Abstract
In an EMS screen for mutations disrupting tracheal development, we identified new alleles of the dalmation (dmt) gene, which had previously been shown to affect peripheral nervous system (PNS) development. Here, we demonstrate that dmt loss results in programmed cell death, disrupting PNS patterning and leading to large gaps in the salivary duct and trachea. Dmt loss results in increased expression of the proapoptotic regulator genes head involution defective (hid) and reaper (rpr), and deletion of these genes or tissue-specific expression of the baculoviral apoptotic inhibitor P35 rescues the dmt defects. dmt is also required to protect cells from irradiation induced expression of hid and rpr during the irradiation resistant stage, which begins as cells become irreversibly committed to their final fates. Thus, we propose that Dmt keeps cells alive by blocking activation of hid and rpr as cells become irreversibly committed.
Keywords: apoptosis, dalmation, heterochromatin, HP1, peripheral nervous system, salivary duct, trachea
INTRODUCTION
From the variety of secretory glands to the vasculature, lungs, and kidneys, tubular organs are indispensable for the viability of multicellular organisms. The Drosophila trachea and salivary gland have proven to be excellent genetically tractable systems for revealing the molecules and mechanisms underlying embryonic tube formation (Kerman et al., 2006). The Drosophila trachea is a network of interconnected tubes that facilitates transport of respiratory gases throughout the embryo. The tracheal primordia are first visible as 10 placodes (or plates) of approximately 40 cells each on both sides of the embryo (Fig. 1A). The primordia invaginate into the underlying mesoderm through a combination of apical cell constriction, cell intercalation and a final radially oriented mitotic division (Isaac and Andrew, 1996; Llimargas and Casanova, 1999; Nishimura et al., 2007). From each internalized tracheal sac, subsets of cells bud and migrate in stereotypical directions to give rise to the six primary tracheal branches (Fig. 1A–C), including the dorsal trunk (DT), which forms the major artery of the trachea along the anterior–posterior (AP) axis, the dorsal branch (DB), which migrates dorsally to oxygenate the heart and dorsal epidermis, the lateral trunk (LT), a more laterally positioned and thinner branch that also connects along the AP axis, the ganglionic branch (GB), which migrates ventrally to oxygenate the central nervous system (CNS), the visceral branch (VB), which oxygenates the viscera, and, finally, the transverse connective (TC), which connects the branches within each segment along the dorsal–ventral (DV) axis.
Fig. 1.
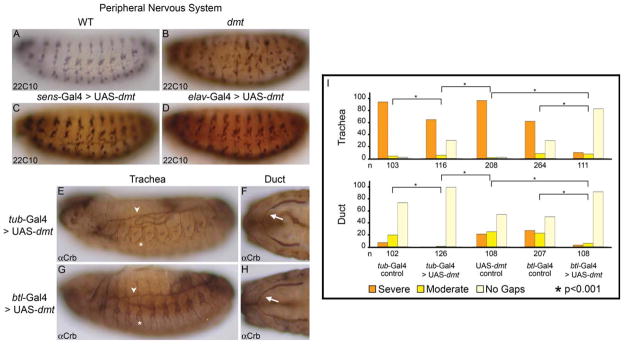
The tracheal dorsal trunk (DT) and salivary ducts of dmtI184/aul (=dmt3999) mutants have frequent gaps. A–C: Tracheal morphogenesis in wild-type embryos is shown. Arrowheads indicate tracheal metamere 4. Inset in middle panel shows different tracheal branches. D–F: Duct morphogenesis in wild-type embryos. Arrows indicate regions where tubes are still closing. G–N: dmtI184/aul3999 mutant trachea and salivary duct have large gaps. Arrowheads indicate gaps in tracheal DT lumena, asterisks indicate shortened tracheal branches and arrows indicate where gaps are present in dmt/aul mutant salivary ducts. All embryos are oriented with anterior to the left. Tracheae are shown in lateral views, salivary ducts in ventral views. O: Tracheal DT and salivary duct phenotypes were quantified either as severe (multiple and/or large gaps), moderate (single small gaps), or no gaps. P: aul mutations were shown to be allelic to dmt through a series of complementation crosses using deficiencies and existing dmt alleles (see also Table 1) and recombination mapping using two nearby P-element insertions, P1 and P2, marked with white+.
Final tracheal differentiation includes the cell rearrangements that allow fusion of the DT and LT with their anterior and posterior neighbors, and the dorsal fusion of the DBs with their corresponding DBs on the opposite side of the embryo (Tanaka-Matakatsu et al., 1996; Uemura et al., 1996; Lee and Kolodziej, 2002; Tanaka et al., 2004). Cells in several branches, including the DB, GB, and VB will slide past one another and remodel apical junctions to allow for the formation of more elongated auto-cellular tubes (Jazwinska et al., 2003; Ribeiro et al., 2004). Finally, terminal cells of many branches will elaborate tracheoles, subcellular lumena that facilitate oxygen delivery to cellular targets (Affolter et al., 1994; Guille-min et al., 1996; Hacohen et al., 1998).
Despite the intricacies of tracheal morphogenesis, tracheal development can proceed entirely normally with only half the number of cells (Beitel and Krasnow, 2000). In experiments where the final round of cell division, which occurs as the primordia are internalizing, is blocked by cyclin A mutations, the trachea form with apparently normal architecture, including having completely intact DTs of normal length and bore size. Moreover, clonal analysis of the early trachea reveals that the ultimate fate of individual cells depends on their relative position in the primordia (and access to signals of varying intensities) and is not regulated by lineage (Samakovlis et al., 1996). Thus, Drosophila organ development appears to be regulative, at least during early stages. Nonetheless, because embryonic tracheal cells have ceased normal divisions by the time the tracheal sac is formed, it is unclear how well cell loss at later stages in tracheal morphogenesis might be tolerated.
The salivary ducts are morphologically similar to trachea, express a subset of the same genes and share a requirement for the transcription factor Trachealess (Trh) for their formation (Isaac and Andrew, 1996). Nonetheless, salivary ducts form by seemingly different mechanisms. The fully formed duct is a Y-shaped tube comprising two narrower posterior tubes, the individual ducts (IDs), which connect to the secretory tubes of the salivary gland, and a single anterior tube, the common duct (CD), which connects IDs to the digestive system (Fig. 1D–F). The duct forms from a contiguous primordium of ~55 cells positioned ventrally to the cells that form the paired secretory tubes of the salivary gland (Fig. 1D). The individual duct cells invaginate following internalization of salivary gland secretory cells. The ducts form by a “wrapping” mechanism, wherein the already polarized sheet of cells curls until its edges meet and seal to form an enclosed tube. Wrapping begins in the individual duct primordia as the last of the secretory cells internalize and continues until the individual duct tubes close at the ventral midline (Fig. 1D,E). Subsequently, the common duct forms a larger bore tube, also by wrapping and beginning with cells attached to both individual duct tubes (Fig. 1F). Although the duct tubes never lose apical–basal polarity, the cells undergo considerable cell rearrangements and shape changes to form this elongated Y-shaped structure (B.E.K. and D.J.A., unpublished observations).
