Summary
Many molecular pathways involved in heart disease have their roots in evolutionarily ancient developmental programs that depend critically on gene dosage and timing. MicroRNAs (miRNAs) modulate gene dosage post-transcriptionally, and among these, the muscle-specific miR-1 is particularly important for developing and maintaining somatic/skeletal and cardiac muscle. To identify pathways regulated by miR-1, we performed a forward genetic screen in Drosophila using wing-vein patterning as a biological assay. We identified several unexpected genes that genetically interacted with dmiR-1, one of which was kayak, encoding a developmentally-regulated transcription factor. Additional studies directed at this genetic relationship revealed a previously unappreciated function of dmiR-1 in regulating the polarity of cardiac progenitor cells. The mammalian orthologue of kay, c-Fos, was dysregulated in hearts of gain- or loss-of-function miR-1 mutant mice in a stress-dependent manner. These findings illustrate the power of Drosophila-based screens to find points of intersection between miRNAs and conserved pathways in mammals.
Introduction
microRNAs (miRNAs) are small non-coding RNA species that regulate mRNA translation or stability and, thereby, protein dosage in eukaryotes (reviewed in Inui et al., 2010). miRNAs interact with their target RNAs primarily through complementary base pairing to the 3′UTRs of the target gene mRNAs. This Watson-Crick pairing, generally involving the 5′ region of the miRNA, results in RNA degradation and/or inhibition of translation (reviewed in Bartel, 2009). Deletion of Dicer, the endonuclease necessary to form miRNAs, results in lethality during gastrulation, implying that miRNAs are necessary for the formation of the body plan (Bernstein et al., 2003). Specific miRNAs play critical roles in myriad processes, including cell fate, cellular differentiation, cancer, apoptosis, and stress responses (reviewed in Croce, 2009; Cordes and Srivastava, 2009; Mendell, 2005; van Rooij et al., 2008). In most cases, the regulatory function of individual miRNAs is achieved by control of multiple steps within a common pathway. Despite significant progress, the function and pathways regulated by most miRNAs remain unknown.
miR-1 is one of the most evolutionarily conserved miRNAs and is highly enriched in heart and muscle. In vertebrates, two distinct loci for the miR-1/miR-133 cluster, likely a result of gene duplication, encode identical mature miR-1s derived from each locus (i.e., miR-1-1 and miR-1-2). Loss of the single dmiR-1 in Drosophila results in cardiac-specification defects in a subset of flies (Kwon et al., 2005) and catastrophic failure of somatic muscle development during the second instar stage (Sokol and Ambros, 2005). In mice, loss of miR-1-2 results in a broad range of abnormalities, including ventricular septal defects, defects in cell-cycle control, and disturbed electrophysiological properties (Zhao et al., 2005; Zhao et al., 2007).
Using a combination of bioinformatic and biochemical approaches, we and others have found transcription factors (Zhao et al., 2005, 2007; Ikeda et al. 2009), receptor ligands (Kwon et al., 2005), (Ivey et al., 2008) ion channels (Yang et al., 2007), anti- apoptotic proteins (Xu et al., 2007), and modifiers of histone acetylation (Chen et al., 2006) that are directly responsive to miR-1 regulation. However, the number of genes known to directly respond to miR-1 or other miRNAs is only a small fraction of the total number of miRNA-sensitive genes and pathways. Therefore, an experimental method to discover pathways regulated by specific miRNAs in vivo would be useful. Uncovering the points of intersection between a particular miRNA and basic cellular processes would allow for a more comprehensive understanding of the miRNA’s function within the context of an intact cell or organ system.
Here, we describe a genome-wide enhancer/suppressor screen using the power of Drosophila genetics to identify cellular pathways regulated by miR-1. We discovered an interaction of miR-1 with numerous factors, including the transcription factor kayak, revealing an unexpected function for dmiR-1 in controlling cell polarity of cardiac progenitors during development. The mammalian orthologue of kayak, c-Fos, was also dysregulated upon manipulation of miR-1 levels in the mouse heart, highlighting the utility of Drosophila genetics for studying miRNA-dependent pathways in mammals.
Results
Genome-Wide Enhancer/Suppressor Screen for Genetic Interaction with miR-1
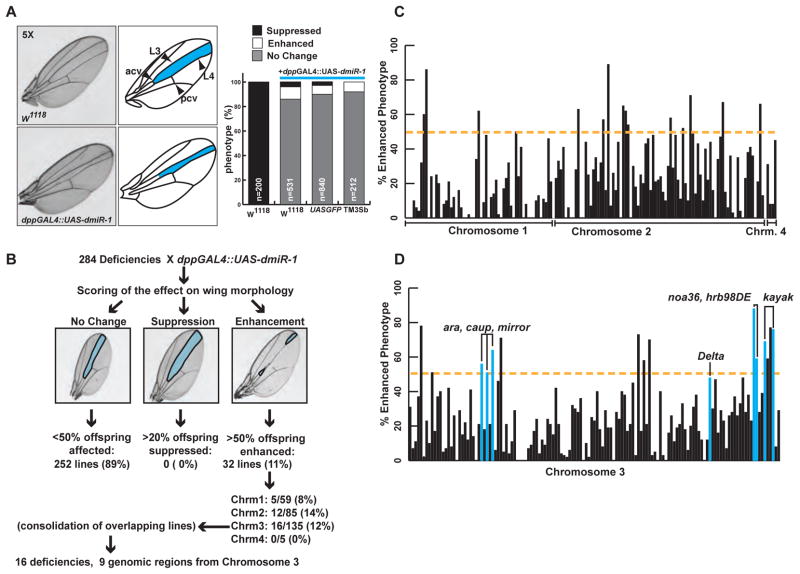
Many of the signals that determine body axis formation and cell fate are operational in the Drosophila melanogaster wing and are evolutionarily conserved between widely divergent species and tissues. Because the fly wing is easy to visualize and is a non-essential organ, it is often used for high-throughput genetic screening. The morphogen decapentaplegic (dpp) is normally expressed in the wing imaginal disc, the precursor of the adult fly wing structure, between the long veins number three and four (L3, L4). We generated a fly line that contains the dpp promoter driving GAL4 (dpp-GAL4) (Bloomington Stock Center) and a GAL4-sensitive upstream activating sequence (UAS) regulating a dmiR-1 transgene (UAS-dmiR-1) on the third chromosome. These mutant animals were viable to adulthood and were fertile, but exhibited a loss of wing tissue between veins L3 and L4, creating a scorable phenotype (Figure 1A).
Figure 1. Misexpression of dmiR-1 in the Drosophila wing results in a distinctive phenotype and identifies genetic partners of dmiR-1.
(A) Upper left: image of a wildtype wing, with pertinent anatomical structures labeled to the right. L3: long vein number 3, L4: long vein number 4, acv: anterior cross vein, pcv: posterior cross vein. Area highlighted in blue: area of expression of the wing disc specific decapentaplegic (dpp) enhancer. Lower left: image of wing from a dpp-GAL4::UAS-dmiR-1 mutant fly. Note narrowing of the L3/L4 inter-vein distance. Left: Incidence of enhancement, and suppression of the wing phenotype seen in dpp-GAL4::UAS-dmiR-1 mutant flies when mated with control fly lines. W1118: wildtype. The lines UAS-GFP and dpp-GAL4::UAS-dmiR-1/TM3Sb (TM3Sb) served as controls. TM3Sb: balancer.
(B) Summary of results from the forward genetic screen.
(C) Distribution of percentage affected offspring from each deficiency, organized by chromosome (1, 2, and 4). Yellow dashed line: point at which 50% of progeny were affected. Those deficiencies that met or exceeded this level were considered strongly positive (p<0.0001).
(D) Distribution of percentage affected offspring from each deficiency, organized by position on chromosome 3, going left to right. Blue bars: deficiencies containing specific genes (labeled) that enhanced the wing phenotype.
