal abscess and a foot abscess, each of which yielded multiple organisms. Patient 3 had a bone culture performed for a diabetic foot osteomyelitis, which yielded S. aureus in addition to C. sordellii. The patient was treated with a first-generation cephalosporin with a good clinical response. The fourth patient had nasal mucosal tissue cultures performed due to chronic rhinosinusitis, which yielded C. sordellii, Klebsiella oxytoca, S. aureus, and multiple other anaerobic bacteria. The patient responded to oral trimethoprim-sulfamethoxazole and topical mupirocin. The fifth patient had cultures obtained from a debridement of infected bone and prosthetic materials that were used to repair a previous fracture. These grew coagulase-negative staphylococci and C. sordellii. Vancomycin was used initially but ampicillin-sulbactam was added in response to the C. sordellii, which was later changed to metronidazole. The patient developed diarrhea 48 hrs into metronidazole therapy, which was stopped. The wrist infection improved with vancomycin alone for 10 weeks. Lastly, patient 6 was a renal transplant recipient with diabetes mellitus, who grew C. sordellii and Serratia marcescens from a culture of a postoperative wound infection of the foot. Ciprofloxacin was used with a good clinical response noted.
Molecular typing of C. sordellii isolates
Detailed molecular studies were performed to determine the relatedness of these six clinical isolates. We used genomic analyses to compare these isolates with a TcsL-negative clinical strain (DA-108) that had been obtained ~15 months prior to the onset of this apparent outbreak and with a TcsL-positive laboratory reference strain (ATCC9714) that is used for quality control purposes in the microbiology laboratory. Each of the six clinical isolates involved in this apparent outbreak were positive for the gene encoding lethal toxin (tcsL; data not shown). PFGE analysis with either of two different restriction enzymes (ApaI or SmaI) yielded two distinct PFGE patterns (ApaI result shown, Fig. 1B): all six clinical isolates involved in this apparent outbreak showed the same PFGE patterns as the ATCC9714 reference strain and only the tcsL-negative clinical isolate DA-108 yielded a second pattern with either restriction enzyme.
Results of Environmental Investigation
During the investigation, it was noted that approximately one month before the first C. sordelli isolate was identified, the microbiology laboratory had changed their procedure for processing anaerobic cultures. Previously, anaerobic specimens were plated within an anaerobic chamber, but during the time of the outbreak, specimens were being processed in a biological safety hood outside of the anaerobic chamber. However, these specimens were inoculated on culture media that was prereduced and stored within the anaerobic chamber. The transport media used for the specimens were from different lots and recovery of C. sordellii was not associated with a single reagent or type of media. The pseudo-outbreak was also temporally associated with an increase in teaching activities where students were frequently handling the reference ATCC9714 strain. No overt contamination events were reported and no breaks in sample handling protocols were observed. No environmental testing was performed during this episode. Nevertheless, thorough disinfection of the anaerobe chamber and replacement of commonly used materials was undertaken, and included a re-emphasis of the proper handling of specimens and control isolates to minimize inadvertent release of spores. In addition, the laboratory switched to the use of individually packaged, sterile, prereduced culture media for all anaerobic cultures. These efforts appear to have eliminated the problem as no further C. sordellii strains have been isolated in the subsequent 8 months.
DISCUSSION
Herein we report a pseudo-outbreak of C. sordellii that occurred in a large academic teaching hospital. These cases were temporally related to changes in specimen processing and an increase in handling of a reference strain used in the clinical microbiology laboratory for quality control and teaching purposes. By molecular PFGE-typing, all of the clinical isolates were identical to this reference strain (ATCC9714). We speculate that this strain contaminated surfaces in the microbiology laboratory work space and contaminated clinical specimens being processed there.
Our investigation did not reveal an exact source for the contamination of anaerobic cultures processed in our laboratory. This pseudo-outbreak was temporally associated with an increase in teaching activities using the ATCC9714 strain, and it is possible that, despite appropriate supervision, inadvertent environmental contamination with clostridial spores occurred. C. sordellii is a prolific spore-forming bacterium, which might have contributed to the problem. In our chamber system, anaerobic gases are circulated by fans that could facilitate dissemination of unintentionally-released spores within the chamber.
Inappropriate treatment of infections can result from pseudo-outbreaks. Patient number 1 received oral clindamycin for six weeks in response to a positive anaerobic sinus culture for C. sordellii. No complications of this therapy were noted. Patient number 5 had a coagulase-negative Staphylococcus infection. Attempts to treat the C. sordellii with oral metronidazole resulted in the development of severe diarrhea that resolved after the medication was stopped.
C. sordellii is not a common cause of true outbreaks. Healthcare providers should maintain a high degree of suspicion for a pseudo-outbreak if an increase in the frequency of isolation of an uncommon pathogen is observed, including C. sordellii.
ACKNOWLEDGMENTS
Financial support. Funding for this work was provided by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation (D.M.A.) and the National Institutes of Health grant HD057176 (D.M.A.).
Footnotes
Conflicts of interest. The authors have no conflicts of interest to report.
REFERENCES
- 1.Aldape MJ, Bryant AE, Stevens DL. Clostridium sordellii infection: epidemiology, clinical findings, and current perspectives on diagnosis and treatment. Clin Infect Dis. 2006;43:1436–46. doi: 10.1086/508866. [DOI] [PubMed] [Google Scholar]
- 2.Popoff MR. Purification and characterization of Clostridium sordellii lethal toxin and cross-reactivity with Clostridium difficile cytotoxin. Infect Immun. 1987;55:35–43. doi: 10.1128/iai.55.1.35-43.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez RD, Wilkins TD. Purification and characterization of Clostridium sordellii hemorrhagic toxin and cross-reactivity with Clostridium difficile toxin A (enterotoxin). Infect Immun. 1988;56:1215–21. doi: 10.1128/iai.56.5.1215-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao Y, Senn T, Opp J, et al. Lethal toxin is a critical determinant of rapid mortality in rodent models of Clostridium sordellii endometritis. Anaerobe. 2009 doi: 10.1016/j.anaerobe.2009.06.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menon A, Stefanova R, Mwangi P, Ruhe J, Hoffman K, Woods GL. A pseudo-outbreak of Clostridium sordellii linked to contamination with a laboratory reference strain.. Annual Meeting of the Society for Healthcare Epidemiology of America; Chicago. 2006.pp. 135–136. [Google Scholar]
- 6.Lynch JM, Anderson A, Camacho FR, Winters AK, Hodges GR, Barnes WG. Pseudobacteremia caused by Clostridium sordellii. Arch Intern Med. 1980;140:65–8. [PubMed] [Google Scholar]
- 7.McGregor JA, Equils O. Response to letter to the editor. Contraception. 2006;74:175–176. [Google Scholar]
- 8.McGregor JA, Soper DE, Lovell G, Todd JK. Maternal deaths associated with Clostridium sordellii infection. Am J Obstet Gynecol. 1989;161:987–95. doi: 10.1016/0002-9378(89)90768-0. [DOI] [PubMed] [Google Scholar]
- 9.Clabots CR, Johnson S, Bettin KM, et al. Development of a rapid and efficient restriction endonuclease analysis typing system for Clostridium difficile and correlation with other typing systems. J Clin Microbiol. 1993;31:1870–5. doi: 10.1128/jcm.31.7.1870-1875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaassen CH, van Haren HA, Horrevorts AM. Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism. J Clin Microbiol. 2002;40:101–4. doi: 10.1128/JCM.40.1.101-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]