To identify new mutations affecting both tracheal and salivary duct morphogenesis and thus likely to have a more general role in tube formation, we screened EMS mutations known to affect tracheal development (Myat et al., 2005) for those also affecting salivary duct formation. Among the mutants identified through this screen was andúril (aul), loss of which results in large gaps in both the trachea and salivary duct. Mapping experiments revealed that aul is allelic to dalmatian (dmt), a gene previously known for its effects on the peripheral nervous system (PNS; Salzberg et al., 1994, 1997), which is disorganized and has fewer cells in dmt mutants.17,18 We show that dmt encodes a novel nuclear protein that localizes to the heterochromatin and functions tissue autonomously. The gaps in the trachea and salivary ducts, as well as loss of peripheral neurons, are due to programmed cell death. Expression of the apoptotic activators reaper (rpr) and head involution defective (hid) are up-regulated in dmt mutants, consistent with our ability to rescue the dmt phenotypes with either a deficiency that deletes these genes or through tissue-specific expression of an apoptotic inhibitor. Finally, we show that Dmt is required to block hid and rpr expression in response to γ-irradiation in later stage, more differentiated, embryos.
RESULTS
anduril/dalmation Mutants Have Gaps in the Trachea and Salivary Duct
In an F4 EMS screen for mutations affecting embryonic trachea and/or salivary gland morphology (Myat et al., 2005), we identified two alleles of an essential gene that disrupted both the trachea and salivary duct. Homozygous mutant embryos exhibited frequent breaks and/or gaps in the DT, often associated with shorter ventral tracheal branches (Fig. 1G,I,O; Table 1). Related defects were seen in mutant salivary ducts, where the individual ducts often had significant gaps or were completely missing (Fig. 1H,J,O; Table 1). Based on these phenotypes, we named the corresponding gene anduril (aul), for the “sword that was broken” (Tolkien, 1954). Staining the same mutants with nuclear markers (αTgo for trachea and αDri for salivary duct) revealed that the breaks in the tracheal and duct lumena corresponded to regions where the cells appeared to be missing (Fig. 1K–N).
TABLE 1.
Phenotypic Analysis of dmt Alleles, Allelic Combinations, and Various Recombinant Lines Used in the Study
| Salivary Duct1 |
Trachea2 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gap(s) in ID |
No gaps | n | Gap(s) in DT |
No Gaps | n | |||
| Severe | Moderate | Severe | Moderate | |||||
| wt | 0% | 0% | 100% | 102 | 0.9% | 14.2% | 84.9% | 106 |
| dmt589 | 0% | 6.6% | 93.4% | 106 | 37.9% | 3.9% | 58.3% | 103 |
| dmt589/dmt3999 | 11.9% | 21.1% | 67.0% | 109 | 77.4% | 1.9% | 20.8% | 106 |
| dmt589/dmtI184 | 10.5% | 17.1% | 72.4% | 105 | 27.0% | 9.0% | 64.0% | 111 |
| dmt589/DfGB104 | 0.9% | 6.6% | 92.5% | 106 | 32.5% | 8.8% | 58.8% | 114 |
| dmt3999 | 25.2% | 18.7% | 56.1% | 123 | 96.3% | 1.9% | 1.9% | 107 |
| dmtI184 | 31.9% | 26.9% | 41.2% | 119 | 35.8% | 14.2% | 50% | 120 |
| dmt3999/dmtI184 | 31.4% | 18.6% | 50.0% | 102 | 57.5% | 6.6% | 35.8% | 106 |
| dmt3999/DfGB104 | 16.8% | 25.2% | 58.0% | 143 | 68.9% | 5.8% | 25.2% | 103 |
| dmtI184/DfGB104 | 19.6% | 19.6% | 60.8% | 102 | 36.7% | 8.3% | 55.0% | 109 |
| DfGB104 | 32.7% | 20.9% | 46.4% | 110 | 97.1% | 1.0% | 1.9% | 103 |
| DfH99, dmtI184 | 5.3% | 15.9% | 78.8% | 113 | 15.6% | 10.8% | 73.5% | 102 |
| dmt3999, btl-GAL43/dmt3999 | 27.1% | 22.7% | 50.2% | 207 | 62.1% | 8.0% | 29.9% | 264 |
| dmt3999, UAS-dmt4/dmt3999 | 21.3% | 25.0% | 53.7% | 108 | 96.6% | 1.4% | 1.9% | 208 |
| UAS-P357; dmt3999/dmt3999, btl-GAL43 | 1.9% | 11.1% | 87.0% | 108 | 29.3% | 13.8% | 56.9% | 123 |
| dmt3999, UAS-dmt4/dmt3999, btl-GAL43 | 2.8% | 5.6% | 91.7% | 108 | 9.9% | 7.2% | 82.9% | 111 |
| dmt3999, tub-GAL45/dmt3999 | 6.9% | 19.6% | 73.5% | 102 | 94.2% | 3.9% | 1.9% | 103 |
| dmt3999, UAS-dmt4/dmt3999, tub-GAL45 | 0.0% | 0.8% | 99.2% | 126 | 64.7% | 5.2% | 30.2% | 116 |
| sens-GAL47; dmt3999/dmt3999, UAS-dmt4 | 6.9% | 8.8% | 84.3% | 102 | 95.1% | 2.0% | 2.9% | 102 |
| elav-GAL48;; dmt3999/dmt3999, UAS-dmt4 | 8.4% | 0.9% | 90.6% | 107 | 97.1% | 1.0% | 1.9% | 103 |
| btl-GAL4 > UAS-hid6 | 11.4% | 26.7% | 61.9% | 105 | 100% | 0% | 0% | 122 |
| btl-GAL4 > UAS-grim7 | 4.0% | 36.6% | 59.4% | 101 | 100% | 0% | 0% | 104 |
| btl-GAL4 > UAS-rpr7 | 0% | 0% | 100% | 100 | 94.2% | 3.8% | 1.9% | 104 |
| btl-GAL4 | 1.0% | 1.0% | 98% | 104 | 10.9% | 8.2% | 80.9% | 110 |
| UAS-grim7 | 0% | 0% | 100% | 104 | 5.7% | 1.9% | 92.5% | 106 |
| UAS-rpr7 | 0% | 0% | 100% | 104 | 5.0% | 5.0% | 90% | 100 |
| UAS-hid6 | 1% | 0% | 100% | 106 | 17.9% | 5.4% | 76.8% | 112 |
The phenotypes were quantified either as severe (missing whole ID and/or gaps in both IDs), moderate (a single gap in one ID), or normal (no gaps).