(See also Supplementary Table 1)
We hypothesized that genetic partners of dmiR-1 could be identified by scoring the wings for loss or gain of intervein distance (“enhancement” or “suppression” of the wing phenotype, respectively). The wing phenotype was sensitive to temperature; higher temperatures (22–25°C) resulted in higher transcriptional activity of GAL4 and more severe loss of wing tissue between L3 and L4 (data not shown). Due to this temperature sensitivity, all assays involving the dpp-GAL4::UAS-dmiR-1 flies were performed at 18°C. The loss of tissue did not appear to be the result of deranged cell size, as the spacing of the hairs, which serves as a crude marker for cell size (Garcia-Bellido et al., 1994), was not significantly changed. Control crosses on wild type backgrounds revealed rates of enhancement or suppression of the wing-vein phenotype among offspring of 8% or 3%, respectively (Figure 1A).
To identity genes that interact genetically with dmiR-1, the dpp-GAL4::UAS-dmiR-1 fly line was crossed with chromosome deficiency lines obtained from DrosDel (Ryder et al., 2007; Cambridge, UK) and Bloomington Stock Center (Bloomington, IN). Each line within these deficiency kits contains a molecularly defined deletion within one of the four Drosophila chromosomes. From 284 deficiency matings (representing approximately 70% of the fly genome), 32 fly lines (11%) had enhanced loss of wing tissue between L3 and L4 in at least 50% of offspring, while 252 lines (89%) had no effect on the dmiR-1-mediated phenotype (Figure 1B). The 32 lines that enhanced the dmiR-1 phenotype were distributed fairly evenly between chromosomes X, 2, and 3 (Figure 1C, D). No progeny reproducibly rescued, or suppressed, the phenotype. Further studies were performed on the 16 fly lines harboring deficiencies on chromosome 3 (Figure 1D). Loci on chromosomes 1, 2, and 4 are the subject of ongoing studies.
To narrow the genomic regions of interest on chromosome 3, we used smaller, overlapping deficiencies. Within these deficiencies, genes implicated in transcriptional regulation, development, cell-cycle control, apoptosis, mRNA processing, Notch signaling, or muscle biology were given the highest ranking. If available, mutant alleles of candidate genes from chromosome 3 were requested from the Bloomington Stock Center and crossed to the dpp-GAL4::UAS-dmiR-1 fly line. Whenever possible, multiple lines containing different mutant alleles were tested to minimize background and allele-specific effects. Mutant genes that, when heterozygous, recapitulated the enhancement of the dpp-GAL4::UAS-dmiR-1 phenotype underwent further study.
Identification of delta and mirror Alleles Validate dmiR-1 Enhancer Screen
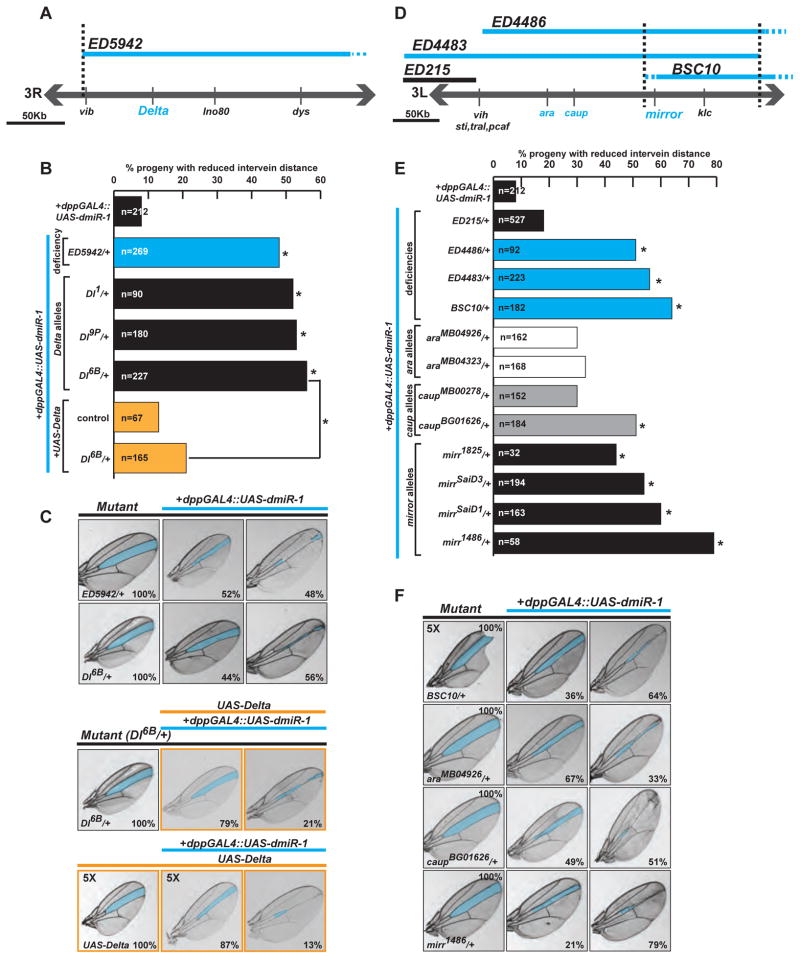
In our screen, the deficiency line Df(3R)ED5942 enhanced the dpp-GAL4::UAS-dmiR-1 phenotype, as ~50% of progeny with the pertinent genotype had narrowing of the intervein area (p<0.01). This deficiency contains the gene delta (Dl) (Figure 2A), which encodes a ligand for Notch, an important regulator of cell fate and differentiation (reviewed in Bray 2006). Crosses between dpp-GAL4::UAS-dmiR-1 flies and flies heterozygous for mutant Dl alleles, enhanced the dpp-GAL4::UAS-dmiR-1 phenotype (Figure 2B,C, and data not shown). Over-expression of wildtype Dl reduced the expressivity of wing vein defects in dpp-GAL4::UAS-dmiR-1 flies in the context of a Dl mutant background, confirming the specificity of the genetic relationship between Dl and dmiR-1 (Figure 2B, C, middle panels).
Figure 2. Validation of the dmiR-1 enhancer/suppressor genetic screen.
(A) Schematic of the deficiency ED5942 spanning delta.
(B) Percentages of affected offspring when specific deficiencies and alleles of delta were mated to the dpp-GAL4::UAS-dmiR-1 line. Dl1, Dl9P, Dl6B are loss of function alleles. Yellow bars depict rescue of enhancement as shown in the center panels of (C). * p-value <0.001.
(C) Panels of representative fly wings with the L3/L4 area highlighted in blue. Control parental mutant lines (ED5942, Dl6B and UAS-delta) shown to the left, with representative wings of pertinent offspring when crossed with the dpp-GAL4::UAS-dmiR-1 line. Wings scored as “no change from dpp-GAL4::UAS-dmiR-1” (center) or “enhanced” (right), with percentages noted in bottom right corner. Yellow edged images: (top) UAS-delta, Dl6B, dpp-GAL4::UAS-dmiR-1 representative fly wings displaying rescue of the phenotype, and the control UAS-delta, dpp-GAL4::UAS-dmiR-1, representative fly wings (bottom).
(D) Schematic of deficiencies containing the Iroquois locus: araucan (ara), caupolican (caup), mirror (mirr). Blue lines: chromosomal location of deficiencies that enhance the dpp-GAL4::UAS-dmiR-1 phenotype in ≥ 50% of progeny.
(E) Percentages of affected offspring when specific deficiencies (as in D), and loss of function alleles of the Iroquois locus (araMB04926, araMB04323, caupMB00278, caupBG01626, mirr1825, mirrSaiD3, mirrSaiD1, mirr1486) were mated to the dpp-GAL4::UAS-dmiR-1 line. * p-value <0.001.
(F) Control parental mutant lines (BSC10, araMB04323, caupBG01626 and mirr1486) shown to the left, with representative wings of pertinent offspring when crossed with the dpp-GAL4::UAS-dmiR-1 line (as in E). Wings scored as “no change” (center) or “enhanced” (right), with percentages noted in bottom right corner.
(See also Supplementary Figure1&2)
The deficiency lines Df(3L)ED4486, Df(3L)ED4483, and Df(3L)BSC10 also enhanced the dpp-GAL4::UAS-dmiR-1 phenotype (Figure 2D,E). Contained within these deficiency lines was the highly conserved Iroquois (Iro) gene family, which encodes homeodomain-containing transcription factors, including mirror (mirr), araucan (ara), and caupolican (caup). Activity of the Iro locus in fly is critical for establishing developmental territories within the organism (reviewed in Cavodeassi et al., 2001). In mice, Iro family members are important for vertebrate cardiac chamber specification (Bao et al., 1999) and cardiac conduction (Costantini et al., 2005). mirr loss-of-function alleles greatly enhanced the dpp-GAL4::UAS-dmiR-1 phenotype, while alleles of ara and caup displayed less robust enhancement (Figure 2E,F, Figure S1).
