Abstract
It is of significant biological interest and medical importance to develop class- and isoform-selective histone deacetylases (HDAC) modulators. The impact of the linker component on HDAC inhibition specificity was revealed, but has not been understood. Herein with Born-Oppenheimer ab initio QM/MM molecular dynamics simulations, a state-of-the-art approach to simulating metallo-enzymes, we have found that the hydroxamic acid remains to be protonated upon its binding to HDAC8, and thus disapproved the mechanistic hypothesis that the distinct zinc-hydroxamate chelation modes between two HDAC subclasses come from different protonation states of the hydroxamic acid. Instead, our simulations suggested a novel mechanism that the chelation mode of hydroxamate with the zinc ion in HDACs is modulated by water access to the linker binding channel. This new insight into the interplay between the linker binding and the zinc chelation emphasizes the importance and gives guidance regarding the linker design for the development of new class-IIa specific HDAC inhibitors.
Histone Deacetylases (HDACs) enzymes, that are responsible for the removal of acetyl groups from acetyl-lysine residues of histones and other cellular important proteins, are central to the regulation of many vital cellular functions.1-3 Inhibition of HDACs has emerged as a highly promising strategy for the development of new therapeutics against cancer and various other human disorders.4-8 A key challenge in HDAC inhibitor design is to control the class- and isoform-selective inhibition.9-13 Most HDAC inhibitors, including two recent FDA-approved anticancer drugs SAHA (suberoylanilide hydroxamic acid) and FK228, can be described by the cap-linker-chelator model.14 As illustrated in Figure 1a, the chelator refers to the zinc binding group, the linker part mimics the aliphatic part of the acetyl-lysine side chain spanning the binding channel, and the cap component interacts with the rim region of the active site cavity. Very recently, a novel chemical phylogenetic analysis9 indicated the linker-chelator motif as the principal component to cluster HDAC inhibitors, and revealed the impact of the linker component on HDAC inhibition selectivity, particularly for class IIa HDAC enzymes. However, no mechanism has been suggested regarding the interplay between the linker binding and the zinc chelation.
Figure 1.
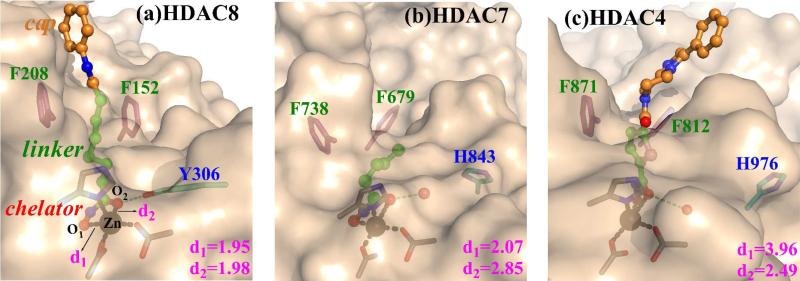
The active site of enzyme-inhibitor complex in HDAC8 (a), HDAC7 (b) and HDAC4 (c). For HDAC8, the inhibitor is SAHA, for HDAC7, it is a truncated-SAHA in the crystal structure, and for HDAC4, it is a SAHA-like hydroxamic Acid (HA). The conserved two phenylalanines are located at the entrance of the pocket. Y306 forms a hydrogen bond with SAHA in HDAC8 but it is replaced by a His in HDAC7/4, in which a crystal water is close enough to form the hydrogen bond with the inhibitor. The oxygen-zinc distances d1/d2 were measured from XRD structures.16,19,20
Among eleven known isoforms of zinc-dependent HDACs in human, structures of three HDAC isoforms in complex with hydroxamate inhibitors have been obtained, including HDAC8, a class I enzyme,15-18 and HDAC4 and HDAC7, two class IIa enzymes.19,20 As shown in Figure 1 and S1, in spite of having almost the same first zinc coordination shell, different hydroxamate-zinc coordination modes are observed: bidentate in HDAC8 (PDB ID 1T6916), monodentate in HDAC7 (PDB ID 3C0Z20), and weakly dentate with zinc in HDAC4 (PDBID 2VQM19). A key distinction between class-I and class-IIa HDACs is that an active site tyrosine residue (Y306 in HDAC8) in the active site, which is conserved in all class-I HDACs, is replaced by a histidine in class IIa enzymes. Experimental studies of HDAC4 and HDAC7 have shown that the mutation of this histidine to tyrosine would significantly increase enzyme activity as well as its binding to hydroxamate LAQ-824.9,19-21 Meanwhile, DFT calculations of zinc complexes have suggested that hydroxamic acid should be deprotonated upon its binding to the zinc ion, resulting the tight bidentate complexation. 22 Thus, a current hypothesis is that the zinc-hydroxamate chelation mode in HDACs is determined by the protonation state of the hydroxamic acid: it is deprotonated in HDAC8 due to the existence of Y306 and thus leading to the bidentate chelation.9,22 Another distinction between class-I and class-IIa HDACs is about different orientations of two conserved phenylalanines around the entrance of the binding pocket: they are bound to the linker component of SAHA with a “sandwich-like” configuration in HDAC8, while it is not the case in either HDAC7 or HDAC4, as illustrated in Figure 1 and S1. Experimental studies have found that mutation of these two corresponding Phe residues in HDAC1 led to an inactive enzyme.23 Thus, an intriguing question to ask is whether and how this distinct structural feature in the linker channel would affect the binding of HDAC inhibitors.
