Abstract
New indoline alkaloid–type compounds which inhibit HCV production by infected hepatoma cells have been identified. These compounds, dimeric-type compounds of previously known inhibitors, display double digit nanomolar IC50 and EC50 values, with cytotoxicity CC50 indexes higher than 36 micromolar, thus providing ample therapeutic windows for further development of HCV drugs.
Hepatitis C virus (HCV) [i], which infects 130 – 170 million people worldwide [ii], is the main cause of liver disease in humans. This single-strand, positive RNA virus encodes ten proteins [iii], all of which are essential for viral infection and propagation. Currently, the only treatment of HCV infection is a 48-week regimen, a combination of interferon and ribavirin, that cures less than half the people infected, depending on viral genotype and strain [iv]. Several of the viral proteins have been targeted for drug development, with the viral NS3 protease [v] and NS5B polymerase [vi] key among them, and several candidates have advanced significantly in the drug pipeline [vii].
Core, the HCV capsid protein, is the most conserved of the ten viral proteins across all HCV genotypes [viii]. Core is responsible for nucleocapsid formation, recruitment of HCV replicase proteins to lipid droplets, and assembly of the viral particles in infected cells. As such, core is an interesting target for HCV drug development distinct from the HCV protease and polymerase enzymes, from which resistant mutants have already emerged [ix]. With combination therapies likely to evolve for HCV treatment [x], inhibitors of core dimerization, and hence nucleocapsid formation, could play a key role.
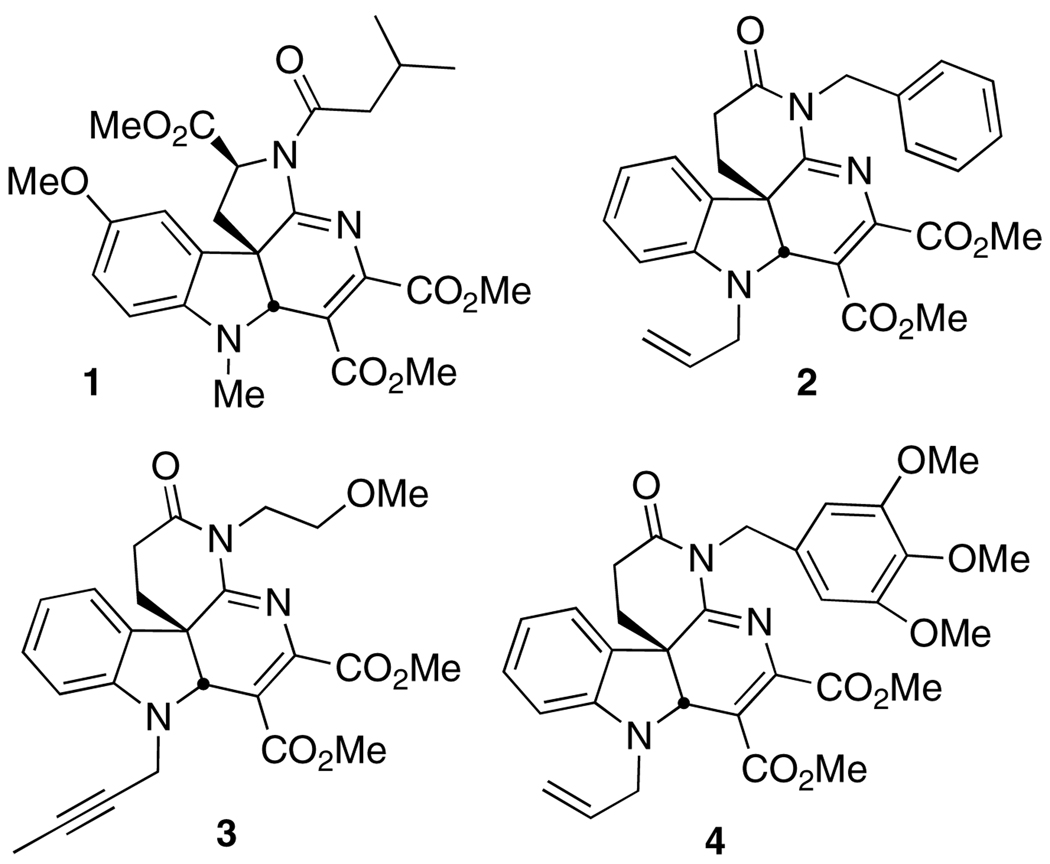
We have been interested in the discovery of new, small molecule inhibitors of core dimerization as lead structures for anti-HCV agents. To this end, we have reported new assays to screen for inhibitors of core dimerization [xi] and of interactions of core with other HCV proteins, including NS3helicase [xii]. Screening of a small molecule library led to the identification of 1 as an initial hit for further elaboration (Figure 1) [xiii]. An additional focused library was prepared, which revealed three new core dimerization inhibitors as racemates (2 – 4) with IC50’s in the single digit µM range. All four inhibitors were also found to inhibit HCV production in infected Huh-7.5 hepatoma cells; only 4 showed levels of cytotoxicity comparable to activity against core dimerization. We now report a successful search for more potent inhibitors of core protein dimerization based on the structures of 2 – 4, which are also effective in blocking HCV proliferation in infected cells.