Identification of Dl and mirr mutant alleles as phenotypic enhancers validated the screen’s methodology since Dl and Delta-like-1 are miR-1 targets in flies and mice, respectively (Kwon et al., 2005; Ivey et al., 2008), and Irx5 is a target of miR-1 in the mammalian heart (Zhao, et al., 2007). Using the Dl and mirr mutants as positive controls for interpretation of affected progeny, all crosses were subsequently scored positive if ~50% of progeny (~6X above background) showed enhancement of the wing phenotype.
Identification of Genetic Partners of dmiR-1
The overlapping deficiencies Df(3R)3450 and Excel6209 enhanced the dmiR-1 phenotype (Figure S2A,B and data not shown). Within these deficiencies, we identified two genes, noa36 and hrb98DE, whose mutant alleles enhanced the phenotype (Figure S2B, C and data not shown). noa36 is predicted to bind metal ions, and hrb98DE acts to regulate alternative mRNA splicing. Bioinformatically, hrb98DE was predicted to contain four dmiR-1 binding sites in its 3′UTR (Stark et al., 2003) that were conserved in Anopheles. To determine if dmiR-1 directly regulates hrb98DE, we co-transfected a GAL4-dmiR-1 construct with a luciferase reporter construct containing 1035 bp of the 3′UTR of hrb98DE into Drosophila Schneider 2 (S2) cells. Luciferase activity was repressed in the presence of dmiR-1, suggesting that hrb98DE was a direct target of dmiR-1 (Figure S2D).
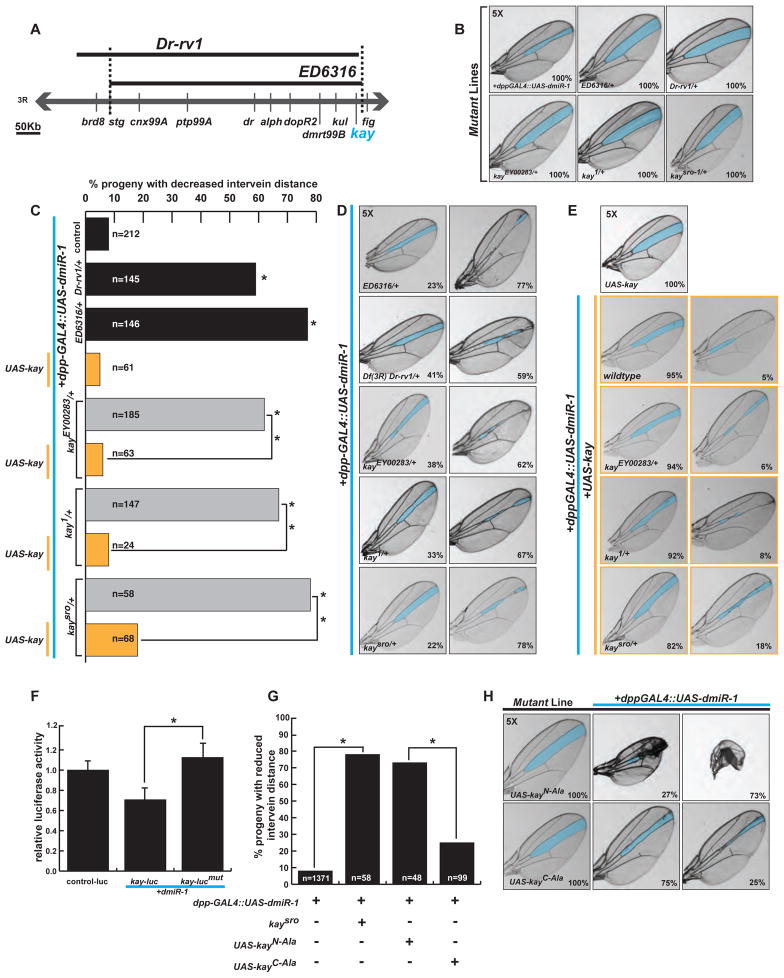
The deficiency lines Df(3R)Dr-rv-1 and Df(3R)ED6316 also enhanced the dpp-GAL4::UAS-dmiR-1 phenotype (Figure 3A–D). Contained within these fly deficiency lines was kayak (kay), which encodes a leucine-zipper-containing transcription factor whose mammalian orthologue is c-Fos. We used several different kay loss-of-function alleles, including a null allele (kay[1]), a hypomorphic recessive lethal allele (kay[sro-1]), a hypomorphic, homozygous viable allele (kay[EY00283]), and a presumed hypomorphic allele (kay[EY01644]). Parental lines of kay heterozygous mutant alleles had normal wing anatomy (Figure 3B). Mating of each of the kay alleles to the dpp-GAL4::UAS-dmiR-1 fly line resulted in enhancement of the wing phenotype (Figure 3C, D).
Figure 3. Kayak genetically interacts with dmiR-1.
(A) Schematic of the deficiencies Df(3R) Dr-rv-1 and Df(3R)ED6316 spanning kayak.
(B) Panels of representative parental mutant fly wings with the L3/L4 area highlighted in blue. Note normal L3/L4 inter-vein distance.
(C) Percentages of affected offspring when specific deficiencies and mutant alleles of kay were mated to the dpp-GAL4::UAS-dmiR-1 line. Yellow bars depict rescue of phenotypic enhancement with expression of kay (UAS-kay) as shown in D. * p-value <0.001.
(D) Representative wings of pertinent offspring when Df(3R) Dr-rv-1 and Df(3R)ED6316, and the loss of function kay alleles (kayEY00283, kay1, kaysro) were crossed with the dpp-GAL4::UAS-dmiR-1 line. Wings scored as “no change” (left) or “enhanced” (right), with percentages noted in bottom right corner.
(E) Images of representative wings from a genetic rescue experiment. Top: control wing from a UAS-kay fly. Lower rows: Matings between UAS-kay, dpp-GAL4::UAS-dmiR-1 flies and flies harboring heterozygous mutant alleles of kay (kayEY01644, kay1, kaysro). Wings were scored as “no change” (left) or “enhanced” (right), with percentages noted in bottom right corner.
(F) Relative luciferase activity with or without the 3′UTR of kay which is inserted into the 3′UTR of a constitutively active luciferase vector in the presence or absence of dmiR-1. Kay-lucmut: luciferase reporter with the two putative dmiR-1 binding sites mutated as shown in Figure S3B. *p<0.05. Error bars are represented as standard error of the mean.
(G) Percentage of affected offspring, comparing kaysro mutants to mutants containing amino- and carboxy-terminal substitutions of serine and threonine residues to alanine. * p-value <0.001.
(H) Photographs of representative wings for data in (G).
(See also Supplementary Figure 3)
To determine if the enhanced phenotype was specific to decreased kay function, we performed a genetic rescue assay by mating the dpp-GAL4::UAS-dmiR-1;kay[sro-1] fly to flies harboring a UAS-kay element (Figure 3C, E). Co-expression of dmiR-1, kay[sro-1], and wild-type kay resulted in rescue of the phenotype, and this was also true for the kay[1] and kay[EY01644] alleles (Figure 3C, E). We concluded that a decrease in kay function specifically enhanced the dpp-GAL4::UAS-dmiR-1 wing phenotype.
To determine if dmiR-1 reduced kay mRNA levels through direct targeting of the kay 3′UTR, we introduced the full 3′UTR into a luciferase reporter vector and transfected this with or without dmiR-1 into fly S2 cells. We observed a dose-dependent reduction in luciferase activity in the presence of dmiR-1, signifying that dmiR-1 likely directly regulates kay levels through the 3′UTR (Figures 3F and S3A). Mutation of two putative dmiR-1 binding sites resulted in loss of dmiR-1 mediated repression, indicating functionality of these sites (Figure 3F, and S3B).