The phenotypes were quantified either as severe (multiple and/or large gaps), moderate (single small gaps), or normal (no gaps).
dmt3999, btl-GAL4 is one recombinant line of btl-GAL4 on third chromosome with dmt3999.
dmt3999, UAS-dmt collectively refers to two different recombinant lines of UAS-dmt on third chromosome with dmt3999 and to one recombinant line of UAS-GFP-dmt on second chromosome with dmt3999. We saw no differences in the percent rescue with the tagged versus untagged versions of UAS-dmt.
dmt3999, tub-GAL4 collectively refers to two different recombinant lines of tub-GAL4 on third chromosome with dmt3999.
UAS-hid refers to two different insertion lines of UAS-hid one on X chromosome and one on the second chromosome.
All four insertions are on the second chromosome.
elav-GAL4 insertion is on the X chromosome.
By complementation tests with deficiencies in cytological region 85E (Fig. 1P; Supp. Table S2, which is available online), the possible candidate gene disrupted in aul mutants was narrowed to approximately 30 genes (horizontally shaded region in Fig. 1P; Supp. Table S1). Meiotic recombination using two closely linked and molecularly mapped P-element insertions marked with the white+ gene (Zhai et al., 2003) with the aid of an ebony (e) mutation on the aul chromosome revealed that aul maps between the two insertions (P1, aul e recombinant 1:7116, P2, aul recombinant 2:6346; vertical shaded region in Fig. 1P). Available mutant alleles for the genes known to map to the same interval as aul were used in complementation crosses with both aul3999 and aul589 alleles. All three tested alleles of dmt failed to complement aul3999 and aul589 (Supp. Table S2), including dmtI184, a chemically induced allele (Salzberg et al., 1994). dmtI184, either homozygous or in trans to aul mutations, led to the same defects in the trachea and salivary duct as observed in aul homozygotes (Table 1). Correspondingly, aul3999, either homozygous or in trans to dmtI184, led to defects in the PNS similar to those previously described for dmt mutations (Fig. 4A,B, data not shown and Salzberg et al., 1994, 1997). Importantly, both aul alleles, as well as dmtI184, had stop codons introduced early in the open reading frame (ORF) of dmt, resulting in significantly truncated proteins (Fig. 2A). Importantly, both tissue-specific and global expression of either green fluorescent protein (GFP) -tagged or untagged dmt rescued the trachea, salivary duct, and PNS defects (see below; Fig. 4C–I; Table 1). We conclude that aul is allelic to dmt and renamed aul3999 and aul589 as dmt3999 and dmt589, respectively.
Fig. 4.
dmt acts tissue autonomously. Global (tub-Gal4) or appropriate tissue-specific (sens-Gal4, elav-Gal4, btl-Gal4) expression of wild-type dmt can rescue the dmt defects. A–D: PNS defects are rescued with PNS expression of dmt. E–H: Global as well as trachea and duct specific expression of dmt using btl-Gal4 rescues defects in the duct and trachea. Refer to Figure 1 for images of the dmt loss-of-function phenotypes in these tissues. I: Quantification of the defects reveals that significant rescue of the dmt tracheal and duct defects is achieved with either global or tissue-specific expression of wild-type dmt constructs.
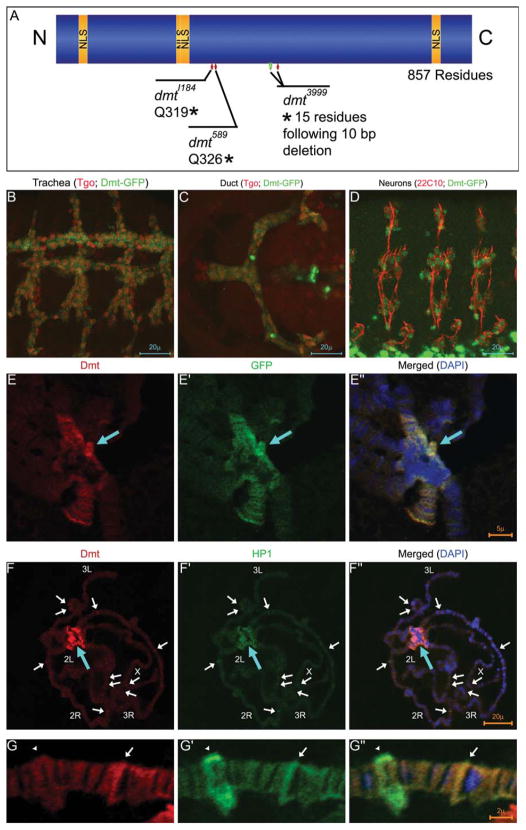
Fig. 2.
Dmt is a novel heterochromatin associated protein. A: A schematic representation of the Dmt protein is shown with the four predicted nuclear localization signals (NLS, gold). The EMS alleles dmtI184 and dmt589 have early stop codons (*) and dmt3999 has a 10 bp deletion that results in a frame shift, introducing 15 novel residues at the C-terminus followed by a stop codon (*). B–D: N-terminal green fluorescent protein (GFP) -tagged Dmt constructs were expressed in specific cell types using btl-Gal4 for the trachea and duct and elav-Gal4 for peripheral neurons. Dmt-GFP localized to nuclei and cytoplasm of trachea and duct and to nuclei of peripheral neurons. E: Dmt-GFP localized strongly to heterochromatic regions (blue arrow) when expressed in the secretory cells of the salivary gland using sage-Gal4. α-Dmt is shown in red and α-GFP in green. F,G: Heterochromatic localization of Dmt is confirmed by colocalization of Dmt (red) and HP1 (green). Dmt also localized to distinct bands along the chromosome arms, some of which overlap with HP1 localization (white arrows in upper panels). A higher magnification view of polytene chromosome shows Dmt and HP1 colocalization (white arrow in lower panels) and a band occupied by HP1 but not Dmt (arrowhead in lower panels).
dmt Encodes a Novel Nuclear Heterochromatin Protein
The dmt ORF encodes a novel protein of 857 residues. It has four predicted nuclear localization signals (NLSs), but no other domains can be identified and no obvious homologues exist outside the Drosophila group (Fig. 2A). The NLS motifs appear to be functional because both N- and C-terminally GFP-tagged Dmt proteins (Dmt-GFP) localized to nuclei when expressed in the trachea, salivary duct, or PNS (Fig. 2B–D). Dmt-GFP was also detected in the cytoplasm of trachea and duct cells and may be due to overexpression of the Dmt-GFP protein in these tissues. Dmt was detected on polytene chromosomes from salivary gland secretory cells induced to express Dmt-GFP, using antibodies directed against either Dmt or GFP (Fig. 2E). Immunostaining for Dmt and GFP colocalized to the chromocenter—the heterochromatic regions—of polytene chromosomes (Fig. 2E). This staining was not detected in control chromosomes from wild-type larvae, which do not express dmt in the secretory cells of the salivary gland (data not shown). Moreover, preimmune serum from the animal in which the antibody against Dmt was raised also did not stain the polytene chromosomes (data not shown), further demonstrating that staining is specific to Dmt. This localization is consistent with the localization of GFP-tagged Dmt to chromatin reported (in data not shown) by others (Goshima et al., 2007).