Figure 1.
Previously reported inhibitors of core dimerization.11
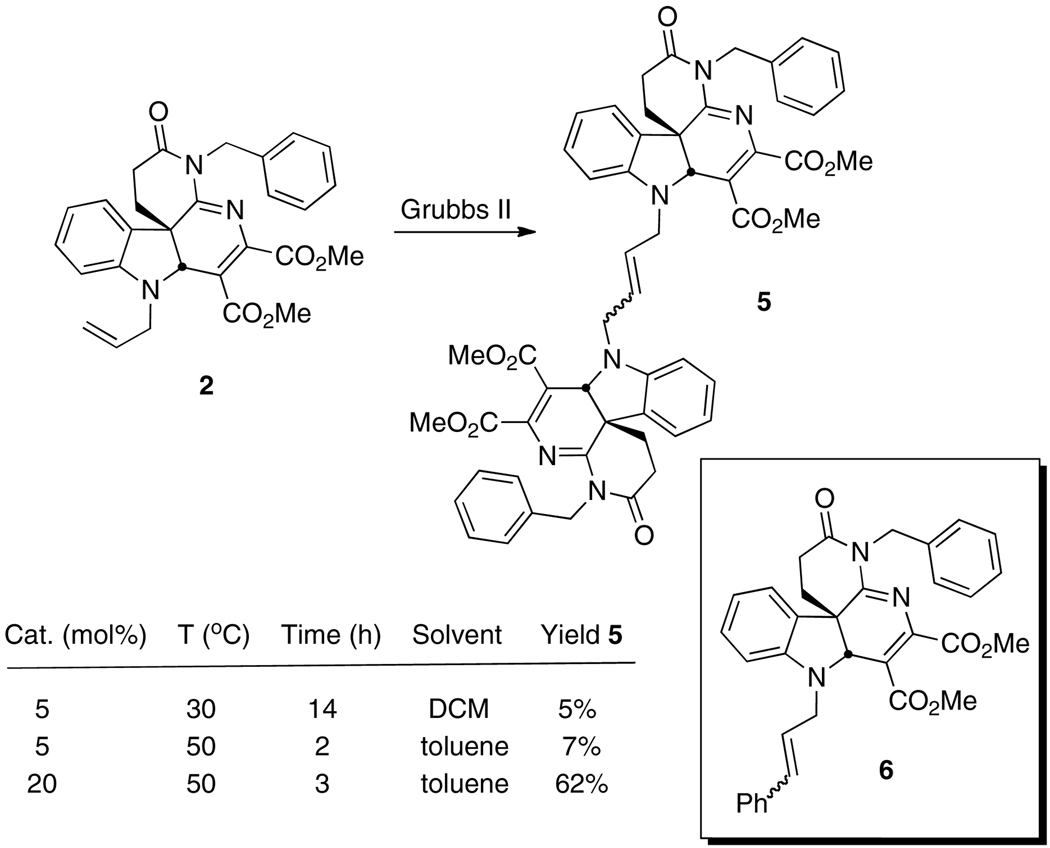
Since inhibition of core dimerization requires blockage of a protein-protein interaction (PPI) [xiv], our initial approach to improve on the activity of 1 – 4 was to prepare a dimer of the most active inhibitor 2 since dimerization of inhibitors of PPI have been shown to be an effective strategy to improve activity [xv,xvi,xvii]. The hope was that a second binding site or “hot spot” might be found on the surface of core for the second heterocycle to occupy that would magnify the dimerization inhibition. It was also understood that since 2 was racemic, dimerization of 2 would produce a mixture of enantiomers and as well as the meso diastereomer which would be likely difficult to separate. Nonetheless, if such a mixture would show improved activity over the original monomer, this would suggest that attachment of the second heterocyclic subunit could be a viable approach to preparing compounds with improved activity.