Genetic Interaction Between dmiR-1 and kay Depends on JNK-Mediated Signals
Kay modulates migration, cell polarity, and directionality cues within developing tissues (Riesgo-Escovar et al., 1997, Zeitlinger et al., 1997, Weber et al., 2008) and kay-null flies are non-viable. In contrast, mammals have multiple kay orthologues (Fra1, Fra2, c-Fos, FosB) that appear to be partially redundant (Fleischmann et al., 2000). As an immediate-early response gene, c-Fos is rapidly up-regulated in the heart under a variety of pathological conditions (Izumo et al., 1988; Larsen et al., 1998) and modulates the genomic response to mechanical and cellular stress. Kay/c-Fos dimerizes with other proteins, such as c-Jun, to form the AP-1 complex. Kay/c-Fos is phosphorylated by both the Ras/MAPK and jun-kinase (JNK) pathways, resulting in distinct transcriptional responses.
We tested proteins within the Ras/MAPK, and JNK signaling pathways for a genetic interaction with dmiR-1. Mutants within the Ras/MAPK or JNK pathway did not genetically interact with dmiR-1 to the same degree as kay in our wing-based assay (data not shown). Gain-of-function alleles predicted to activate Ras/MAPK, including one involving the epidermal-like growth factor receptor (EGFR), also did not consistently enhance the wing vein phenotype (data not shown).
To test these pathways in an alternative fashion, we utilized fly lines containing transgenes encoding mutants of kay that were resistant to phosphorylation by either JNK or MAPK kinases (gifts from D. Bohmann, Univ. of Rochester). These UAS-based fly lines have putative JNK and RAS-MAPK phosphorylation sites in kay mutated to alanine residues, allowing segregation of JNK from RAS/MAPK signaling (Ciapponi et al., 2001). The mutant UAS-kay[Pan-Ala], which has all putative threonine (T) or serine (S) residues involved in phosphorylation mutated to alanine (A), resulted in high lethality when crossed to the dpp-GAL4::UAS-dmiR-1 line (data not shown). The UAS-kay[C-Ala] mutant harboring alanine substitutions of RAS-MAPK-sensitive S/T residues in the carboxy-terminal region only mildly enhanced the dpp-GAL4::UAS-dmiR-1 phenotype (Figure 3G, H bottom row). In contrast, UAS-kay[N-Ala], containing alanine substitutions of JNK-sensitive residues, recapitulated the loss of function alleles of kay, implying that phosphorylation of the JNK-sensitive residues was important for kay function in this assay (Figure 3G, H top row). These results provided evidence that JNK activity might be critical to the observed genetic interaction between kay and dmiR-1.
dmiR-1 Expression in the Wing Imaginal Disc Results in Trichome Misalignment
Dl, mirr, and kay have been implicated as members of the genetic network governing planar cell polarity (PCP) in Drosophila (Cooper and Bray, 1999; Fanto and Mlodzik, 1999; Weber et al., 2008). Cell polarity signals flow through the transmembrane receptor frizzled (fz), which associates with dishevelled (dsh) and strabismus/van gogh (Vang), to activate the JNK signaling cascade (Boutros et al., 1998; Weber et al., 2000). Core proteins within the PCP signaling cascade are highly conserved, and PCP in mammals is central to fundamental processes, such as gastrulation and organogenesis (reviewed in Veeman et al., 2003, Wallingford et al., 2002, Zallen, 2007). In flies, PCP generates the stereotypical proximal-to-distal orientation of hairs and bristles over the ectoderm and, when disturbed, results in a whorled bristle pattern (Classen et al. 2005).
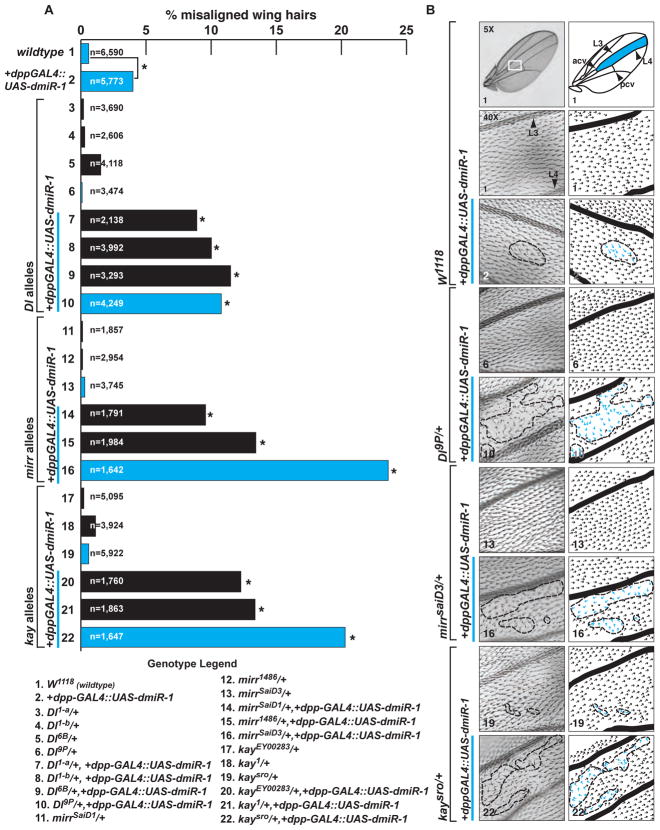
We hypothesized that kay, Dl and mirr might have been identified in our screen in part due to a common functionality in the generation of cell polarity. To test this, we counted the number of misaligned hairs, or trichomes, in a standard location (boundaries marked in the anterior-posterior (A-P) dimension by L3 and L4 and in the proximal-distal (P-D) dimension by the posterior cross vein and anterior cross vein). Interestingly, in dpp-GAL4::UAS-dmiR-1 flies, ~4% of wing hairs were not oriented in the normal P-D orientation compared to 0.6% seen in wild type animals (Figure 4). To determine if these results were specific to dmiR-1, the dpp-GAL4 fly line was mated to a UAS-miR-7 fly line (gift of E. Lai, Rockefeller University) to generate the control fly line dpp-GAL4::UAS-dmiR-7. The dpp-GAL4::UAS-dmiR-7 fly line had numbers of misaligned trichomes closer to wild type animals (0.18%), supporting the notion that dmiR-1 specifically interferes with the generation of PCP (data not shown). We found that heterozygosity of Dl, mirr, or kay mutant alleles, along with dmiR-1 expression, significantly increased the numbers of misaligned wing hairs when compared to wildtype or dpp-GAL4::UAS-dmiR-1 flies (Figure 4 and S4A). In addition to being misaligned, many hairs were abnormally foreshortened and thickened (data not shown). Over-expression of Dl through the use of two UAS-Dl lines did not rescue the alignment defect (Figure S4B). In contrast, overexpression of kay (UAS-kay) rescued the wing PCP defects induced by expression of dmiR-1 in the kay heterozygous background (Figure S4C).
Figure 4. Enhancement of dmiR-1 PCP defects by haploinsufficiency of delta, mirror and kayak.
(A) Graphic of percentage of misaligned wing hairs with n= number of hairs counted, representing a minimum number of five individual animals. Genotypes detailed below. * p<0.001. Blue bars denote genotypes represented by photographs in (B).
(B) Photographs of representative adult fly wings of noted genotypes with schematic of hair alignment (right). Top: W1118 (wildtype) wing with pertinent anatomical structures noted to right. White box: low magnification view of location where hair alignment was determined. Areas with misaligned wing hairs are outlined in a dashed line, with individual hairs of normal proximal-distal orientation (black) or misoriented (blue) indicated. Genotype noted in bottom left corner.
(See also Supplementary Figure 4 and 5)
dmiR-1 Genetically Interacts with Members of the Core PCP Pathway
To determine if trichome alignment defects might reflect dmiR-1 regulation of core PCP proteins, we tested two members of this cascade that are bioinformatically predicted to be direct targets of dmiR-1: van gogh (vang) and prickle (pk) (Stark et al., 2003), as well as their upstream effector, dishevelled (dsh). When we tested alleles of dsh, vang and pk in the intervein wing assay, we found that multiple alleles of these genes genetically interacted with dmiR-1 (Figure S5A, right). Examination of the directionality of the wing hairs also demonstrated that multiple alleles of dsh, vang and pk enhanced the effect of dmiR-1 on PCP (Figure S5A, left). Interestingly, unlike our results for Dl and kay, the degree of effect on intervein distance for pk alleles did not predict the severity of PCP defects. These results may indicate that the loss of intervein distance may be mechanistically distinct from the loss of PCP. Testing of the 3′UTRs of both vang and pk in S2 cells indicated that pk, but not vang, is a direct target of dmiR-1 (Figure S5B). We observed a dose-dependent reduction in pk-luciferase activity in the presence of dmiR-1, signifying that dmiR-1 likely directly regulates pk levels through the 3′UTR (Figure S5C). The latter results are consistent with dmiR-1 regulating PCP on multiple levels (Figure S5D) and highlight the ability of our screen to identify genetic pathways regulated by dmiR-1.
