Abstract
Affinity-purification coupled to mass spectrometry (AP-MS) represents a powerful and proven approach for the analysis of protein-protein interactions. However, the detection of true interactions for proteins that are commonly considered background contaminants is currently a limitation of AP-MS. Here using spectral counts and the new statistical tool, Significance Analysis of INTeractome (SAINT), true interaction between the serine/threonine phosphatase 5 (PP5) and a chaperonin, heat shock protein 90 (Hsp90), is discerned. Furthermore, we report and validate a new interaction between PP5 and an Hsp90 adaptor protein, stress-induced phosphoprotein 1 (STIP1; HOP). Mutation of PP5, replacing key basic amino acids (K97A and R101A) in the tetratricopeptide repeat (TPR) region known to be necessary for interactions with Hsp90, abolished both the known interaction of PP5 with Cdc37 and the novel interaction of PP5 with STIP1. Taken together, the results presented demonstrate the usefulness of label-free quantitative proteomics and statistical tools to discriminate between noise and true interactions, even for proteins normally considered as background contaminants.
Keywords: Protein interactions, Hsp90, protein phosphatase, PP5, affinity purification-mass spectrometry, contaminant filtering, SAINT
1. Introduction
Affinity purification coupled to mass spectrometry (AP-MS) is widely used for the identification of interaction partners for proteins of interest. This approach has been used for the identification of interaction partners for kinases, phosphatases and other molecules in yeast and human cells [1–3]. The identification of true interactions occurring in “a sea” of background contaminants is not trivial. To distinguish true interactions from contaminants, several groups have developed approaches to directly reduce background, such as performing sequential purification steps (e.g. Tandem Affinity Purification [4]). However, the lengthy multi-step purification processes required have major drawbacks. Notably, both weakly and transiently associated proteins are lost.
As an alternate approach, single step purifications can be performed. However, the background is often much higher with this approach. With either a single- or multi-step approach, typically a first step in the identification of true interactors is the removal of proteins that bind to the affinity matrix alone (or to another negative control). In addition, proteins that associate with a high percentage of different baits (“frequent flyers”) are also removed. These filtering methods are classically applied following the analysis of binary data (indicating the presence or absence of a protein). For example, high-throughput studies have systematically removed proteins co-purified in an arbitrarily determined percentage of the analyses (e.g. 5% in the Ho et al. 2002 study; [5]). Besides the obvious problem that the frequency filters are arbitrarily chosen, it is possible that a true high-abundance interactor for a given protein of interest (or bait) is also detected in lower abundance with other baits in negative control runs. For these reasons, obtaining a quantitative measure for the presence of the given hit or prey protein across all purifications may assist in determining the likelihood that the interaction is indeed significant. Quantitative approaches using stable isotopes have been successfully used to identify true interactions between molecules of interest (reviewed in Gingras et al, 2007;[6]), but these techniques are often costly and not necessarily amenable to the analysis of all samples. In recent years, the use of spectral counts (which are easily extracted from mass spectrometry data) as a proxy for abundance measurement has gained widespread use (e.g. [7]). We and others have made use of this quantitative information to help identify true interaction partners from background contaminants [3, 8, 9].
Recently, we have developed a generalized computational approach for Significance Analysis of INTeractome (SAINT). SAINT was first used for the analysis of a global yeast kinase and phosphatase interactome [3], and then extended as a generalized model including negative controls to networks of various scales [10]. The method utilizes label-free quantitative data, such as spectral counts, to assign a confidence value to individual protein-protein interactions. SAINT performs semi-supervised analysis using data from control purifications and constructs separate distributions for true and false interactions to derive the probability of a bona fide protein-protein interaction.
To fully demonstrate the power of statistical analysis of spectral count distributions, we decided to analyze a challenging test-case. The serine/threonine phosphatase 5 (PP5; PPP5C) offered such an example, since PP5 is expressed ubiquitously in human tissues and is known to interact with the chaperonin Hsp90 (one of the major “frequent fliers” in AP-MS data). PP5 belongs to the PPP-family of enzymes and shares a common mechanism for mediating the hydrolysis of phosphoprotein substrates with PP1-PP6 [11]. However, unlike PP1, PP2A and PP4, which obtain regulation and substrate specificity via interactions with scaffold, regulatory and substrate targeting subunits encoded by separate genes, a single gene encodes the PP5 catalytic domain along with unique N- and C-terminal domains that regulate both interactions with other proteins and catalytic activity [11–13]. Therefore, determining the biologically relevant interactions for PP5 is also needed to help understand the roles of PP5 in normal biology and human disease [14].