In order to elucidate the origin of the observed distinct zinc-hydroxamate chelation mode between class-I and class-IIa HDACs, we have employed Born-Oppenheimer ab initio QM/MM molecular dynamics,24-30 a state-of-the-art approach to simulating metallo-enzymes. It provides a first-principle description of interactions and dynamics of the zinc active site, while explicitly taking account of heterogeneous and fluctuating enzyme environment. The QM sub-system, including the zinc ion, the inhibitor, residues in the first coordination shell, and two histidines in the active site, was treated by B3LYP functional with Stuttgart ECP/basis set (SDD31) for the zinc atom and 6-31G* basis set for all other atoms. This level of QM treatment28,30,32-35 has been extensively tested and employed successfully to describe zinc coordination shell. The QM/MM boundaries were described by the pseudobond approach with the improved parameters.36 For each prepared enzyme system as well as umbrella sampling at each window along the reaction coordinate, 25 ps B3LYP(SDD,6-31G*) QM/MM MD simulations at 300 K were carried out with modified Q-Chem37 and Tinker38 programs.
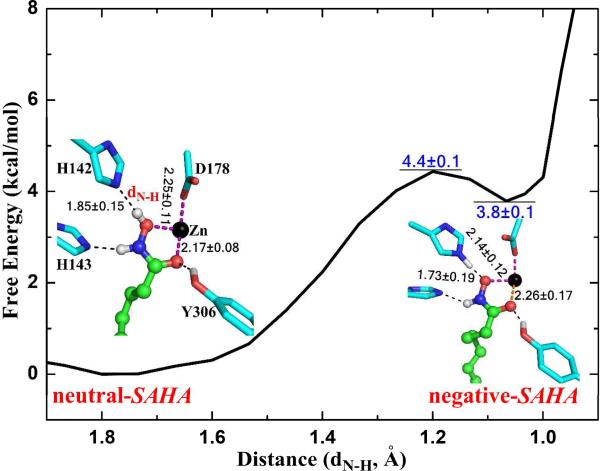
Our first task is to examine whether the hydroxamic acid is deprotonated by His142 upon its binding to HDAC8 which leads to its bidendate chelation mode.22 Here we employed ab initio QM/MM MD simulations with umbrella sampling to directly compute the free energy profile of this proton transfer process to determine the free energy difference between these two states, as shown in Figure 2. We can see that the deprotonated-SAHA state, in which the proton is transferred to His142, is about 3.8 kcal/mol less stable than the neutral-SAHA state. This indicates that the hydroxamic acid, which is neutral in the aqueous environment, remains to be protonated upon its binding to HDAC8 in spite of the existence of Y306. Moreover, we found that the zinc-hydroxamate chelation mode in HDAC8 is not dependent on the protonation state of SAHA, which is bidendate in both states as illustrated in Figure 2.
Figure 2.
The free energy profile for the proton transfer from SAHA to His142. The distance between H142:N and SAHA:H (dN-H) was chosen as the reaction coordinate. The statistical error is estimated by averaging the free energy difference between 5~15ps and 15~25ps.