Dysregulation of dmiR-1 and kay in Fly Heart Results in Cell Polarity Defects
The artificial fly wing assay proved effective in discovering genetic interactions with dmiR-1, including those involving the kay and PCP pathways. dmiR-1 is normally expressed in cardiac and somatic muscle, so we investigated whether the genetic relationships regarding PCP identified in the wing were present in areas of endogenous expression, in the absence of overexpression. The fly heart, or dorsal vessel, is segmentally patterned with a defined number of myocardial and pericardial cells, and expresses dmiR-1 early in development (Kwon et al., 2005; Sokol & Ambros, 2005). In the dorsal vessel, Slit, a glycoprotein secreted from the medial surface of cardioblasts, acts as a polarity marker and signal by interacting with its receptor, roundabout (robo), to assure that cardioblasts migrate properly to the midline (Qian et al., 2005; Santiago-Martinez et al., 2006, MacMullin and Jacobs, 2006).
To determine if dmiR-1 functions to regulate cell polarity within the fly heart, we analyzed dmiR-1 null fly embryos (Δ/ΔdmiR-1) at stage 16. Immunostaining of these embryos for the cardioblast marker Neuromancer (Nmr) and for Slit revealed that 77% of embryos had misaligned cardioblasts and abnormally diffuse secretion of Slit from the medial and lateral sides of cardioblasts (Figure 5B, F). Parallel studies in embryos heterozygous for dmiR-1 revealed 8% affected embryos, similar to wild type embryos (Figure 5F). To determine if supra-physiologic levels of dmiR-1 affected cardiac polarity, we used the tinCΔ4-GAL4 driver (Lo and Frasch, 2001) to increase the levels of dmiR-1 specifically within the fly heart. The resultant tinCΔ4-GAL4::UAS-dmiR-1 embryos collected at 29° or 18°C also had diffuse Slit secretion, indicating that the dosage of dmiR-1 within the fly heart was critical for normal cell polarity and cardioblast alignment (Figure 5C,G). Immunostaining for α-Spectrin, a second cardioblast polarity marker, in the same genotypes as described above was notable for disruption of the normal apical-lateral expression pattern, providing further evidence that cell polarity within the embryonic heart was disrupted (Figure S6 B,C,F,G).
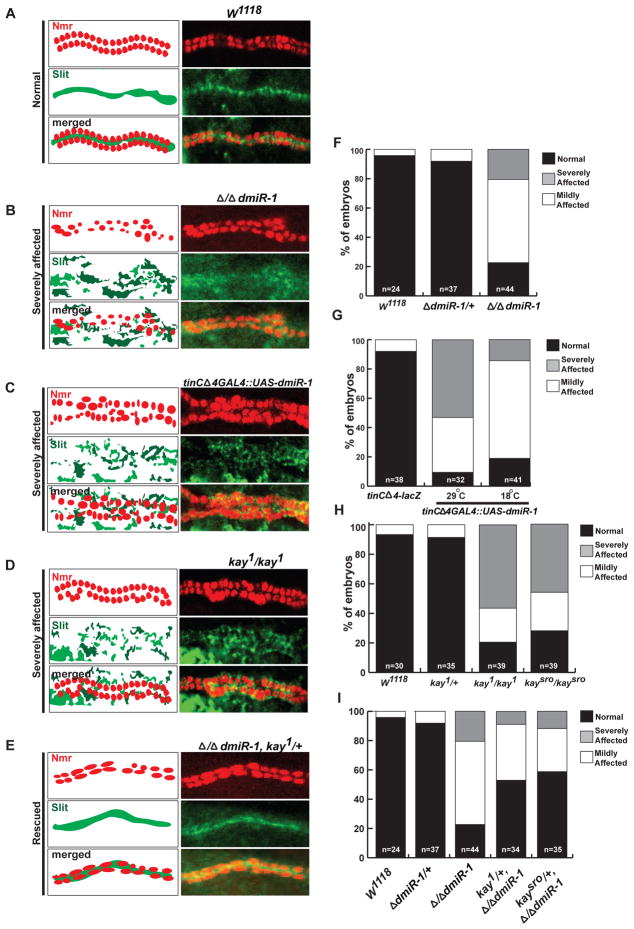
Figure 5. dmiR-1 influences cell polarity in the Drosophila heart.
(A) Schematic of the expression pattern for Neuromancer (Nmr, red) and Slit (green) in wildtype (W1118) Drosophila hearts (left). Confocal images showing Nmr and Slit localization within the fly heart in wildtype (W1118) stage 16 embryos (right).
(B) Schematic (left) and confocal (right) images of homozygous-null dmiR-1 stage 16 embryos, with Nmr and Slit expression in red and green, respectively.
(C) Schematic (left) and confocal (right) images of tinC Δ4GAL4::UASdmiR-1 stage 16 embryos, with Nmr and Slit expression in red and green, respectively.
(D) Schematic (left) and confocal (right) images of homozygous mutant kay1 stage 16 embryos, with Nmr and Slit expression in red and green, respectively.
(E) Schematic (left) and confocal (right) images of homozygous-null dmiR-1, kay1/+ stage 16 embryos, with Nmr and Slit expression in red and green, respectively.
(F) Quantification of panel (B) with normal (black), mildly (white), or severely (grey) affected embryos, according to genotype. Scoring was based on the degree of misalignment of the cardioblasts and diffuse expression of Slit.
(G) Quantification of panel (C) with normal (black), mildly (white), or severely (grey) affected embryos, according to genotype and temperature.
(H) Quantification of panel (D) with normal (black), mildly (white), or severely (grey) affected embryos, according to genotype.
(I) Quantification of panel (E) with normal (black), mildly (white), or severely (grey) affected embryos, according to genotype.
(See also Supplementary Figure 6 and 7)
We then determined if kay mutants also displayed cell specification or polarity abnormalities. We harvested homozygous kay[1] and kay[Sro-1] stage 16 embryos and immunostained for Nmr, Dmef2 (muscle progenitors), Tin (muscle progenitors), or Eve (pericardial cell) (Figure S7). We detected no significant differences in the numbers of cardioblasts or pericardial cells per hemi-segment compared to wildtype. However, when we immunostained the homozygous kay[1] and kay[Sro-1] embryos for Nmr and Slit to evaluate the morphology of the heart tube, we found evidence of cardioblast misalignment and diffuse, but not reduced, expression of Slit from the medial and lateral sides of the cardioblasts (Figure 5D, H, data not shown). Immunostaining of homozygous kay[1] and kay[Sro-1] stage 16 embryos with α-Spectrin also revealed loss of the normal apical-lateral expression pattern of α-Spectrin (Figure S6D,H and data not shown).These results indicated that kay function was necessary for normal dorsal vessel formation and cell polarity of cardioblasts but was not important for cell specification.
Since kay was a direct target of repression by dmiR-1 and genetically interacted in the wing vein assay, we tested whether the heterozygosity of kay could mitigate the loss of dmiR-1 in the developing heart. We found decreased kay activity from two different kay loss-of-function alleles reduced the penetrance of the Δ/Δ dmiR-1 heart phenotype with respect to the diffuse medial and lateral expression of Slit (Figure 5E, I, data not shown). Furthermore, immunostaining of these doubly mutant embryos for α-Spectrin demonstrated that the normal apical-lateral expression pattern was partially restored in the Δ/ΔdmiR-1 embryos heterozygous for kay (Figure S6 E, I). These results suggested that a portion of the dosage effects of dmiR-1 in the heart are due to abnormal kay activity.