Attempts to dimerize 2 through metathesis gave disappointingly low yields unless 20 mol% of Grubbs II catalyst was employed (Scheme 1). Under optimal conditions, a 62% yield of dimer 5 was obtained as a mixture of E- and Z-isomers of the racemate and the meso diastereomer, along with 6 (15%), resulting from metathesis with the benzylidene ligand of the catalyst.
Scheme 1.
Dimerization of 2 through metathesis.
Due to the difficulty in purification, the poor yields in the subsequent non-chemoselective hydrogenation of the olefinic double bond of 5, as well as the relatively poor yield of 5 without the use of a large amount of catalyst, an alternative approach was explored.
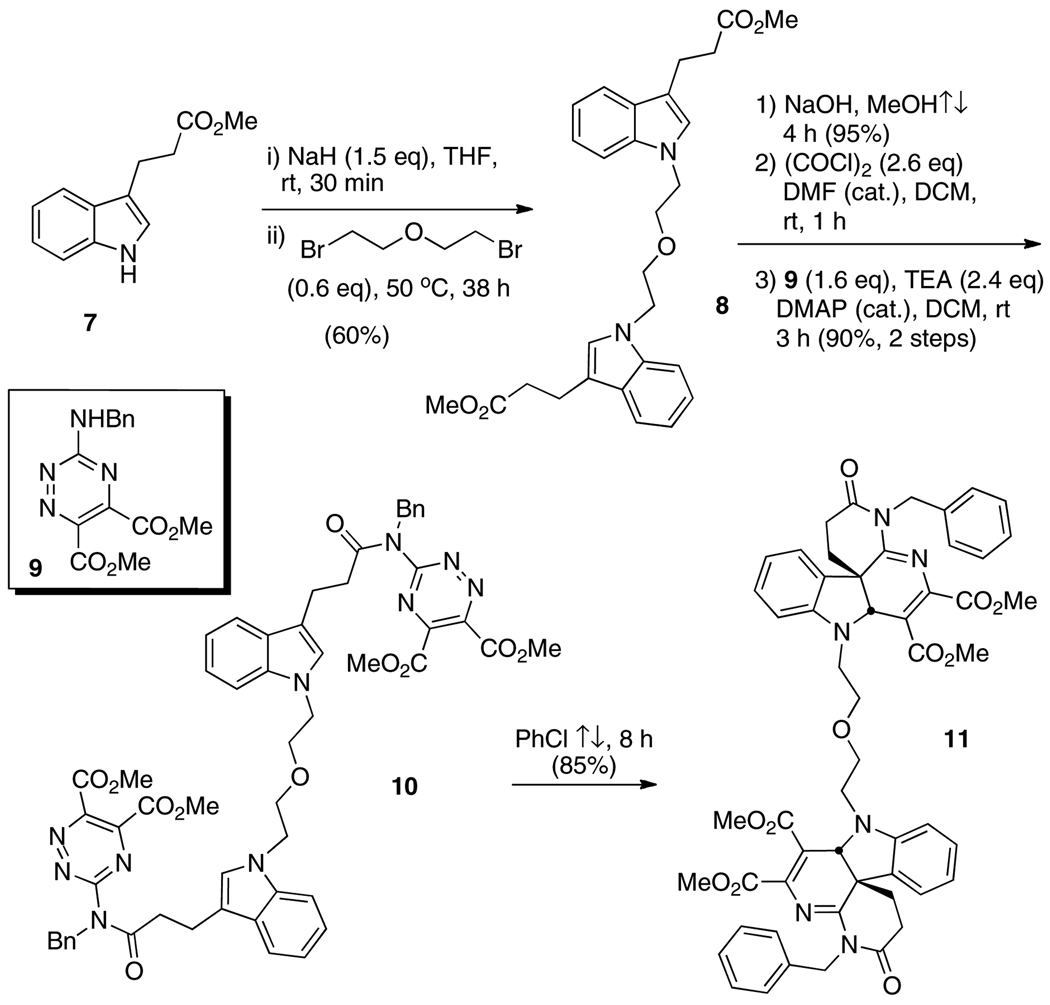
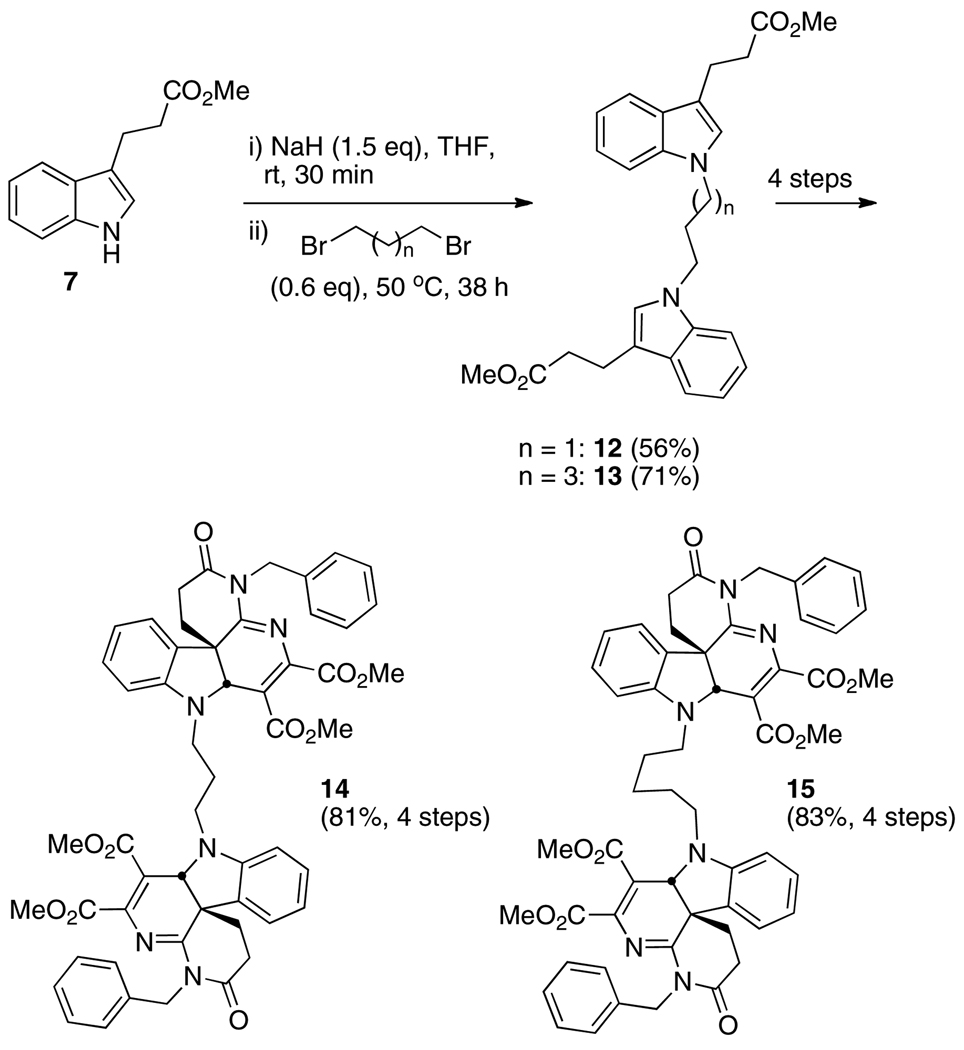
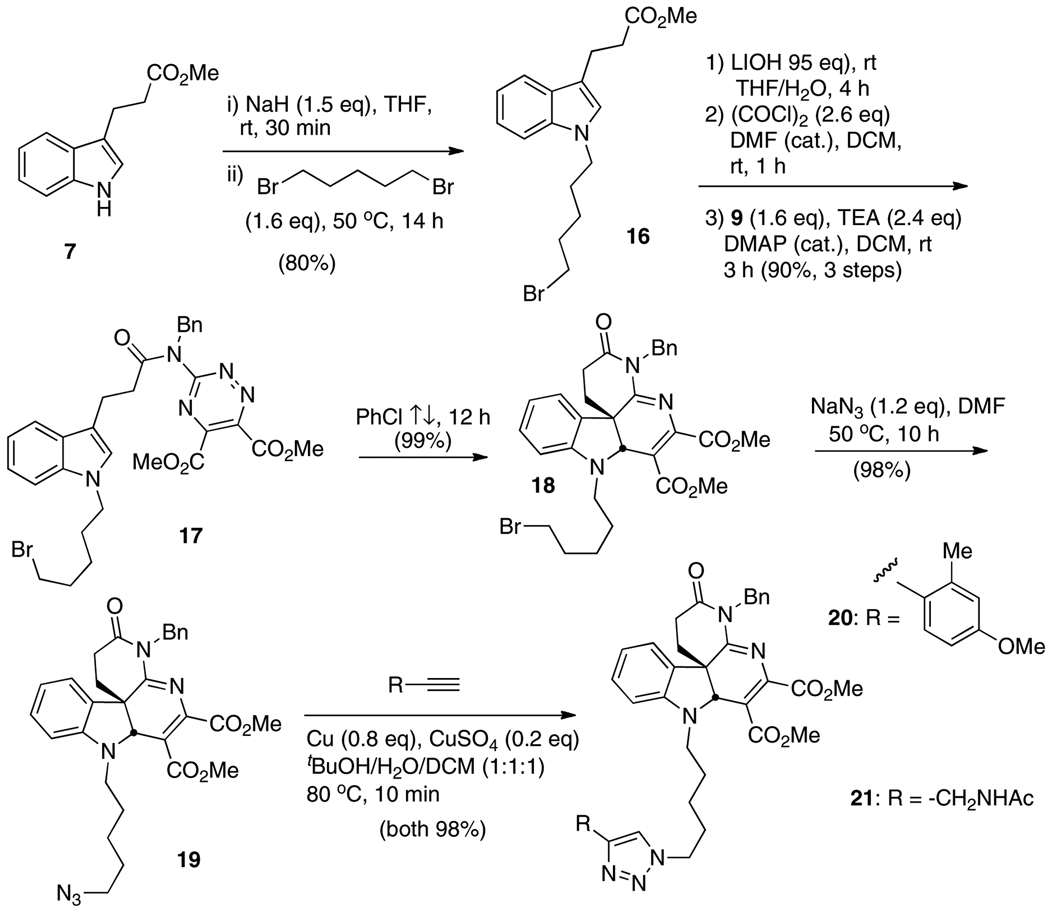
Dimer 11 was ultimately prepared by first linking two indolylpropionate esters 7 with dibromoethyl ether, producing the symmetric dimer 8 (60%, Scheme 2). Basic hydrolysis of the esters, conversion to the bis-acid