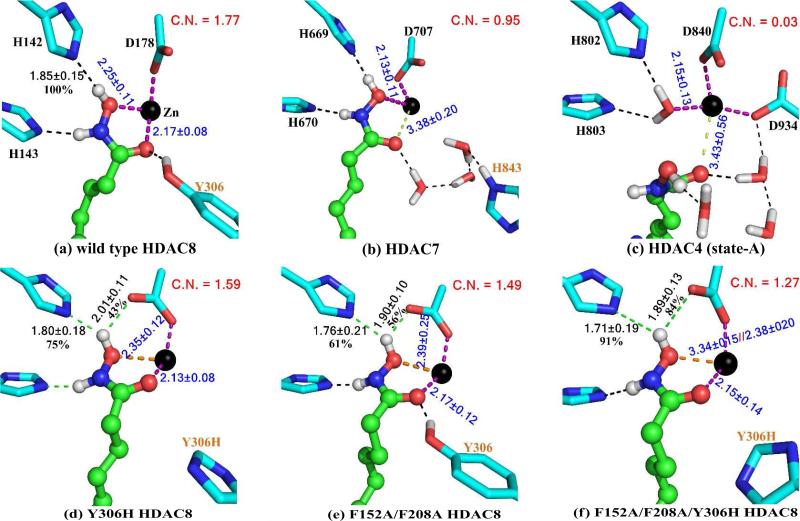
Then we carried out ab initio QM/MM MD simulations of HDAC7 and HDAC4, two class–IIa enzymes, with the very similar computational protocol as previously employed in simulating HDAC8.28 The resulted hydroxamate chelation modes for all three HDACs are illustrated in Figure 3, and the distributions of the distances from the oxygen atoms (O1 and O2) of hydroxamate to the zinc atom are summarized in Figure S2. It is very encouraging that our QM/MM MD simulations reproduce the coordination configurations in the crystal structures of wild type HDAC8 and HDAC7 very well (shown in Figure S1). Considering the distance between O2 and Zn in HDAC4 from our simulation (state A, 3.43 ± 0.56 Å) is longer than that in the crystal structure (2.49 Å), we set up another QM/MM MD simulation on HDAC4 (state B, see Figure S3), in which the Zn-O2 distance was first restrained at the 2.49 Å during the first 3 ps QM/MM MD simulation and then followed by another ~20 ps simulations without any restraints. The resulted trajectory shows that the originally bound water leaves the first zinc coordination shell and the hydroxamate is monodentate with Zn2+. Nevertheless, both simulations indicate a 4-fold zinc coordination shell in the HDAC4 complex. The widespread distribution of Zn-O distances indicates a loose binding of hydroxamate in HDAC4 (see Figure S2).
Figure 3.
Illustration of zinc chelation modes and the hydrogen bond network for each model from our QM/MM MD simulations. C.N. means the Coordination Number between zinc and hydroxamate. C.N. is 1 if Zn-N ≤ 2.15 Å, equals 0 if Zn-N ≥ 2.40 Å, and is a linear scalar between 0 and 1 if Zn-N is between 2.15 and 2.40 Å. Similarly, the values of 2.20 and 2.60 are used for Zn-O, respectively. These values are chosen based on very recent analysis on databases of zinc enzyme structures database.39
So far, our simulations of wild type HDACs confirm the experimental structural finding that in spite of having the same coordinating ligands, different HDACs can have distinct zinc-hydroxamate chelation modes. To find out the origin of such difference, we further carried out ab initio QM/MM MD simulations on three SAHA-HDAC8 mutants, i.e., single mutant Y306H, double mutant F152A/F208A, and triple mutant F152A/F208A/Y306H. The results in Figure 3 indicate that although these mutations have little effect on the coordination interactions between zinc and amino acid residues, they do lead to significantly different zinc-hydroxamate chelation modes, especially for the triple mutant F152A/F208A/Y306H. For Y306H single mutant, the average coordination distance between SAHA-O1 and zinc is elongated by 0.1 Å and its zinc-hydroxamate coordination number is reduced by about 0.2; For F152A/F208A double mutant, not only the average coordination distance is longer, but also the fluctuation is significantly larger which indicates a more flexible and weaker zinc-hydroxamate coordination; For the triple mutant, the average value and the fluctuation of the coordination distance between SAHA-O1 and zinc have significantly increased and its distribution curve in Figure S2 clearly displays two distinct peaks indicating that the SAHA chelation becomes monodentate to some extent. Thus our simulations confirm that the existence of the active site tyrosine residue would strengthen the zinc-hydroxamate binding, which is consistent with experimental mutation results.9,19-21 However, our results indicate that this residue alone would not determine the chelation mode of the hydroxamate.