To further investigate the localization of Dmt, we stained the Dmt-expressing polytene chromosomes with an antibody against Heterochromatin-associated Protein-1 (HP1) and with αDmt, simultaneously. The antisera colocalized to the centromeric heterochromatin as well as to overlapping bands along the euchromatic chromosome arms (Fig. 2F,G). HP1 is a nonhistone chromosomal protein that plays roles in heterochromatin formation, gene silencing, telomere capping, and, as recently shown, control of gene expression (Fanti and Pimpinelli, 2008). HP1, when bound to euchromatic regions, can spread along the chromosome and silence gene expression (Danzer and Wallrath, 2004). Both heterochromatin formation and epigenetic gene silencing are achieved at least in part by interactions between HP1, histone methyltransferase, and histone H3 methylated at Lys 9 (Fanti and Pimpinelli, 2008). The colocalization of Dmt with HP1 to the heterochromatin suggests that Dmt and HP1 may have similar or overlapping functions.
dmt Is Expressed to High Levels in the Nervous System
Mutations in dmt lead to disorganization of the PNS, fewer PNS neurons, and the presence of small round cells in the ectoderm that stain darkly with the neuronal marker 22C10 (Salzberg et al., 1994; Fig. 4A,B). Consistent with the PNS phenotypes that have been described, dmt mRNA was detected to high levels in the PNS (Fig. 3). dmt was also detected to high levels in the CNS and in the posterior spiracle (Fig. 3). Dmt RNA levels were notably lower in the salivary gland secretory cells, which are visible on the embryo surface at embryonic stage 10. Surprisingly, despite the highly penetrant defects in the trachea and salivary duct, dmt transcript levels in both tissues were relatively low (Fig. 3). This finding suggested either that dmt acts nonautonomously in these tissues or that the low level of dmt observed in these tissues is sufficient.
Fig. 3.
dmt is robustly expressed in the nervous system. A–L: An RNA antisense probe to dmt (blue; left panels) or to both dmt (blue) and trh (pink; right panels) reveals high level expression in the peripheral nervous system (PNS) and central nervous system (CNS), but not trachea or salivary gland or duct (white arrows). A,B,G,H: The dmt transcripts were detected at high levels in most cells of stage 10 embryos, with reduced levels in the salivary gland secretory cells and in cells that express trh. C,D,I,J: The dmt transcripts were detected to high levels in the peripheral neuron precursors (black arrowhead) and ventral nerve cord (white asterisks) of late stage 12 embryos; however, dmt expression is low in the trh-expressing tracheal cells. E,F,K,L: The dmt transcript levels remain high in the peripheral neurons (white arrowheads) and CNS (asterisks) in embryonic stages 13 and 14, whereas dmt levels remain low in the trh expressing cells of the trachea (arrowheads in E,F) and duct (arrows in K,L). Note that, although the dmt levels in the trachea and duct are lower than in the CNS and PNS, the levels are higher than in the yolk.
dmt Acts Tissue Autonomously
To learn about the tissue-specific requirements for dmt function, the Gal4-UAS expression system (Brand and Perrimon, 1993) was used to express full-length untagged Dmt or Dmt-GFP either in the whole embryo or in specific tissues. Expression of dmt in the PNS using sens-Gal4 or elav-Gal4 rescued the PNS defects (Fig. 4C,D); however, gaps were still observed in the trachea (Supp. Fig. S1A,C; Table 1), arguing against a nonautonomous role for Dmt in the PNS causing the tracheal defects. sens-Gal4 and elav-Gal4 driven expression of UAS-dmt also rescued the salivary duct defects (Supp. Fig. S1B,D; Table 1) but, because both drivers are expressed to low levels in the salivary duct (data not shown), this finding also does not support a nonautonomous role for Dmt. On the other hand, global expression of dmt using a tub-Gal4 driver rescued the defects in the trachea and salivary duct (Fig. 4E,F,I; Table 1) and, consistent with a tissue autonomous function for dmt, trachea- and duct-specific expression of dmt using btl-Gal4 rescued the defects in both tissues (Fig. 4G–I; Table 1). Therefore, Dmt is required tissue autonomously in the PNS, trachea, and salivary duct for their proper organization.
dmt Mutants Show Increased Levels of Apoptosis
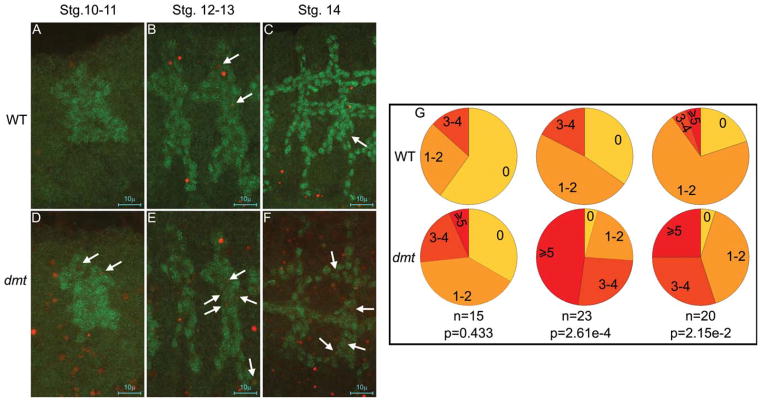
The gaps observed in both the dorsal trunk and individual salivary ducts, the shorter ventral tracheal branches (Fig. 1G–N), as well as the decrease in peripheral neurons (Fig. 4A,B) in dmt mutants could be explained by cell loss through apoptosis. Consistent with this possibility, TUNEL (terminal deoxynucleotidyl transferase–mediated deoxyuridinetriphosphate nick end-labeling) staining revealed a general increase in cell death in dmt mutant embryos relative to wild-type (Fig. 5). The number of apoptotic cells in the developing trachea in dmt mutants and wild-type embryos at different embryonic stages was determined. During early stages of development, subsequent to invagination but before primary branch migration when tracheal cells are still relatively plastic in their fate and organization (embryonic stages 10 and 11), we observed a slight (but not statistically significant) increase in the number of apoptotic tracheal cells in dmt mutants relative to wild-type (Fig. 5A,D,G, left panels). During stages 12 and 13, when the primary branches of the trachea form and cells are more committed in their fate and spatial organization, the number of apoptotic cells per trachea increased significantly in dmt mutants (Fig. 5B,E,G, middle panels). A significantly increased level of apoptosis in the trachea was also observed at later stages, subsequent to dorsal trunk fusion (Fig. 5C,F,G, right panels). In conclusion, there was an increase in the number of apoptotic cells in the whole embryo in dmt mutants with a significant increase in tracheal cell apoptosis at later stages.