chloride, then acylation of triazine 9 as previously described in the preparation of monomer 2 [13] all proceeded uneventfully, yielding dimer 10 (86% over three steps). Double inverse electron demand Diels-Alder cycloadditions in refluxing chlorobenzene produced the target dimer 11. Beginning with the 3- and 5-carbon tethers, 1,3-dibromopropane and 1,5-dibromopentane, dimers 14 and 15 were similarly prepared in 45% and 59% overall yields, respectively (5 steps each beginning with 7, Scheme 3). In each of the tethering reactions with the respective dibromides, it was imperative to keep the number of equivalents of dibromide relatively low (0.6 eq) to avoid monoalkylations (vide supra). Dimers 11, 14, and 15 were also mixtures of enantiomers and the meso diastereomer.
Scheme 2.
Preparation of dimer 11 with an ether linkage between the monomeric subunits.
Scheme 3.
Preparation of dimers 14 and 15 with all-carbon tethers linking the monomeric subunits.
In the core dimerization assay, all three dimers were active, with dimeric mixture 11 having an IC50 value of 98 nM, more than an order of magnitude greater than that observed for the monomer 2 (Table 1). None of the dimers showed significant cytotoxicity (CC50’s > 36 µM. Whole cell assays with Huh-7.5 hepatoma cells infected with HCV 2a strain J6/JFH-1 were treated with increasing concentrations (0.001 – 100 µM) of the dimers to assess their effect on HCV propagation as previously described [11,13]. The known NS3/NS4A protease inhibitor BILN2061 was included as a positive control. The EC50’s were calculated at an early (T1) and late (T2) stage. Sensitivity to the nature of the tether linking the dimers is readily apparent, with 14 emerging as the best inhibitor (EC50 T1: 88 nM, T2: 735 nM). Dimeric mixture 15 was also quite active in early stage inhibition of HCV infectivity (T1 EC50 90 nM), but with a dramatic loss of activity at late stage (T2 EC50 29.9 µM). The main drawbacks with these dimers are their high MW and cLogP values (Table 1).
Table 1.