Regulation of c-Fos and c-Jun Activity by miR-1 in the Stressed Mammalian Heart
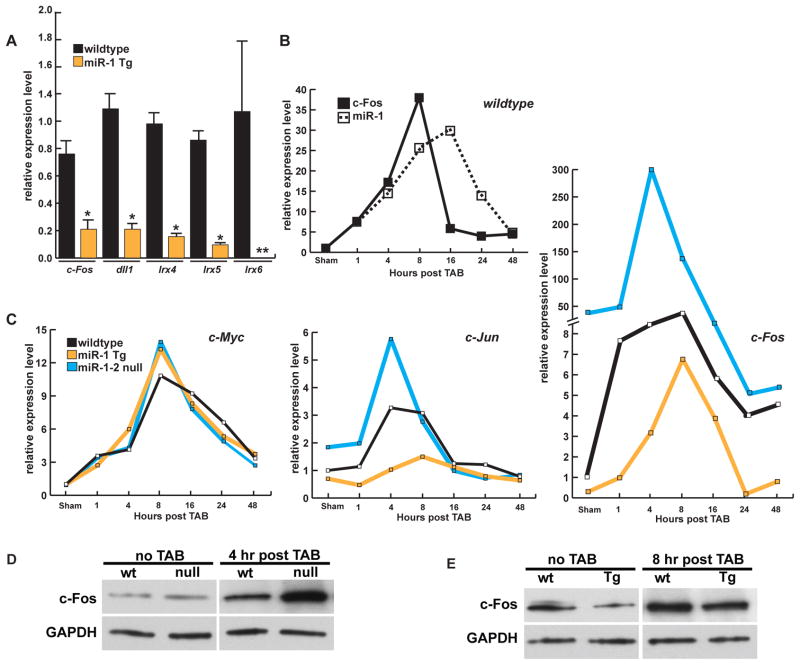
Our screen was based on the hypothesis that interrogation of the Drosophila genome for genetic partners of dmiR-1 would yield insights into mammalian cardiac and skeletal muscle biology. To test our most promising candidates, we performed quantitative PCR (q-PCR) on cardiac mRNA obtained from 8-week-old mice overexpressing miR-1 by threefold in the post-natal heart under control of the α-MHC promoter (α-MHC-miR-1) (Zhao et al., 2005; Zhao et al., 2007). We found a statistically significant reduction in mRNA levels of c-Fos, Delta-like-1, Irx4, -5, and -6 in miR-1 transgenic mice (Figure 6A). These results provided evidence that our wing-based screen identified several genetic partners of miR-1 that could be extrapolated to the mammalian heart, including some that are direct targets such as Delta-like-1 and Irx5 (Ivey et al., 2008; Zhao et al., 2007).
Figure 6. miR-1-2 limits stress-induced activation of the kay orthologue, c-Fos, in mouse hearts.
(A) Quantification of mRNA levels for mouse homologues of genes found to have a genetic interaction with dmiR-1 in wildtype (black) or α-MHC-miR-1 transgenic (Tg) mice (yellow). *p=<0.05. dll1:delta-like-1, Irx4,5,6: Iroquois family 4,5,6. All values normalized to GAPDH levels. ** not detectable. n=4 animals for each time point. Error bars are represented as standard error of the mean.
(B) Time course of c-Fos or miR-1 expression after thoracic aortic banding (TAB) by qPCR in wildtype mice.
(C) Time course of c-myc (left), c-Jun (middle) and c-Fos (right) expression as determined by qPCR analysis after TAB. Wildtype animals (black), α-MHC-miR-1 transgenic mice (yellow) and miR-1-2 null (blue) mice are indicated.
(D–E) Western blots of mouse hearts at time points of maximal c-Fos induction in miR-1-2 null (ko) (D) or α-MHC-miR-1 transgenic (Tg) animals (E) after TAB.
c-Fos is an immediate early response gene and is transiently upregulated in the first few hours after cardiac stress, such as that induced by pressure overload (Komoro et al., 1990, Schunkert et al., 1991). To determine how c-Fos and miR-1 mRNA levels were regulated in the first few hours of increased afterload, we examined left ventricular gene expression from wild type mice after thoracic aortic banding (TAB) at 1, 4, 8, 16, 24, and 48 hours post-procedure. c-Fos mRNA and miR-1 levels rose dramatically, with peak miR-1 levels coinciding with a precipitous decline in c-Fos mRNA levels in ventricular tissue (Figure 6B).
To determine if the rise of miR-1 levels participated in the negative regulation of the immediate early gene response and might contribute to the rapid decrease in c-Fos expression, we performed TAB experiments on miR-1-2 null mice. miR-1-2 null animals have a 50% reduction in the amount of mature miR-1 due to the presence of the redundant miR-1-1 allele (Zhao et al., 2007). The temporal pattern for c-Fos expression was shifted from a normal peak at 8 hours, to a peak 4 hours post-procedure, and more significantly, c-Fos levels were ~6-fold higher in miR-1-2 mutants than in wild type animals (Figure 6C, right panel). Conversely, transgenic mice overexpressing miR-1 in cardiomyocytes had greater than 6-fold lower c-Fos mRNA levels after TAB compared to wild type (Figure 6C, right panel). A similar pattern was observed for c-Jun expression (Figure 6C, center panel). Interestingly, there were no significant differences in the levels or timing of expression for c-Myc (Figure 6C, left panel). Western blots performed on hearts harvested at the time points of maximal induction in miR-1-2 null or miR-1 transgenic animals confirmed our q-PCR results (Figure 6D,E). To determine if the dysregulation of c-Fos was specific to the myocyte population, we performed immunohistochemistry on heart sections from wildtype and transgenic miR-1 overexpressing mice. The number of cardiomyocytes that expressed c-Fos, which was largely nuclear (Figure S8A), was significantly decreased in transgenic mice overexpressing miR-1 in cardiomyocytes (α-MHC-miR-1). Unlike the direct targeting of kayak by dmiR-1 in flies, the 3’-UTR of c-Fos was not responsive to miR-1 (Fig. S8B), suggesting conservation of the pathway regulated by miR-1, but divergence in the precise targets by which the miRNA exerts its biologic effects.
Discussion
This study highlights the utility of using Drosophila to conduct genome-wide studies to identify genetic pathways in which miRNAs function. Using an in vivo biological assay, followed by genetic and biochemical studies, we uncovered several candidate genes subject to dmiR-1 regulation. Importantly, we identified previously validated targets, which served as positive controls. Furthermore, we identified miR-1 responsive genes and pathways that functioned to regulate PCP during fly cardiogenesis and to appropriately limit the mammalian heart’s response to stress.
We purposefully chose a very high threshold (enhancement of phenotype in ~50% of progeny) as a cut-off for further evaluation, and therefore likely excluded many additional biologically relevant genetic partners of dmiR-1. As our studies relied on the wing-specific dpp-GAL4 system, the interrogated genes only encompass those that are normally expressed in the inter-vein region between L3 and L4. Because many of the pathways that determine cell fate, position, and differentiation are used reiteratively in divergent tissues, the genome-wide screen with an artificial wing assay revealed genes relevant to cardiovascular biology. It would be of interest to use heart- and muscle-specific expression assays to perform analogous in vivo studies with muscle- or heart-based phenotypes as a primary endpoint. The results presented here highlight the results obtained from one genetic locus. Other loci of interest are the subject of on-going studies.
dmiR-1 Regulates Cardiac Cell Polarity in Drosophila
In addition to the loss of intervein distance, we found that dmiR-1 over-expression also resulted in an increase in the frequency of misoriented wing hairs, consistent with defects in PCP. This led us to find that cell polarity within the Drosophila heart was also influenced by the dosage of dmiR-1 and that aberrant dmiR-1 activity resulted in defects in cardioblast alignment and the normal polarization of Slit and α-Spectrin expression in cardioblasts. These results suggest dmiR-1 functions to generate or maintain cell polarity in the fly heart. It would be of interest to determine if miR-1 modulation of PCP occurs primarily through modulation of pk and JNK/kay/c-Fos, or whether miR-1-mediated modulation of cytoskeletal proteins also plays a key role.