Fig. 5.
More cells undergo apoptosis in dmt mutants. A–F: Apoptag (TUNEL [terminal deoxynucleotidyl transferase–mediated deoxyuridinetri-phosphate nick end-labeling]; red) and αTgo (green) staining reveals a general increase in cell death as well as an increase in tracheal-specific cell death at multiple stages in dmt mutants compared with wild-type embryos. G: The number of apoptotic cells in tracheal metamere 4 was counted in each embryo and quantified as percentages of metameres falling into each group: no apoptosis (0), 1–2, 3–4, and 5 or more cells undergoing apoptosis.
Inhibition of Apoptosis Rescues dmt Phenotype
To learn the relative contribution of programmed cell death to the dmt phenotypes, we asked if blocking apoptosis could restore normal PNS, tracheal, and duct morphology. In Drosophila, the proapoptotic proteins Hid, Grim, and Rpr function as gatekeepers regulating programmed cell death by antagonizing the Inhibitor of Apoptosis Protein (IAP; Steller, 2008). Thus, apoptosis can be inhibited in the whole embryo using Df(3L)H99, a deficiency that deletes hid, grim, and rpr. In Df(3L)H99 and dmt recombinants, the number of dorsal trunks and individual ducts with gaps was significantly reduced, the ventral tracheal branches were longer, and the PNS was more organized (Fig. 6A,B,E,F; Table 1). Consistent with the tissue-autonomous role of dmt, when apoptosis was inhibited specifically in the trachea and salivary duct by expression of the baculoviral anti-apoptotic protein P35 under the control of btl-Gal4, the defects in these tissues were also largely rescued (Fig. 6C,D,F; Table 1).
Fig. 6.
Repression of proapoptotic genes by Dmt is required to keep cells alive. A,B,E: Global inhibition of apoptosis by Df(3L)H99 rescued tracheal (A), duct (B), and peripheral nervous system (PNS; E) defects. C,D: Tissue specific inhibition of apoptosis through expression of P35 rescued trachea (C) and duct (D) defects. F: Quantification of the defects reveals significant rescue of both tracheal and duct defects. The phenotypes were quantified as described in Figure 1. G–L: Ectopic expression of three proapoptotic genes—hid (G,H), rpr (I,J), or grim (K,L)—mimicked dmt phenotypes in trachea and duct. Germ-band retraction failure of hid-expressing embryos (top left panel) is possibly related to the expression of btl-Gal4 in midline cells. M: The ectopic expression phenotypes were quantified as described in Figure 1.
Next, we asked if expression of the proapoptotic genes could, on its own, induce a dmt phenotype. Indeed, expression of hid, grim, or rpr under the control of btl-Gal4 caused gaps of varying severity in the dorsal trunk (Fig. 6G,I,K,M). hid and grim expression led to the most severe defects, with gaps in the dorsal trunk as well as disintegration of some of the trachea (Fig. 6G,K,M). Expression of rpr also resulted in frequent gaps in the dorsal trunk (Fig. 6I,M). Similarly, expression of hid and grim, but not rpr, in the duct resulted in loss of one or both individual ducts (Fig. 6H,J,L,M). The differential effects of the proapoptotic genes in trachea versus duct may reflect real differences in the apoptotic potential of the proapoptotic genes in the two tissues or may simply be a consequence of differences in relative expression levels. The latter hypothesis is supported by two observations: 1) The duct defects were observed only when the experiments were carried out at higher temperatures, which are known to increase Gal4 activity. 2) Of the two hid expression lines used in these assays, the line that caused more severe tracheal defects also caused more frequent duct defects. Based on both the rescue experiments and the dmt phenocopies observed with tissue-specific expression of the proapoptotic genes, we conclude that the dmt mutant phenotypes in the PNS, trachea, and salivary duct are largely due to increased apoptosis in these tissues.
To learn if loss of dmt increased cell death through derepression of the proapoptotic genes, we assayed expression of hid, grim, and rpr in wild-type and dmt mutants. Indeed, whole-mount in situ hybridizations revealed that both hid and rpr mRNA levels were notably higher in dmt mutants than in wild-type at all stages (Fig. 7, columns 1,3), whereas grim mRNA levels were comparable (data not shown). We conclude that Dmt keeps embryonic cells alive by blocking expression of hid and rpr.
Fig. 7.
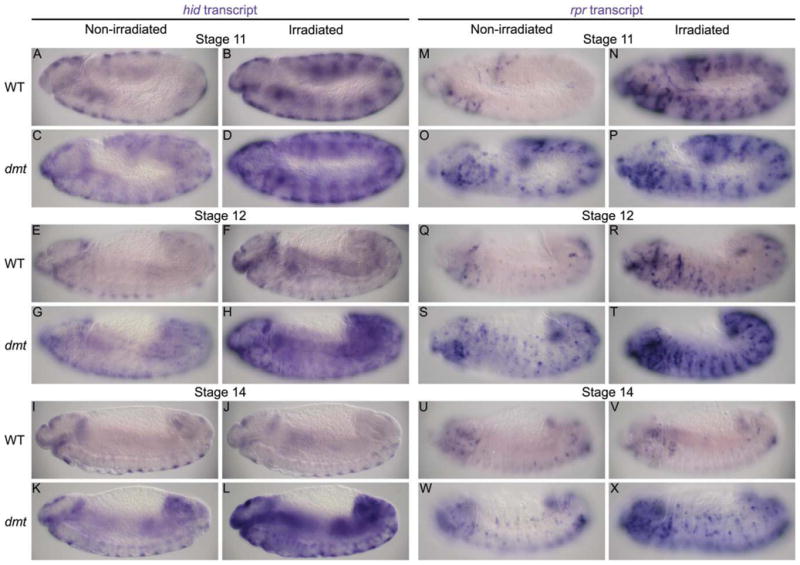
Levels of proapoptotic gene expression are increased throughout development in dmt mutants and fail to decrease in irradiated late stage dmt embryos. A–X: Transcript levels of the proapoptotic genes hid and rpr are significantly higher in dmt mutants (C,G,K,O,S,W) than in wild-type (A,E,I,M,Q,U) at all stages of embryonic development. At early stages, hid and rpr levels are higher in irradiated wild-type and dmt embryos (B,D,N,P) than in nonirradiated embryos (A,C,M,O). The transcript levels in irradiated wild-type embryos are lower and similar to nonirradiated levels at stage 12 (E,F,Q,R) and are comparable to nonirradiated levels at stage 14 (I,J,U,V). Transcript levels are significantly higher in irradiated dmt embryos (D,H,L,P,T,X) than nonirradiated embryos (C,G,K,O,S,W) at all stages. All panels are lateral views. Due to differences in expression patterns between hid and rpr, different focal planes of similarly staged embryos are shown.