Summary of Bioactivities.
| Compd | IC50 (µM)a |
T1-EC50 (µM)b |
T2-EC50 (µM)c |
CC50 (µM)d |
MW | cLogP |
|---|---|---|---|---|---|---|
| 1e | 9.3 | 14.8 | 22.2 | >320 | 513 | 4.7 |
| 2e | 1.4 | 2.3 | 3.2 | 127.2 | 485 | 6 |
| 3e | 2.0 | 4.9 | 0.7 | >320 | 465 | 4.6 |
| 4e | 2.0 | 3.8 | 3.0 | 5.3 | 575 | 5.3 |
| 11 | 0.098 | 0.48 | 2.5 | >100 | 960 | 10.2 |
| 14 | 2.9 | 0.088 | 0.735 | >36 | 930 | 10.5 |
| 15 | 0.72 | 0.09 | 29.9 | >36 | 958 | 10.1 |
| 20 | 0.092 | 1.4 | 1.1 | >100 | 702 | 7.9 |
| 21 | 0.341 | 26.2 | 56.2 | 150 | 653 | 4.4 |
| BILN2061f | N/A | 0.072 | 0.071 | 5.3 | - | - |
Values are means of three experiments.
Values are means of three experiments. T1 corresponds to initial 72 hr culture of cells in presence of inhibitors. See ref. 11,13.
Values are means of three experiments. T2 corresponds to second 72 hr culture of fresh cells, infection resulting from virus secreted in T1. See ref. 11,13.
Values are means of three experiments. 50% Cytotoxic conc. Vs Huh-7.5 hepatoma cells.
Data from ref. 13.
Known inhibitor of the HCV NS3/NS4A protease [xviii].
With the validation of tethering a second heterocyclic unit to the basic active scaffold as an approach to enhanced activity against core dimerization as well as inhibition of HCV production in infected cells, two dimer “mimics” were then prepared in an effort to discover inhibitors equally potent to 11 and 14, but without the meso diastereomer contamination (Scheme 4). To this end, alkylation of 7 with 1,5-dibromopentane (1.6 eq) produced the monobromide 16 in 80% yield, with only a small amount of tethered dimer 13 (9%). Conversion to the corresponding acid chloride and attachment of triazine 9 proceeded smoothly under standard conditions, as did the cycloaddition to give 18. No unwanted chemistry of this primary alkyl bromide disrupted the strategy. Azide displacement to 19, then Cu-catalyzed dipolar cycloadditionxix with appropriate alkynes produced racemic triazoles 20 and 21, with the cycloadditions both occurring in 98% yield.
Scheme 4.
Preparation of tethered bis-heterocycles 20 and 21 via Click dipolar addition chemistry.
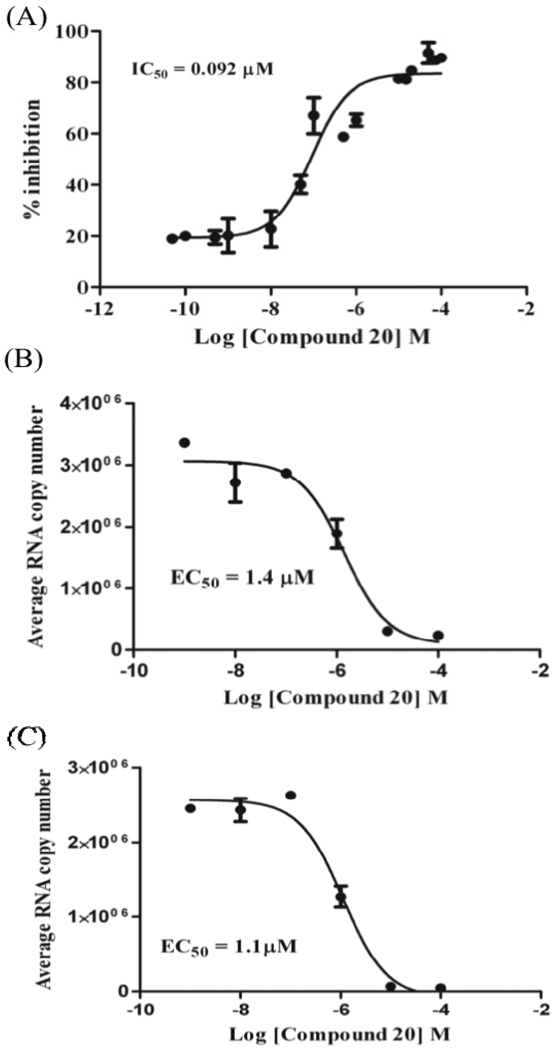
Both 20 and 21 proved to be excellent inhibitors of core dimerization, with sub-micromolar IC50’s (92 and 341 nM, respectively); the activity of 20 was comparable to that of dimer 11. Equally important, the cytotoxicities of 20 and 21 were in the high µM range, and the whole cell activity of 20 remained in the single digit µM region. The main drawback to be addressed is that the most effective of these new tethered bis-heterocycles (20, Figure 1) also has the highest MW (702) and the highest cLogP value (7.9).