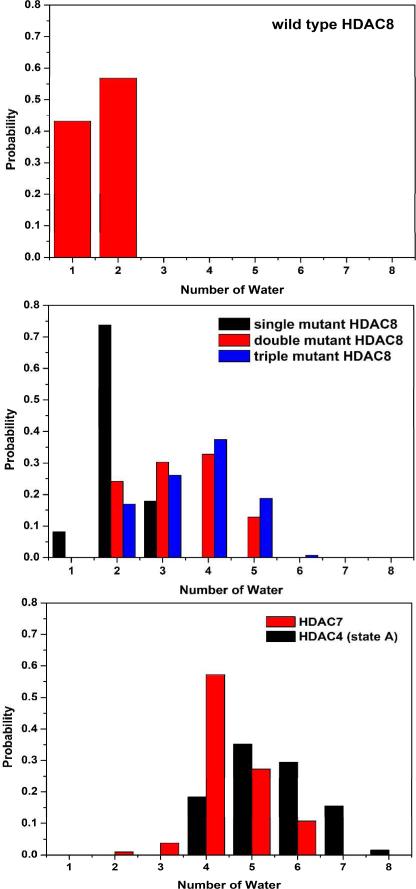
By further analyzing all QM/MM MD simulations of HDACs, we find that the more water molecules inside the binding pocket, which mostly stay in the linker binding channel, the more likely for the monodentate zinc-hydroxamate binding. From Figure 4, we can see that there are only one or two waters in the binding pocket of the wild type HDAC8 and two in Y306H HDAC8, but in average four in F152A/F208A and Y306H/F152A/F208A HDAC8. For HDAC7 and HDAC4 models, there can be up to six water molecules. We can also see that in double/triple mutant HDAC8, wild type HDAC7 or HDAC4, there exists at least one stable hydrogen bonded water chain in the linker channel along which the waters enter into the zinc active site, as shown in Figure 5, and S4/S5. With the presence of more water molecules in the binding pocket, the dielectric constant would be increased and thus in turn it would lead to weaker electrostatic interactions between zinc and ligands in the binding site. Considering that Zn2+ has a saturated electronic configuration of d10, its coordination with ligands is dominated by electrostatic interactions and thus would be weakened by the presence of more water molecules. Meanwhile, due to the presence of two carboxyl groups in the first coordination shell of HDACs, the flexibility of its zinc coordination has been previously found to be much smaller than that in other zinc enzymes40 while mostly comes from its chelation with the non-amino-acid ligand28. Thus this would explain that different water molecules in the active site of HDACs would affect the binding mode of the hydroxamate in spite of having the very same first coordination shell. Furthermore, Figure S6 indicates that the calculated ESP charge on the zinc ion is reduced when more water molecules are in the binding pocket, leading to even weaker zinc ligand interactions.
Figure 4.
Number of water molecules in the binding pocket of each model during the last 20 ps QM/MM MD trajectory.
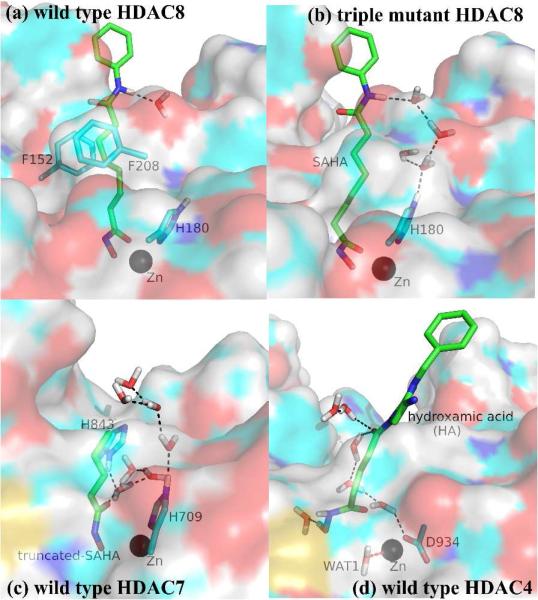
Figure 5.