Dmt Function and Irradiation Resistance in Older Embryos
Early Drosophila embryos are highly sensitive to radiation-induced apoptosis, whereas older, more differentiated, embryos become refractory to radiation-induced cell death. This transition in radiation sensitivity depends on epigenetic silencing (or blocking) of a cis-acting irradiation-responsive enhancer region (IRER) that regulates expression of both hid and rpr (Zhang et al., 2008). The IRER forms a heterochromatin-like structure during the transition from early stages, when rpr and hid are robustly activated in response to irradiation, to late stages, when the two genes become transcriptionally refractory to irradiation (embryonic stage 12). During this transition, the IRER becomes resistant to DNaseI treatment and enriched for trimethylated histones, HP1, and Polycomb-group protein binding. The coincidence in timing of the increased tracheal cell death in dmt mutants, the corresponding increase in rpr and hid expression throughout the embryo, and the localization of Dmt to heterochromatin suggested that Dmt could play a role in blocking irradiation-induced proapoptotic gene expression in older embryos. To test this idea, we compared levels of rpr and hid expression in early and late wild-type and dmt mutant embryos exposed to death gene-inducing levels of γ-irradiation. Levels of hid and rpr transcripts were higher in irradiated dmt embryos than in nonirradiated dmt embryos throughout development (Fig. 7). At the early irradiation-sensitive stage (stage 10), both wild-type and dmt irradiated embryos showed elevated levels of hid and rpr expression (Fig. 7A–D,M–P). During the transition to resistance stage (stage 12), the levels of both transcripts in irradiated wild-type embryos were reduced and more comparable to levels in nonirradiated wild-type embryos (Fig. 7E,F,Q,R), whereas levels of hid and rpr remained quite high in irradiated dmt embryos (Fig. 7G,H,S,T). By the resistant stage (stage 14), when the hid and rpr levels were quite similar between irradiated and nonirradiated wild-type embryos (Fig. 7I,J,U,V), hid and rpr levels were significantly elevated in irradiated dmt embryos (Fig. 7K,LW,X). These findings reveal that Dmt is required to prevent hid and rpr expression in response to cytotoxic stimuli in older embryos.
DISCUSSION
Our phenotypic characterization of dmt defects reveals that a primary job of the Dmt nuclear protein is to keep cells alive. Defects in all of the tissues we have analyzed, that is, PNS, trachea, and salivary duct, are rescued by tissue-specific dmt expression, revealing that Dmt is required cell (tissue) autonomously. Dmt localizes to chromatin and appears to keep cells alive by keeping at least two genes encoding activators of programmed cell death, hid and rpr, transcriptionally silenced. Transcriptional repression of hid and rpr appears to be a major job of Dmt because simply removing these genes (and silencing them by their absence) can fully restore the tissues in which Dmt function is required. Indeed, our demonstration that tissue-specific expression of the death activators alone can phenocopy loss of dmt puts the nail in the coffin, so to speak. Importantly, loss of dmt sensitizes late embryos to irradiation-induced cell death by allowing expression of the death activators hid and rpr, emphasizing Dmt’s pivotal role in choosing life versus death in response to cytotoxic stimuli.
Deletion of all three Drosophila IAP antagonists hid, grim, and rpr inhibits almost all apoptosis in Drosophila (White et al., 1994), whereas ectopic expression of these genes leads to increased apoptosis (Steller, 2008). Thus, transcriptional control of these genes is a key step in controlling cell death and is used by a variety of apoptotic signals, including the moulting hormone ecdysone, cell stress, developmental signals, irradiation, and tissue-specific transcription factors (Steller, 2008). One such tissue-specific transcription factor is Fkh, which is required to keep salivary gland cells alive early in development (Myat and Andrew, 2000) and whose prolonged expression during early stages of metamorphosis controls the timing of larval salivary gland death (Cao et al., 2007). In the absence of fkh, the differentiating cells of the salivary gland express higher levels of hid and rpr and are lost by programmed cell death (Myat and Andrew, 2000). Similar to what has been shown for Dmt, deletion of the death activators in fkh mutants prevents this premature salivary gland cell death, although in the case of fkh, it does not rescue other aspects of tissue morphogenesis. Nonetheless, PNS, trachea and salivary duct cells are not alone in requiring active processes to stay alive, suggesting a general requirement for all differentiated embryonic cells to actively keep death at bay and to perhaps do so through the transcriptional silencing of the death genes. Indeed, it is worth noting that the common duct, which is relatively unaffected by the loss of dmt, expresses low levels of Fkh (D.J.A., unpublished observations). Perhaps this low level of Fkh expression protects the common duct progenitors from apoptosis in the absence of dmt function.
Given the large number of signals and transcription factors that regulate hid and rpr expression, it is not surprising that these genes have large regulatory regions composed of several enhancer and silencer regions (Steller, 2008; Zhang et al., 2008). These control regions not only have binding sites for known transcription factors, but have recently been shown to be under epigenetic control (Zhang et al., 2008). Silencing of an upstream enhancer element that controls both rpr and hid expression through heterochromatin formation limits transcription of these genes in response to irradiation (Zhang et al., 2008). Heterochromatization of the IRER occurs during the transition from early (irradiation sensitive) to late (irradiation resistant) stages. Several histone-modifying enzymes are required for this process because mutations in the corresponding genes delay the blocking of hid and rpr expression in response to irradiation. Here, we show that Dmt is also required to block hid and rpr activation in response to irradiation. Furthermore, our finding that Dmt localizes to heterochromatin suggests that Dmt could have a direct role in this process.
The timing of the transition in the cellular response to cytotoxic stimuli is very intriguing because it overlaps with the stage when many of the cells of the Drosophila embryo become irreversibly committed. The transition to committed fates is quite evident in the trachea, where it is known that if the number of tracheal cells is reduced by two-fold, or even doubled, at early stages when the cells are first internalizing, the trachea develops quite normally (Beitel and Krasnow, 2000); however, as we have shown, tracheal cell loss at later stages either by mutations in dmt or by expression of proapoptotic genes cannot be tolerated. Later tracheal cell loss results in significant gaps in the DT and shortened tracheal branches. Therefore, it is likely that epigenetic silencing of rpr and hid evolved to set a high barrier to the activation of death promoting genes in committed cells, whose loss would significantly impact organismal viability. Therefore, we propose that Dmt may be a key factor that binds to the IRER to initiate or promote its heterochromatization, limiting transcriptional activation of both rpr and hid, and thus preventing the death of the organism that would result from the loss of essential cells. Dmt localizes to heterochromatin and to several sites on the euchromatic arms, many of which are also bound by HP1 (Fig. 2). Dmt loss leads to increased levels of both hid and rpr expression (Fig. 7) and to increased rates of death that, in the trachea, where we have directly measured dying cells, becomes significantly higher during embryonic stage 12 (Fig. 5), coincident with when the development of resistance of hid and rpr transcriptional activation in response to irradiation. We propose that the timing of Dmt action and IRER heterochromatization is linked to the developmental stage when a large number of the cells become irreversibly committed to specific developmental programs and, consequently, indispensable for the survival of the organism.