In the current study, the use of the SAINT algorithm allows us to discern true interactions between PP5 and Hsp90. Interestingly, the most significant interactor for PP5 is an Hsp90 co-chaperone, stress-induced phosphoprotein 1 (STIP1; also called HOP or STI1). STIP1 mediates interactions between Hsp90 and heat shock protein 70 (Hsp70); consistent with this, Hsp70 was also recovered in our analysis. The Hsp90 co-chaperone Cdc37 (cell division cycle 37 homolog) was also recovered in the PP5 purification, albeit in lower amount than STIP1. Analysis of the interactions mediated by Hsp90 allowed us to conclude that PP5 exhibits preference for STIP1 as compared to Cdc37.
2. Material and methods
2.1 Expression plasmids
Human PP5 (NM_006247.2) was amplified by PCR, incorporating EcoRI/ NotI sites along with a C-terminal FLAG sequence (MDYKDDDDK) by adding the appropriate sequence in the synthetic primers. PP5-FLAG was then cloned into pcDNA3 (pcDNA3-PP5-FLAG). Along with wild-type PP5, a PP5-FLAG containing a mutated TPR-domain (K97A and R101A) was generated using Stratagene QuikChange II Site-Directed Mutagenesis (pcDNA3-K97A-R101A-PP5-FLAG; ΔTPR-PP5-FLAG). All constructs were sequenced in their entirety. pcDNA3-FLAG-PP4 (which encodes the PPP4C phosphatase) was described previously [15] HSP90AA1 was amplified from BC023006 and cloned into the AscI/NotI sites of the vector pcDNA5-FRT-FLAG. pcDNA5-FLAG was constructed by subcloning the HindIII/XhoI cassette from pcDNA3-FLAG [15] into pcDNA5-FRT-TO (Invitrogen). An internal EcoRI site was removed by mutagenesis. HSP90AA1 was excised from the FLAG vector and shuttled into pcDNA5-FRT-eGFP. pcDNA5-FRT-eGFP was constructed by subcloning the HindIII/AscI cassette from pcDNA3-eGFP into pcDNA5-FRT-TO. CDC37 (DQ892174) and STIP1 (DQ893295) Gateway entry clones were cloned by recombination into vector pDEST pcDNA5/FRT/TO-eGFP (a kind gift from K. Colwill and T. Pawson).
2.2 Cell lines and Stable Expression
HEK293 cells, passage 15, were transfected in a 6 well format with the indicated plasmids (~0.3 μg pcDNA3-PP5-FLAG, 0.3 μg pcDNA3-K97A-R101A-PP5-FLAG). G418 treatment was used to select for stable transformed cell lines expressing PP5-FLAG, and Western analysis was used to determine the relative expression of PP5 (endogenous) and PP5-FLAG. Cell expressing the desired constructs were subcloned and monitored for expression levels. Cell lines expressing low levels of PP5-FLAG expression were chosen for furtherer studies. Flp-In T-REx 293 cells were co-transfected with 0.2 μg of pcDNA5-FLAG-HSP90AA1 and 2 μg of pOG44, using lipofectamine PLUS (Invitrogen), according to the manufacturer’s instructions, and selected in 200 μg/ml hygromycin. Stable cell clones were selected and the protein expression was induced by 1μg/ml tetracycline for 24 hours.
For mass spectrometric analysis, cells were grown in 15 cm plates (using 10–12 plates per treatment group for PP5 and 4 plates for HSP90AA1). Prior to harvest, the plates were washed twice with 10 mL of ice cold PBS. Cells were then harvested and collected in 0.75 mL of ice cold PBS by centrifugation at 1500 × g at 4°C for 5 minutes. Cell pellets were resuspended and washed 3 times in ice cold PBS. After the final wash, the excess PBS was removed and the pellet was frozen in liquid nitrogen and stored at −80°.