Comparison of the binding pockets in different models. A stable hydrogen-bonded water chain was observed during the QM/MM MD simulation with (b), (c) and (d) models.
Different number of water molecules inside the binding pocket observed in distinct HDACs can be ascribed into the gating effect of two Phe residues sitting around the pocket entrance. For HDAC7 and HDAC4, since the aromatic rings of both phenylalanines are away from the inhibitor and pointing toward to the protein surface, the channel is wide enough to allow more water molecules to enter the pocket and form stable hydrogen bonds with the inhibitor and Zn-bound ligands. However, for wild type HDAC8, F152/F208 and the linker constitute a “sandwich-like” conformation to block the channel and prevent water entering the binding pocket. Volume calculations indicate that the binding pocket is 2225 ± 184 Å3 in the wild type HDAC8 (Figure S7), much smaller than those in HDAC7 and HDAC4, which are 4137 ± 206 and 4360 ± 262 Å3, respectively. But the channel in HDAC8 (11.6 ± 1.3 Å) is deeper than the one in HDAC7 (7.3 ± 0.6 Å) and HDAC4 (6.4 ± 1.0 Å), indicating that the channel in HDAC8 wild type is much narrower. Even though the Y306H mutation of HDAC8 enlarges the vacant space of the pocket, the channel is still blocked by the conserved F152/F208 and the linker part of SAHA. Only after the F152A/F208A mutation, the channel blockage is cleared and extra room is given to allow more water molecules to enter the pocket. For the HDAC8 triple mutant F152A/F208A/Y306H (see Figure S5), its entrance becomes significantly wider than that of the wild type, the space (3718 ± 223 Å3) is enlarged and the channel becomes less deeper (10.1 ± 0.8 Å), which yields a binding pocket resembling those of HDAC7 and HDAC4. These results demonstrate the important gating effect of F152/F208 in HDAC8, and provide further support for our new mechanistic suggestion that the zinc-hydroxamate coordination mode in HDACs is modulated by water access to the linker binding channel.
In summary, our ab initio QM/MM MD simulations do not support the mechanistic hypothesis that the distinct zinc-hydroxamate chelation modes between two HDAC subclasses come from different protonation states of the hydroxamic acid. Instead, our simulation results have suggested one novel mechanism regarding the interplay between the linker binding and the zinc chelation: the zinc-hydroxamate coordination mode in HDACs can be modulated by water access to the linker binding channel. This implies that for the development of new class-IIa specific inhibitors, one intriguing direction to explore would be to design a linker component to block the access of water molecules into the binding pocket.
Supplementary Material
Acknowledgment
This work was supported by Chinese NSF (20733002 & 20873105) and the Ministry of Science and Technology (2011CB808504), American NIH (R01-GM079223), NSF (CHE-CAREER-0448156, TeraGrid resources). We thank NYU-ITS for providing computational resources and Dr. Shenglong Wang for computing support.
Footnotes
Supporting Information Available: Computational details; Figure S1-S7; complete refs 16 and 37. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Cole PA. Nat. Chem. Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith BC, Denu JM. BBA-Gene Regul. Mech. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haberland M, Montgomery RL, Olson EN. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kazantsev AG, Thompson LM. Nat. Rev. Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 6.Paris M, Porcelloni M, Binaschi M, Fattori D. J. Med. Chem. 2008;51:1505–1529. doi: 10.1021/jm7011408. [DOI] [PubMed] [Google Scholar]
- 7.Bolden JE, Peart MJ, Johnstone RW. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 8.Minucci S, Pelicci PG. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 9.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Nat. Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertrand P. Eur. J. Med. Chem. 2010;45:2095–2116. doi: 10.1016/j.ejmech.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 11.Bieliauskas AV, Pflum MKH. Chem. Soc. Rev. 2008;37:1402–1413. doi: 10.1039/b703830p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estiu G, Greenberg E, Harrison CB, Kwiatkowski NP, Mazitschek R, Bradner JE, Wiest O. J. Med. Chem. 2008;51:2898–2906. doi: 10.1021/jm7015254. [DOI] [PubMed] [Google Scholar]