In mammals, cell polarization is emerging as an important process during the formation of the four-chambered heart and, in particular, the outflow tract of the heart. For example, mutations in the PCP genes van gogh-like-2 (Vangl2/Looptail) (Phillips et al., 2005, 2008), scribbled (Scrib/Circletail) (Phillips et al., 2007) and dishevelled 2 (Hamblet et al., 2002) cause ventricular septal defects and outflow tract alignment defects. These defects have been partly attributed to aberrant migration of myocardial cells into the endocardial cushions. Interestingly, 50% of miR-1-2 mutant embryos have ventricular septal defects that result in embryonic lethality (Zhao et al., 2007), and it will be interesting to determine if this is due to a PCP defect.
miR-1 and the Myocardial Response to Stress
From our pool of candidates, we chose to narrow our focus to the conserved genetic interaction between miR-1 and the immediate-early gene, c-Fos. As c-Fos and c-Jun are induced by TAB, these transcription factors might mediate some of the adaptive transcriptional responses to stress. Unlike earlier studies that documented the down-regulation of miR-1 days after myocardial remodeling (Care et al., 2007, Sayed et al., 2007), we examined the relationship between miR-1 and the immediate-early response genes induced within hours of banding the aorta. miR-1, c-Fos, and c-Jun were normally activated by TAB within a few hours, and in mice lacking miR-1-2, the initial upregulation of c-Fos and c-Jun was exaggerated in response to stress.
Our findings implicate miR-1 in the initial dampening of stress response genes and in governing sets of genes used during periods of pressure overload. Serum response factor (SRF) is a direct upstream regulator of both c-Fos and miR-1, suggesting that SRF activates the transcriptional mediators of the immediate early gene response and, in a negative feedback loop, limits the extent of the stress response by activating miR-1 expression. The 3’UTR of SRF (Care et al. 2007) and Elk-1 (K. Cordes, unpublished results), both necessary factors for c-Fos induction, are not subject to direct regulation by miR-1. Therefore, the mechanism by which miR-1 modulates c-Fos levels in mammalian heart remains to be determined. However, previous studies have indicated that JNK signaling is activated during hypertrophic stimuli (Ramirez et. al 1997, Wang et al., 1998) and in particular, is critical to the activity of TCF/Elk-1 on the c-Fos promoter (Cavigelli et al., 1995). Therefore, it is possible that in the murine heart, miR-1 modulation of JNK activity may indirectly affect c-Fos levels through changes in the phosphorylation of TCF/Elk-1 (Fig. S8C). It is intriguing that over millions of years of evolution the networks regulated by a miRNA can be conserved, but by virtue of the myriad targets of a miRNA, the points at which a miRNA can impinge on the network may diverge. Analogous to what we observe here, miR-1, directly targets the cardiac developmental transcription factor, Hand2, in mice but not in flies, yet regulates cardiogenesis in both species (Zhao et al., 2005; Kwon et al., 2005; Han et al., 2006). It will be interesting to determine if this is a common feature in the evolutionary development of miRNA-mediated gene regulation.
Experimental procedures
Drosophila Stocks
Flies with genomic deletions (deficiencies) were ordered from Bloomington Stock Center (Bloomington, IN) and DrosDel (Cambridge, U.K.). The following fly lines were generously provided by their creators: ΔdmiR-1 (C. Kwon, Gladstone Institute, San Francisco), UAS-dmiR-7 (Eric Lai, Sloan-Kettering Institute, New York). Fly lines not specifically mentioned in the text were obtained from the Bloomington Stock Center. Wildtype control flies are W1118. Overexpression of transgenes was achieved using the UAS-GAL4 system (Brand and Perrimon, 1993). The following Gal4 and UAS lines were used: dpp-GAL4 (Bloomington Stock Center), tinCΔ4-GAL4 (R. Bodmer, Burnham Institute, La Jolla, CA), UAS-kay[N-Ala] , UAS-kay[C-Ala] (D. Bohmann, University of Rochester, New York).
Wing Hair Polarity Scoring
Flies were anesthetized with CO2 and euthanized with isopropanol. Wings were removed and mounted on histology slides with Canada Balsam (Sigma). Hairs were counted at 20X magnification with ImagePro software (MediaCybernetics), and misaligned hairs were scored manually. A minimum of five animals per genotype were assayed.
Immunohistochemistry
Antibody staining was performed as described (Han et al., 2002). Cy3- or FITC-conjugated secondary antibodies (Jackson Labs) were used for fluorescent confocal microscopy. All the secondary antibodies were used at 1:200. Embryos with fluorescent staining were mounted in VectaShield (Vector Laboratories) and preparations were analyzed using Zeiss LSM510 and Biorad MRC-1024MP confocal microscopes. The following primary antibodies were used in this study: rabbit anti-Tinman, 1:1000 (Venkatesh et al., 2000); rabbit anti-β-galactosidase, 1:2000 (Invitrogen); mouse anti-β-galactosidase, 1:500 (Sigma); rabbit anti-DMef2, 1:2000 (Lilly et al., 1995); rabbit anti-Nmr, 1:500 (Leal et al., 2009); mouse anti-Slit 1:500 (Hybridoma Bank, Univ. of Iowa); rabbit anti-Eve, 1:300 (Frasch et al., 1987).c-Fos 1:200 (Santa Cruz) and α-Actinin antibody 1:800 (Sigma) were used for immunohistochemistry of mouse heart sections.
Thoracic Aortic Banding Model
The protocol was approved by institutional guidelines (University of California, San Francisco). Mice (8 weeks old) were anesthetized with 2.4% isoflurane/97.6% oxygen and placed in a supine position on a heating pad (37°C). Animals were intubated with a 19G stump needle and ventilated with room air with a MiniVent Type 845 mouse ventilator (Hugo Sachs Elektronik-Harvard Apparatus, Germany; stroke volume 250 μl, respiratory rate 120 breaths per minute). Pressure overload was induced by TAB. A 3-mm left-sided thoracotomy was created at the second intercostal space. The transverse aortic arch was ligated (7-0 Prolene) between the innominate and left common carotid arteries with an overlying 27-gauge needle. The needle was removed, leaving a discrete region of stenosis. The chest was closed, and the left-sided pneumothorax was evacuated. Some mice were subjected to a sham operation in which the aortic arch was visualized but not banded. Peri-operative (24-hour) mortality was <10%.
Quantitative Real-Time PCR
For mouse q-PCR, RNA was extracted by TRizol method (Invitrogen) from individual hearts. q-PCR was performed using the Superscript III first-strand synthesis system (Invitrogen). q-PCR was performed using the ABI 7900HT (TaqMan, Applied Biosystems), per the manufacturer’s protocols. Optimized primers from Taqman Gene Expression Array were used. miRNA RT was conducted using the Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems). miRNA q-RT-PCR was performed per the manufacturer’s protocols by using primers from Taqman MicroRNA Assays (Applied Biosystems). Expression levels were normalized to GAPDH expression and U6 (microRNA q-PCR).
Western Blots
Western blots were performed as described (Zhao et al., 2005). Rabbit anti-Fos (Cell Signaling) and rabbit anti-GAPDH (Santa Cruz) were used at 1:1000 dilutions for western blots. All western blots were quantified using Alpha imager software from Alpha Innovations.
Statistics
Analysis of the rates of enhancement or suppression in the genomic screen were analyzed using the chi-squared test. Luciferase assays, q-PCR, and quantification of immunohistochemistry were analyzed using the one-way student t-test. Error bars are represented as standard error of the mean. Sample numbers were indicated in corresponding figures.
S2 Luciferase Assays
S2 cells were maintained at room temperature in Schneider’s Drosophila Medium (Gibco) supplemented with 10% FBS. Effectene Transfection Reagent (QIAGEN) was used for each transfection according to QIAGEN’s protocol. For each well of a twenty four-well plate, 200 ng of pGL3-target (e.g., kay) 3′UTR, pUAS-dmiR-1, pMT-GAL4 and 50 ng of Renilla were transfected, along with vector only as a negative control. Luciferase assays were performed as described with the Dual-Luciferase Reporter Assay System (Promega).