In conclusion, several dimers of the previously reported core dimerization inhibitor 2 have been prepared, and been shown to be more effective core dimerization and HCV inhibitors than the original lead compound, though with cellular activity which declines with increasing incubation time, perhaps due to compound instability in the cellular assays. All compounds were stable upon storage. Using click chemistry,xx two racemic tethered bis-heterocycles (20 and 21) were also prepared and shown to be even more effective inhibitors of core dimerization as well as inhibitors of HCV production in infected hepatoma cells. Further studies to (i) probe the nature of the interaction of these inhibitors with core, (iii) resolve and screen the enantiomers of 20 and 21; and (iii) prepare an additional focused library of analogues of 20 and 21 are underway. Completion of these studies should allow for a better SAR understanding.
Figure 1.
Dose-response analyses of 20 using Core106 ALPHA screen assay.11 The compound was dosed from 0 to 100 µM; IC50 and EC50’s were calculated using a non-linear regression ‘Log[inhibitor] vs response’ with four points per concentration. (A) IC50; (B) T1-EC50; (C) T2-EC50.
Acknowledgments
The Scripps-Florida group thanks Prof. Weissmann and Dr. T. Tellinghuisen (Department of Infectology, The Scripps Research Institute-Florida) for helpful discussions and assistance with the HCV studies, the State of Florida for start-up funds, and the NIH (1X01MH085709-01 and 1R21NSO66411 (ADS) for grant support. The Boston University group thanks Professors John Porco and Aaron Beeler for helpful discussions, and NIGMS CMLD initiative (P50 GM067041) for financial support. We (CMLD-BU) are also grateful to the National Science Foundation for supporting the purchase of the NMR (CHE 0619339) and HRMS (CHE 0443618) spectrometers used in this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- i.For some reviews of hepatitis C: Bartenschlager R, Lohmann V. J. Gen. Virol. 2000;81:1631. doi: 10.1099/0022-1317-81-7-1631. Giannini C, Brechot C. Cell Death Diff. 2003;10:S27. doi: 10.1038/sj.cdd.4401121. Simmonds P. J. Gen. Virol. 2004;85:3173. doi: 10.1099/vir.0.80401-0. Alter MJ. World J. Gastroenterol. 2007;13:2436. doi: 10.3748/wjg.v13.i17.2436..
- ii.Lavanchy D. Liver Int. 2009;29:74. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- iii.(a) Rosenberg S. J. Mol. Biol. 2001;313:451. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]; (b) Dubuisson J. World J. Gastroenterol. 2007;13:2406. doi: 10.3748/wjg.v13.i17.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- iv.(a) Cristina J, Moreno-del Pilar M, Moratorio G. Virus Res. 2007;127:185. doi: 10.1016/j.virusres.2007.02.023. [DOI] [PubMed] [Google Scholar]; (b) Pagliaccetti NE, Robek MD. Viruses. 2010;2:1589. doi: 10.3390/v2081589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- v.For reviews: Chen KX, Njoroge FG. Curr. Opin. Investig. Drugs. 2009;10:821. Chary A, Holodniy M. Rev. Rec. Clin. Trials. 2010;5:158. doi: 10.2174/157488710792007293..
- vi.For reviews: Burton JR, Jr, Everson GT. Clin. Liver Dis. 2009;13:453. doi: 10.1016/j.cld.2009.05.001. Powdrill MH, Bernatchez JA, Goette M. Viruses. 2010;2:2169. doi: 10.3390/v2102169. For recent reports: Ruebsam F, Tran CV, Li L-S, Kim SH, Xiang AX, Zhou Y, Blazel JK, Sun Z, Dragovich PS, Zhao J, McGuire HM, Murphy DE, Tran MT, Stankovic N, Ellis DA, Gobbi A, Showalter RE, Webber SE, Shah AM, Tsan M, Patel RA, LeBrun LA, Hou HJ, Kamran R, Sergeeva MV, Bartkowski DM, Nolan TG, Norris DA, Kirkovsky L. Bioorg. Med. Chem. Lett. 2009;19:451. doi: 10.1016/j.bmcl.2008.11.048. Wang P, Chun B-K, Rachakonda S, Du J, Khan N, Shi J, Stec W, Cleary D, Ross BS, Sofia MJ. J. Org. Chem. 2009;74:6819. doi: 10.1021/jo901345j..