2.3 FLAG affinity purification and mass spectrometric analysis
FLAG-affinity purification was performed essentially as described [2], with the following modifications: detergent concentration in the lysis buffer was 0.5% NP-40; the lysis buffer was added at 4 ml/g (wet cell pellet), and cells were subjected to passive lysis (30 minutes) followed by one freeze-thaw cycle. Beads were washed three times in lysis buffer and three times in 50 mM ammonium bicarbonate. Samples were eluted with ammonium hydroxide, lyophilized in a speed-vac, resuspended in 50 mM ammonium bicarbonate (pH 8 – 8.3), and incubated at 37°C with trypsin overnight. The ammonium bicarbonate was evaporated. The samples were resuspended in HPLC buffer A (2% acetonitrile, 0.1% formic acid) and then directly loaded onto capillary columns packed in-house with Magic 5 μm, 100A, C18AQ. MS/MS data was acquired in data-dependent mode (over a 2 hr acetonitrile 2 – 40% gradient) on a ThermoFinnigan LTQ, equipped with a Proxeon NanoSource and an Agilent 1100 capillary pump. *.mgf files were generated from ThermoFinnigan *.RAW files. The searched database was human RefSeq (version 37). *.mgf files were searched with the Mascot search engine using the following parameters: partial trypsin digestion (allowing for one missed cleavage site); asparagine deamidation and methionine oxidation were set as variable modifications. The fragment mass tolerance was 0.6 Da (monoisotopic mass), and the mass window for the precursor was +/− 3 Da. Mascot results were parsed for further analysis into a software developed at the Samuel Lunenfeld Research Institute (ProHits: [16]). Simple filters, based on the frequency of detection of a given hit across multiple purifications of similarly tagged and expressed baits were performed using the ProHits interface. Essentially, the database used consists of >200 FLAG-tagged baits, each expressed in HEK293 cells (or derivatives). Frequency filters were utilized to visualize the PP5 hits: at each of the cutoffs selected (e.g. 5%, 10%, 20%, etc.), the list of the PP5 interaction partners detected in >2 of the biological replicates was manually inspected.
2.4 SAINT analysis
SAINT converts the label free quantification, such as spectral counts, for each prey protein identified in a purification of a bait into the probability of true interaction between the two proteins. For PP5 data, four biological replicates were used for each bait (wt-PP5, ΔTPR-PP5, HSP90AA1) alongside five negative control runs, consisting of a cell line expressing the FLAG tag alone. SAINT calculates scores differently depending on the availability of negative control purifications, and thus the implementation for spectral count data incorporating control purification data was used (details are described in Choi et al. [10]). The probability score was first computed for each prey in independent biological replicates separately (iProb). Then the final probability score for a pair of bait and prey proteins was calculated by taking the average of the probabilities in individual replicates (AvgP); final results with AvgP ≥0.5 were further inspected. For the analysis of the PP5 dataset, we used a simple averaging model (AvgP), which sums individual iProb (individual probabilities) and divides this number by the number of biological replicates performed. The selected cutoff of AvgP ≥0.5 ensures that the interaction has been detected with high probability in at least two of the four replicates (or with moderate probability in all four replicates).
2.5 Validation of interactions by IP/Western
Endogenous PP5 and PP5-FLAG were detected as described previously [17]. Transient transfection in 293T cells was performed in 6 well dishes using previously described methods [2]. Post-transfection (48h), cells were harvested and rinsed in PBS. Cells were lysed and the FLAG-tagged protein was immunoprecipitated from 1 mg cell extract using anti-FLAG M2 agarose beads. Immunoprecipitates were resolved by SDS-PAGE, and the proteins were transferred onto membranes. Immunoblots were performed as described in [2], using the following antibodies [anti-FLAG M2 monoclonal (SIGMA F3165), anti-GFP (Roche 11814460001) and ECL Mouse IgG, HRP-Linked Whole Antibody from sheep (GE Healthcare, NA931)].
3. Results
3.1 Generation of the interaction data set
To generate an interaction dataset for the human PP5 phosphatase, PP5 was tagged at its C-terminus with a FLAG epitope, and the construct was stably expressed in human HEK293 cells. Cells expressing moderate amounts of PP5-FLAG proteins (Supplementary Figure 1) were harvested and four biological replicates were analyzed by mass spectrometry. Because two basic amino acids (K97 and R101) located in a near N-terminal TPR region of PP5 are known to mediate interactions between PP5 and the chaperone Hsp90 [18], a mutated form of PP5 in which these key amino acids were converted to alanine (ΔTPR- PP5-FLAG) was similarly expressed and harvested (n = 4). In parallel, we performed analysis with 5 samples of cells expressing the FLAG tag alone.