Of interest, dmt was independently identified in two recent RNAi screens in S2 cells for genes affecting mitotic spindle association (Goshima et al., 2007; Somma et al., 2008). Down-regulation of dmt resulted in premature sister chromatid separation and subsequent defects in chromosome segregation. A role in cell division may explain both the enhanced expression of rpr and hid and cell loss observed at early stages in dmt mutants; this function of Dmt is, nonetheless, unlikely to play a major role in the apoptotic cell death observed in late stages of the trachea because these cells have ceased dividing several hours before significant increases in apoptosis are observed. Similar arguments can be made for the PNS and salivary duct. Nonetheless, our findings that Dmt both localizes to centromeric heterochromatin and regulates gene expression leaves open direct and indirect roles for Dmt in centromere attachment during metaphase of actively dividing cells, especially because key components of the mitotic machinery have been shown to be regulated by epigenetic mechanisms (Wen et al., 2008).
EXPERIMENTAL PROCEDURES
Fly Strains and Genetics
Flies were grown and maintained on standard cornmeal/agar medium. The fly lines used in this study and their sources are listed in Supp. Table S1. The UAS-Gal4 expression system (Brand and Perrimon, 1993) was used for tissue-specific expression of transcripts. The dmt ORF was PCR amplified from cDNA clone LD40216 and cloned into pTW, pTGW, or pTWG, UAS expression vectors containing the Gateway recombination cassette (Drosophila Genomics Resource Center) to produce UAS-dmt (untagged) and UAS-GFP-dmt (N-terminally tagged), and UAS-dmt-GFP (C-terminally tagged), respectively. Germline transformations were performed as described previously (Rubin and Spradling, 1983) using w1118 embryos as the DNA recipients. The statistical significance (P values) of the relative frequencies of defects in the different genotypes was calculated using an online chi-square test calculator (Preacher, 2001).
Sequence Analysis of dmt Mutations
To map the molecular lesions in the dmt EMS alleles, genomic DNA was isolated from adult flies heterozygous for the dmt alleles (dmtI184, dmt3999, dmt589) and, as a control, for an unrelated mutation generated in the same screen (pnt737; Myat et al., 2005) over the same balancer chromosome, TM3-Ubx-lacZ. Overlapping regions of dmt were amplified by PCR, sequenced and analyzed for allelic differences. The sequences of the PCR primers used to amplify the genomic DNA for this analysis as well as for the generation of UAS-Dmt lines and Dmt antiserum are available upon request.
Immunohistochemistry and In Situ Hybridization
Embryos were fixed and stained as described (Reuter et al., 1990) using the following antibodies: mouse Crb (1:100; Developmental Studies Hybridoma Bank [DHSB]; Iowa City, IA), mouse Tgo (1:10; DHSB), mouse 22C10 (1:50; DSHB), rat Dri (1:20,000; Haberman et al., 2003), rabbit GFP (1:40,000; Molecular Probes; Eugene, OR), mouse β-gal (1:5,000; Promega; Madison, WI). Apoptotic cells were identified by TUNEL staining performed as described previously (Reim and Frasch, 2005) using a mouse TGO antibody (1:2; DSHB) to mark tracheal cells. Appropriate secondary antibodies conjugated to Biotin (Vector Labs; Burlingame, CA) or to fluorescein isothiocyanate, rhodamine, or Alexa fluorophores (Molecular Probes; Carlsbad, CA) were used at a 1:500 dilution. Embryonic dmt, trh, hid, grim, and rpr mRNAs were detected by whole-mount in situ hybridization as described (Lehmann and Tautz, 1994). Embryos were visualized by Nomarski optics using a Zeiss Axiophot microscope or by confocal optics using a Zeiss Axiovert 200 microscope equipped with Zeiss 510 confocal module. Homozygous mutant embryos were identified by lack of βGal or GFP expression from the balancer chromosomes. Three-dimensional projection images of 1-μm-thick fluorescent images were generated using Zeiss LSM Software (Zeiss; Brighton, MI).
Embryo Irradiation
An overnight collection of wild-type, dmt3999, and dmtI184 embryos were treated with 27 Gy γ-ray on collection plates. After a 30-min recovery, irradiated and nonirradiated embryos were processed for in situ hybridization to detect hid, rpr, and grim mRNAs, as described above.
Generation of Dmt Antibody
A fragment of the dmt cDNA LD40216 encoding the N-terminal 386 residues of Dmt was PCR amplified and cloned into the pTrcHisB expression vector (Invitrogen; Carlsbad, CA). Protein was expressed in E. coli and purified in inclusion bodies (Rio et al., 1986). Rat polyclonal antiserum was generated against 1 mg of renatured inclusion body protein administered over several weeks according to company recommendations (Cocalico Biologicals; Reamstown, PA).
Polytene Chromosome Staining
Both untagged and N-terminally tagged UAS-Dmt constructs were expressed in larval salivary gland cells using a salivary gland-specific sage-Gal4 driver (A. Vaishnavi and D.J.A., unpublished observations). Oregon R larvae were used as a negative control for Dmt and GFP staining. Late third instar larval salivary gland chromosomes were prepared as described (Andrew and Scott, 1994), except that the second fixation was 50% glacial acetic acid. Dmt preimmune and immune antiserum was used at a dilution of 1:200, the mouse monoclonal GFP antibody (Molecular Probes; Carlsbad, CA) was used at a dilution of 1:1,000 and a rabbit HP1 polyclonal antibody (DHSB) was used at a dilution of 1:100. All secondary fluorescent antibodies (Molecular Probes; Carlsbad, CA) were used at a dilution of 1:200.
Supplementary Material
Acknowledgments
Grant sponsor: NIH; Grant number: RO1 DE012873.
We thank the Bloomington Stock Center, J. Nambu, M. Acar, and H. Bellen for fly lines and the Developmental Studies Hybridoma Bank for antibodies. We thank C. Hanlon for her help during the final stages of the project. We thank T. Murphy for providing the clones for Gateway cloning of both tagged and untagged Dmt constructs. We thank Y. Zhang for his assistance in the irradiation of fly embryos. We thank N. Lin and L. Zhou on helpful discussions on IRER. We thank P. Devreotes, D. Montell, and G. Seydoux for thoughtful contributions at various stages of this project. Finally, we thank S. Chung, R. Fox, C. Hanlon, and A. Ismat and two anonymous reviewers for critical reading of the manuscript. D.J.A. was funded by the NIH.