- vii. http://www.hcvdrugs.com.
- viii.Strosberg AD, Kota S, Takahashi V, Snyder JK, Mousseau G. Viruses. 2010;2:1734. doi: 10.3390/v2081734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ix.(a) Courcambeck J, Bouzidi M, Perbost R, Jouirou B, Amrani N, Cacoub P, Pepe G, Sabatier JM, Halfon P. Antiviral Therapy. 2006;11:847. [PubMed] [Google Scholar]; (b) De Francesco R, Carfi A. Adv. Drug. Dil. Rev. 2007;59:1242–1262. doi: 10.1016/j.addr.2007.04.016. [DOI] [PubMed] [Google Scholar]
- x.Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N, Rossaro L, Anderson FH, Jacobson IM, Rubin R, Koury K, Pedicone LD, Brass CA, Chaudhri E, Albrecht JK. Lancet. 2010;376 doi: 10.1016/S0140-6736(10)60934-8. [DOI] [PubMed] [Google Scholar]
- xi.Kota S, Scampavia L, Spicer T, Beeler AB, Takahashi V, Snyder JK, Porco JA, Jr, Hodder P, Strosberg AD. ASSAY Drug Dev. Tech. 2010;8:96. doi: 10.1089/adt.2009.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xii.Mousseau G, Kota S, Takahashi V, Frick DN, Strosberg DA. J. Gen. Virol. 2011;92:101. doi: 10.1099/vir.0.023325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xiii.Wei W, Cai C, Kota S, Takahashi V, Ni F, Strosberg AD, Snyder JK. Bioorg. Med. Chem. Lett. 2009;19:6926. doi: 10.1016/j.bmcl.2009.10.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xiv.For reviews of small molecule inhibitors of dimer-dimer interactions: Arkin MR, Wells JA. Nat. Rev. Drug Disc. 2004;3:301. doi: 10.1038/nrd1343. Fletcher S, Hamilton AD. Curr. Top. Med. Chem. 2007;7:922. doi: 10.2174/156802607780906735. Blazer LL, Neubig RR. Neuropsychopharm. Rev. 2009;34:126. doi: 10.1038/npp.2008.151..
- xv.Examples of small molecule dimers as inhibitors of protein-protein interactions: Melchiorre C, Andrisano V, Bolognesi ML, Budriesi R, Cavalli A, Cavrini V, Rosini M, Tumiatti V, Recanatini M. J. Med. Chem. 1998;41:4186. doi: 10.1021/jm9810452. Cavalli A, Bolognesi ML, Capsoni S, Andrisano V, Bartolini M, Margotti E, Cattaneo A, Recanatini M, Melchiorre C. Angew. Chem. Int. Ed. 2007;46:3689. doi: 10.1002/anie.200700256..
- 16.Some examples of synthetic dimers with inhibitory activities: Cholody WM, Hernandez L, Hassner L, Scudiero DA, Djurickovic DB, Michejda CJ. J. Med. Chem. 1995;38:3043. doi: 10.1021/jm00016a007. Hadden MK, Blagg BSJ. Bioorg. Med. Chem. Lett. 2007;17:5063. doi: 10.1016/j.bmcl.2007.07.014. For a review: Hadden MK, Blagg BS. Anticanc. Agt. Med. Chem. 2008;8:807. doi: 10.2174/187152008785914743..
- xvii.For a review of synthetic dimers that promoted protein-protein interactions: Gestwicki JE, Mainec PS. Comb. Chem. High Throughput Scr. 2007;10:667. doi: 10.2174/138620707782507296..
- xviii.Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P, Bos M, Cameron DR, Cartier M, Cordingley MG, Faucher AM, Goudreau N, Kawai SH, Kukolj G, Lagace L, LaPlante SR, Narjes H, Poupart M-A, Rancourt J, Sentjens RE, St. George R, Simoneau B, Steinmann G, Thibeault D, Tsantrizos YS, Weldon SM, Yong C-L, Llinas-Brunet M. Nature. 2003;426:186. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
- xix.Appukkuttan P, Dehaen W, Fokin VV, Van der Eycken E. Org. Lett. 2004;6:4223. doi: 10.1021/ol048341v. [DOI] [PubMed] [Google Scholar]
- 20.Kolb HC, Finn MG, Sharpless KB. Angew. Chem. Int. Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]