Affinity-purification coupled to mass spectrometric analysis was performed on a ThermoFinnigan LTQ mass spectrometer. The data was searched using the Mascot search engine, as detailed in Materials and Methods. Data was analyzed with our laboratory information management system for interaction proteomics, ProHits [16], and multiple sample comparison analysis was performed. The list of interactors without any filters yielded a total of 503 “raw interactions” for wt-PP5-FLAG and ΔTPR-PP5-FLAG (Supplementary Table I). As previously observed, determining which of these proteins specifically interact with PP5, and which do not, was difficult.
As an initial approach to determine the true interaction partners for PP5, we first applyed a simple frequency filter cut-off. For this, we also looked at the recovery of each of the different proteins across multiple analyses using the same protocol for different projects (see Methods). However, with this approach the known partner of PP5, Hsp90, was observed in ~70% of all AP-MS; using a frequency filter of 5% essentially removed Hsp90 and all putative interactions (not one of the remaining proteins was detected in more that two of the biological replicates; not shown).
3.2 Significance Analysis of the INTeractome (SAINT analysis)
Next, the MS results were subjected to SAINT analysis (see Figure 1 for an overview of the approach), using a version of the SAINT algorithm that was modified from its initial application (i.e. the analysis of yeast kinase and phosphatase interaction networks [3]). This version of SAINT [10] incorporates negative control runs into the model. The SAINT model also takes into consideration data from biological replicates, essentially increasing the confidence for interactions that are detected repeatedly in the analyses of multiple biological replicates.
Figure 1. Schematics of the experimental and analytical pipeline.
A) wt-PP5-FLAG, ΔTPR-PP5-FLAG or FLAG alone cells lines were generated, and subjected to AP-MS analysis in four biological replicates. The grey circle represents a non-specific interaction partner which associates to the FLAG alone, the orange circle represents an interaction partner for the wt and mutant PP5 which is not recovered in the FLAG alone purifications. The blue circle represents a protein that strongly interacts with wt but not ΔTPR PP5, yet can also be detected in lower abundance in purifications of the FLAG alone. B) Schematic representation of the spectral counts for the blue, orange and grey proteins across the four biological replicates. C) Typical results obtained using simple binary contaminant filtering, such as frequency filters or removal of all hits identified in the FLAG alone sample. While the orange and grey proteins are successfully identified as specific and background, respectively, the blue protein is erroneously labeled as a contaminant, resulting in a false-negative identification. D) SAINT utilizes a semi-supervised mixture model of the spectral count distribution of each protein across the negative control runs (blue line) and provides probability values that each bait-prey interaction is real.
Application of SAINT led to the recovery of seven interaction partners with an AvgP ≥0.5 with wt-PP5-FLAG. Application of the statistical tools recapitulated the previously reported interaction of wt-PP5 with Hsp90. HSP90AA1 and HSP90AB1 were recovered with an AvgP of 0.69 and 0.84, respectively (Table I and Supplementary Table I). The average spectral count distribution of HSP90AB1 across all purifications of wt-PP5 was ~56, as compared to ~6 for FLAG alone. In addition, in accordance with published data, mutations in the TPR repeat domains prevented this interaction, and the average spectral counts were ~2 for ΔTPR-PP5.
Table I. AP-MS data with ≥0.5 AvgP SAINT value.
Indicated baits and prey (HUGO gene names) with AvgP. Spectral indicates the spectral counts in each of the biological replicates. AvgP is the average of the individual probabilities. Control indicates the number of spectral counts in control runs (See Supplemental Table I for unfiltered data).