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Affolter M, Montagne J, Walldorf U, Groppe J, Kloter U, LaRosa M, Gehring WJ. The Drosophila SRF homolog is expressed in a subset of tracheal cells and maps within a genomic region required for tracheal development. Development. 1994;120:743–753. doi: 10.1242/dev.120.4.743. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Scott MP. Immunological methods for mapping protein distributions on polytene chromosomes. Methods Cell Biol. 1994;44:353–370. doi: 10.1016/s0091-679x(08)60923-1. [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Krasnow MA. Genetic control of epithelial tube size in the Drosophila tracheal system. Development. 2000;127:3271–3282. doi: 10.1242/dev.127.15.3271. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cao C, Liu Y, Lehmann M. Fork head controls the timing and tissue selectivity of steroid-induced developmental cell death. J Cell Biol. 2007;176:843–852. doi: 10.1083/jcb.200611155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer JR, Wallrath LL. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 2004;131:3571–3580. doi: 10.1242/dev.01223. [DOI] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Curr Opin Genet Dev. 2008;18:169–174. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin K, Groppe J, Ducker K, Treisman R, Hafen E, Affolter M, Krasnow MA. The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development. 1996;122:1353–1362. doi: 10.1242/dev.122.5.1353. [DOI] [PubMed] [Google Scholar]
- Haberman AS, Isaac DD, Andrew DJ. Specification of cell fates within the salivary gland primordium. Dev Biol. 2003;258:443–453. doi: 10.1016/s0012-1606(03)00140-4. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Isaac DD, Andrew DJ. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 1996;10:103–117. doi: 10.1101/gad.10.1.103. [DOI] [PubMed] [Google Scholar]
- Jazwinska A, Ribeiro C, Affolter M. Epithelial tube morphogenesis during Drosophila tracheal development requires Piopio, a luminal ZP protein. Nat Cell Biol. 2003;5:895–901. doi: 10.1038/ncb1049. [DOI] [PubMed] [Google Scholar]
- Kerman BE, Cheshire AM, Andrew DJ. From fate to function: the Drosophila trachea and salivary gland as models for tubulogenesis. Differentiation. 2006;74:326–348. doi: 10.1111/j.1432-0436.2006.00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kolodziej PA. The plakin Short Stop and the RhoA GTPase are required for E-cadherin-dependent apical surface remodeling during tracheal tube fusion. Development. 2002;129:1509–1520. doi: 10.1242/dev.129.6.1509. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA, editors. Drosophila melanogaster: practical uses in cell and molecular biology. Vol. 44. San Diego: Academic Press; 1994. p. 755. [Google Scholar]
- Llimargas M, Casanova J. EGF signalling regulates cell invagination as well as cell migration during formation of tracheal system in Drosophila. Dev Genes Evol. 1999;209:174–179. doi: 10.1007/s004270050241. [DOI] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ. Fork head prevents apoptosis and promotes cell shape change during formation of the Drosophila salivary glands. Development. 2000;127:4217–4226. doi: 10.1242/dev.127.19.4217. [DOI] [PubMed] [Google Scholar]
- Myat MM, Lightfoot H, Wang P, Andrew DJ. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev Biol. 2005;281:38–52. doi: 10.1016/j.ydbio.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Inoue Y, Hayashi S. A wave of EGFR signaling determines cell alignment and intercalation in the Drosophila tracheal placode. Development. 2007;134:4273–4282. doi: 10.1242/dev.010397. [DOI] [PubMed] [Google Scholar]
- Preacher KJ. Calculation for the chi-square test: An interactive calculation tool for chi-square tests of goodness of fit and independence. 2001 Available from http://www.quantpsy.org.
- Reim I, Frasch M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development. 2005;132:4911–4925. doi: 10.1242/dev.02077. [DOI] [PubMed] [Google Scholar]
- Reuter R, Panganiban GEF, Hoffmann FM, Scott MP. Homeotic genes regulate the spatial expression of putative growth factors in the visceral mesoderm of Drosophila embryos. Development. 1990;110:1031–1040. doi: 10.1242/dev.110.4.1031. [DOI] [PubMed] [Google Scholar]
- Ribeiro C, Neumann M, Affolter M. Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol. 2004;14:2197–2207. doi: 10.1016/j.cub.2004.11.056. [DOI] [PubMed] [Google Scholar]
- Rio DC, Laski FA, Rubin GM. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell. 1986;44:21–32. doi: 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Vectors for P element-mediated gene transfer in Drosophila. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg A, D’Evelyn D, Schulze KL, Lee JK, Strumpf D, Tsai L, Bellen HJ. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- Salzberg A, Prokopenko SN, He Y, Tsai P, Pal M, Maroy P, Glover DM, Deak P, Bellen HJ. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: mutations affecting embryonic PNS development. Genetics. 1997;147:1723–1741. doi: 10.1093/genetics/147.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samakovlis C, Hacohen N, Manning G, Sutherland DC, Guillemin K, Krasnow MA. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development. 1996;122:1395–1407. doi: 10.1242/dev.122.5.1395. [DOI] [PubMed] [Google Scholar]
- Somma MP, Ceprani F, Bucciarelli E, Naim V, De Arcangelis V, Piergentili R, Palena A, Ciapponi L, Giansanti MG, Pellacani C, Petrucci R, Censi G, Verni F, Fasulo B, Goldberg ML, Di Cunto F, Gatti M. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 2008;4:e1000126. doi: 10.1371/journal.pgen.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008;15:1132–1138. doi: 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, Hayashi S. Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development. 1996;122:3697–3705. doi: 10.1242/dev.122.12.3697. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Takasu E, Aigaki T, Kato K, Hayashi S, Nose A. Formin3 is required for assembly of the F-actin structure that mediates tracheal fusion in Drosophila. Dev Biol. 2004;274:413–425. doi: 10.1016/j.ydbio.2004.07.035. [DOI] [PubMed] [Google Scholar]
- Tolkien JRR. The lord of the rings. London: Allen and Unwin; 1954. [Google Scholar]
- Uemura T, Oda H, Kraut R, Hayashi S, Kotaoka Y, Takeichi M. Zygotic Drosophila E-cadherin expression is required for processes of dynamic epithelial cell rearrangement in the Drosophila embryo. Genes Dev. 1996;10:659–671. doi: 10.1101/gad.10.6.659. [DOI] [PubMed] [Google Scholar]
- Wen H, Andrejka L, Ashton J, Karess R, Lipsick JS. Epigenetic regulation of gene expression by Drosophila Myb and E2F2-RBF via the Myb-MuvB/dREAM complex. Genes Dev. 2008;22:601–614. doi: 10.1101/gad.1626308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila [see comments] Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- Zhai RG, Hiesinger PR, Koh TW, Verstreken P, Schulze KL, Cao Y, Jafar-Nejad H, Norga KK, Pan H, Bayat V, Greenbaum MP, Bellen HJ. Mapping Drosophila mutations with molecularly defined P element insertions. Proc Natl Acad Sci U S A. 2003;100:10860–10865. doi: 10.1073/pnas.1832753100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lin N, Carroll PM, Chan G, Guan B, Xiao H, Yao B, Wu SS, Zhou L. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev Cell. 2008;14:481–493. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.