- 13.Olsen CA, Ghadiri MR. J. Med. Chem. 2009;52:7836–46. doi: 10.1021/jm900850t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sternson SM, Wong JC, Grozinger CM, Schreiber SL. Organic Lett. 2001;3:4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 15.Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, Steinkuhler C, Di Marco SP. Natl. Acad. Sci. USA. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Somoza JR, et al. W. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Vannini A, Volpari C, Gallinari P, Jones P, Mattu M, Carfi A, De Francesco R, Steinkuhler C, Di Marco S. Embo Rep. 2007;8:879–884. doi: 10.1038/sj.embor.7401047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowling DP, Gantt SL, Gattis SG, Fierke CA, Christianson DW. Biochemistry. 2008;47:13554–13563. doi: 10.1021/bi801610c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bottomley MJ, Lo Surdo P, Di Giovine P, Cirillo A, Scarpelli R, Ferrigno F, Jones P, Neddermann P, De Francesco R, Steinkuhler C, Gallinari P, Carfi A. J. Biol. Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, McLean TH, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. J. Biol. Chem. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahm A, Paolini C, Pallaoro M, Nardi MC, Jones P, Neddermann P, Sambucini S, Bottomley MJ, Lo Surdo P, Carfi A, Koch U, De Francesco R, Steinkuhler C, Gallinari P. P. Natl. Acad. Sci. USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang DF, Helquist P, Wiest O. J. Org. Chem. 2007;72:5446–5449. doi: 10.1021/jo070739s. [DOI] [PubMed] [Google Scholar]
- 23.Weerasinghe SVW, Estiu G, Wiest O, Pflum MKH. J. Med. Chem. 2008;51:5542–5551. doi: 10.1021/jm800081j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu P, Wang S, Zhang Y. J. Am. Chem. Soc. 2008;130:3806–3813. doi: 10.1021/ja075896n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu P, Wang SL, Zhang YK. J. Am. Chem. Soc. 2008;130:16721–16728. doi: 10.1021/ja807269j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke ZH, Zhou YZ, Hu P, Wang SL, Xie DQ, Zhang YK. J. Phys. Chem. B. 2009;113:12750–12758. doi: 10.1021/jp903173c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang SL, Hu P, Zhang YK. J. Phys. Chem. B. 2007;111:3758–3764. doi: 10.1021/jp067147i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu R, Hu P, Wang S, Cao Z, Zhang Y. J. Chem. Theory Comput. 2010;6:337–343. doi: 10.1021/ct9005322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou YZ, Wang SL, Zhang YK. J. Phys. Chem. B. 2010;114:8817–8825. doi: 10.1021/jp104258d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu RB, Wang SL, Zhou NJ, Cao ZX, Zhang YK. J. Am. Chem. Soc. 2010;132:9471. doi: 10.1021/ja103932d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dolg M, Wedig U, Stoll H, Preuss H. J. Chem. Phys. 1987;86:866–872. [Google Scholar]
- 32.Sousa SF, Fernandes PA, Ramos MJ. Biophys. J. 2005;88:483–494. doi: 10.1529/biophysj.104.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sousa SF, Fernandes PA, Ramos MJ. J. Am. Chem. Soc. 2007;129:1378–1385. doi: 10.1021/ja067103n. [DOI] [PubMed] [Google Scholar]
- 34.Xiao CY, Zhang YK. J. Phys. Chem. B. 2007;111:6229–6235. doi: 10.1021/jp068657f. [DOI] [PubMed] [Google Scholar]
- 35.Corminboeuf C, Hu P, Tuckerman ME, Zhang YK. J. Am. Chem. Soc. 2006;128:4530–4531. doi: 10.1021/ja0600882. [DOI] [PubMed] [Google Scholar]
- 36.Zhang YK. J. Chem. Phys. 2005;122:024114. doi: 10.1063/1.1834899. [DOI] [PubMed] [Google Scholar]
- 37.Shao Y, et al. Q-Chem, version 3.0. Pittsburgh, PA: 2006. [Google Scholar]
- 38.Ponder JW. TINKER, Software Tools for Molecular Design, Version 4.2. 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamames B, Sousa SF, Tamames J, Fernandes PA, Ramos MJ. Proteins: Struct., Funct., Bioinf. 2007;69:466–475. doi: 10.1002/prot.21536. [DOI] [PubMed] [Google Scholar]
- 40.Papoian GA, DeGrado WF, Klein ML. J. Am. Chem. Soc. 2003;125:560–569. doi: 10.1021/ja028161l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.