Supplementary Material
Acknowledgments
We thank E. Lai, G. Rubin, and D. Bohmann for the fly lines as noted in the text, and F. Biao for provision of selected fly stocks from the DrosDel collection. We thank I.S. Kathiriya for critically reviewing the manuscript, and K. Cordes for sharing unpublished results. We thank all the members from the Srivastava lab for helpful discussion. I.N.K. is supported by 5K08HL079260-4. L.Q. is a Scholar of the California Institute for Regenerative Medicine (CIRM) and is a Lynda and Stewart Resnick Fellow. D.S. was supported by grants from NHLBI/NIH, the CIRM, and the Younger Family Foundation. This work was supported by NIH/NCRR grant (C06 RR018928) to the Gladstone Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nature Medicine. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- Cavigelli M, Dolfi F, Claret FX, Karin M. Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. The EMBO Journal. 1995;14:5957–5964. doi: 10.1002/j.1460-2075.1995.tb00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L, Jackson DB, Mlodzik M, Bohmann D. Drosophila Fos mediates ERK and JNK signals via distinct phosphorylation sites. Genes Dev. 2001;15:1540–1553. doi: 10.1101/gad.886301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classen AK, Anderson KI, Marois E, Eaton S. Hexagonal packing of Drosophila wing epithelial cells by the planar cell polarity pathway. Dev Cell. 2005;9:805–817. doi: 10.1016/j.devcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Cooper MT, Bray SJ. Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature. 1999;397:526–530. doi: 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- Cordes KR, Srivastava D. MicroRNA regulation of cardiovascular development. Circ Res. 2009;104:724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, Pierce SA, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanto M, Mlodzik M. Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature. 1999;397:523–526. doi: 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- Fleischmann A, Hafezi F, Elliott C, Remé CE, Rüther U, Wagner EF. Fra-1 replaces c-Fos-dependent functions in mice. Genes Dev. 2000;14:2695–2700. doi: 10.1101/gad.187900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bellido A, Cortes F, Milan M. Cell interactions in the control of size in Drosophila wings. Proc Natl Acad Sci USA. 1994;91:10222–10226. doi: 10.1073/pnas.91.21.10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblet NS, Lijam N, Ruiz-Lozano P, Wang J, Yang Y, Luo Z, Mei L, Chien KR, Sussman DJ, Wynshaw-Boris A. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 2002;129:5827–5838. doi: 10.1242/dev.00164. [DOI] [PubMed] [Google Scholar]
- Han Z, Fujioka M, Su M, Liu M, Jaynes JB, Bodmer R. Transcriptional integration of competence modulated by mutual repression generates cell-type specificity within the cardiogenic mesoderm. Dev Biol. 2002;252:225–240. doi: 10.1006/dbio.2002.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Yi P, Li X, Olson EN. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development. 2006;133:1175–1182. doi: 10.1242/dev.02285. [DOI] [PubMed] [Google Scholar]
- Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated Calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumo S, Nadal-Ginard B, Mahdavi V. Protooncogene induction and reprogramming of cardiac gene expression produced by pressure overload. Proc Natl Acad Sci USA. 1988;85:339–343. doi: 10.1073/pnas.85.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro I, Kaida T, Shibazaki Y, Kurabayashi M, Katoh Y, Hoh E, Takaku F, Yazaki Y. Stretching cardiac myocytes stimulates protooncogene expression. J Biol Chem. 1990;265:3595–3598. [PubMed] [Google Scholar]
- Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci USA. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen TH, Skar R, Frotjold EK, Haukanes K, Greve G, Saetersdal T. Regional activation of the immediate-early response gene c-Fos in infarcted rat hearts. Int J Exp Pathol. 1998;79:163–172. doi: 10.1046/j.1365-2613.1998.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal SM, Qian L, Lacin H, Bodmer R, Skeath JB. Neuromancer1 and Neuromancer2 regulate cell fate specification in the developing embryonic CNS of Drosophila melanogaster. Dev Biol. 2009;325:138–150. doi: 10.1016/j.ydbio.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech Dev. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- MacMullin A, Jacobs JR. Slit coordinates cardiac morphogenesis in Drosophila. Dev Biol. 2006;293:154–164. doi: 10.1016/j.ydbio.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4:1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- Phillips HM, Hildreth V, Peat JD, Murdoch JN, Kobayashi K, Chaudhry B, Henderson DJ. Non-cell-autonomous roles for the planar cell polarity gene Vangl2 in development of the coronary circulation. Circ Res. 2008;102:615–623. doi: 10.1161/CIRCRESAHA.107.160861. [DOI] [PubMed] [Google Scholar]
- Phillips HM, Murdoch JN, Chaudhry B, Copp AJ, Henderson DJ. Vangl2 acts via RhoA signaling to regulate polarized cell movements during development of the proximal outflow tract. Circ Res. 2005;96:292–299. doi: 10.1161/01.RES.0000154912.08695.88. [DOI] [PubMed] [Google Scholar]
- Phillips HM, Rhee HJ, Murdoch JN, Hildreth V, Peat JD, Anderson RH, Copp AJ, Chaudhry B, Henderson DJ. Disruption of planar cell polarity signaling results in congenital heart defects and cardiomyopathy attributable to early cardiomyocyte disorganization. Circ Res. 2007;101:137–145. doi: 10.1161/CIRCRESAHA.106.142406. [DOI] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Slit and Robo Control Cardiac Cell Polarity and Morphogenesis. Curr Biol. 2005;15:2271–2278. doi: 10.1016/j.cub.2005.10.037. [DOI] [PubMed] [Google Scholar]
- Ramirez MT, Sah VP, Zhao XL, Hunter JJ, Chien KR, Brown JH. The MEKK-JNK pathway is stimulated by alpha1-adrenergic receptor and ras activation and is associated with in vitro and in vivo cardiac hypertrophy. J Biol Chem. 1997;272:14057–14061. doi: 10.1074/jbc.272.22.14057. [DOI] [PubMed] [Google Scholar]
- Riesgo-Escovar JR, Hafen E. Drosophila Jun kinase regulates expression of decapentaplegic via the ETS-domain protein Aop and the AP-1 transcription factor DJun during dorsal closure. Genes Dev. 1997;11:1717–1727. doi: 10.1101/gad.11.13.1717. [DOI] [PubMed] [Google Scholar]
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, et al. The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Martinez E, Soplop NH, Kramer SG. Lateral positioning at the dorsal midline: Slit and Roundabout receptors guide Drosophila heart cell migration. Proc Natl Acad Sci USA. 2006;103:12441–12446. doi: 10.1073/pnas.0605284103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circulation research. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- Schunkert H, Jahn L, Izumo S, Apstein CS, Lorell BH. Localization and regulation of c-Fos and c-Jun protooncogene induction by systolic wall stress in normal and hypertrophied rat hearts. Proc Natl Acad Sci USA. 1991;88:11480–11484. doi: 10.1073/pnas.88.24.11480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease: the sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT. A second canon. Functions and mechanisms of beta-catenin-independent Wnt signaling. Dev Cell. 2003;5:367–377. doi: 10.1016/s1534-5807(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Venkatesh TV, Park M, Ocorr K, Nemaceck J, Golden K, Wemple M, Bodmer R. Cardiac enhancer activity of the homeobox gene tinman depends on CREB consensus binding sites in Drosophila. Genesis. 2000;26:55–66. [PubMed] [Google Scholar]
- Wallingford JB, Fraser SE, Harland RM. Convergent extension: the molecular control of polarized cell movement during embryonic development. Dev Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- Wang Y, Su B, Sah VP, Brown JH, Han J, Chien KR. Cardiac hypertrophy induced by mitogen-activated protein kinase kinase 7, a specific activator for c-Jun NH2-terminal kinase in ventricular muscle cells. J Biol Chem. 1998;273:5423–5426. doi: 10.1074/jbc.273.10.5423. [DOI] [PubMed] [Google Scholar]
- Weber U, Paricio N, Mlodzik M. Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development. 2000;127:3619–3629. doi: 10.1242/dev.127.16.3619. [DOI] [PubMed] [Google Scholar]
- Weber U, Pataki C, Mihaly J, Mlodzik M. Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev Biol. 2008;316:110–123. doi: 10.1016/j.ydbio.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- Zallen JA. Planar polarity and tissue morphogenesis. Cell. 2007;129:1051–1063. doi: 10.1016/j.cell.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Kockel L, Peverali FA, Jackson DB, Mlodzik M, Bohmann D. Defective dorsal closure and loss of epidermal decapentaplegic expression in Drosophila fos mutants. EMBO J. 1997;16:7393–7401. doi: 10.1093/emboj/16.24.7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.