| Bait | Prey | Spectral | Control | AvgP |
|---|---|---|---|---|
| PPP5C_WT | STIP1 | 84|44|30|26 | 0|0|0|0|0 | 1 |
| PPP5C_WT | PPP5C | 48|96|45|44 | 0|0|0|0|0 | 1 |
| PPP5C_WT | HSPA8 | 33|40|19|47 | 26|26|20|20|18 | 0.94 |
| PPP5C_WT | HSPA1B | 59|50|42|81 | 36|25|24|18|17 | 0.9 |
| PPP5C_WT | HSP90AB1 | 61|53|33|76 | 9|8|7|4|4 | 0.84 |
| PPP5C_WT | HSP90AA1 | 61|41|31|54 | 10|9|7|7|4 | 0.69 |
| PPP5C_WT | CCT4 | 0|4|2|1 | 0|0|0|0|0 | 0.67 |
| PPP5C_WT | CDC37 | 5|3|0|0 | 0|0|0|0|0 | 0.5 |
| PPP5C_MUT | PPP5C | 108|42|42|22 | 0|0|0|0|0 | 1 |
| HSP90AA1 | STIP1 | 49|96|81|77 | 0|0|0|0|0 | 1 |
| HSP90AA1 | HSP90AB1 | 2265|1771|1406|716 | 9|8|7|4|4 | 1 |
| HSP90AA1 | HSP90AA1 | 2608|2222|1676|853 | 10|9|7|7|4 | 1 |
| HSP90AA1 | HSPA1B | 292|240|177|122 | 36|25|24|18|17 | 1 |
| HSP90AA1 | HSPA8 | 189|224|197|124 | 26|26|20|20|18 | 1 |
| HSP90AA1 | CDC37 | 41|23|11|29 | 0|0|0|0|0 | 1 |
| HSP90AA1 | FKBP4 | 16|21|8|38 | 0|0|0|0|0 | 0.99 |
| HSP90AA1 | STUB1 | 20|28|9|14 | 0|0|0|0|0 | 0.99 |
| HSP90AA1 | TOMM34 | 2|4|3|6 | 0|0|0|0|0 | 0.99 |
| HSP90AA1 | FKBP5 | 3|7|24|5 | 0|0|0|0|0 | 0.99 |
| HSP90AA1 | GIGYF2 | 2|2|2|4 | 0|0|0|0|0 | 0.97 |
| HSP90AA1 | FKBP8 | 3|6|10|1 | 0|0|0|0|0 | 0.95 |
| HSP90AA1 | TUBB2B | 167|118|215|73 | 37|31|29|0|0 | 0.94 |
| HSP90AA1 | SSBP1 | 4|8|6|10 | 1|0|0|0|0 | 0.94 |
| HSP90AA1 | TUBB | 207|167|301|88 | 66|49|40|40|38 | 0.93 |
| HSP90AA1 | TUBA1A | 287|190|190|80 | 23|27|45|33|0 | 0.93 |
| HSP90AA1 | TUBB2C | 173|130|302|86 | 57|43|42|40|37 | 0.87 |
| HSP90AA1 | DNAJA1 | 2|4|12|1 | 2|1|0|0|0 | 0.66 |
| HSP90AA1 | RPAP3 | 1|0|4|3 | 0|0|0|0|0 | 0.66 |
| HSP90AA1 | PDRG1 | 1|3|3|0 | 0|0|0|0|0 | 0.64 |
| HSP90AA1 | DNAJA2 | 2|4|4|3 | 2|1|1|0|0 | 0.63 |
| HSP90AA1 | FASN | 2|2|1|0 | 0|0|0|0|0 | 0.63 |
| HSP90AA1 | TTC4 | 1|1|0|3 | 0|0|0|0|0 | 0.61 |
| HSP90AA1 | HSPA4 | 0|1|1|5 | 0|0|0|0|0 | 0.6 |
| HSP90AA1 | POLR2E | 2|1|2|3 | 2|0|0|0|0 | 0.57 |
| HSP90AA1 | IRS4 | 0|1|9|8 | 1|0|0|0|0 | 0.54 |
| HSP90AA1 | TUBAL3 | 9|7|4|9 | 3|0|0|0|0 | 0.52 |
The most significant interaction partner of wt-PP5 was STIP1 (also called HOP; AvgP = 1), which is an Hsp90 co-chaperonin. STIP1 was recovered with an average spectral count of ~39 in the PP5 samples, but never detected in the FLAG alone samples or the TPR mutant. The Hsp70 proteins HSPA8 and HSPA1B were recovered with an AvgP of 0.94 and 0.9, respectively. Other likely interactors for wt-PP5-FLAG include CCT4 (AvgP 0.67) and the Hsp90 co-chaperonin Cdc37 (AvgP 0.5). These interactions are all likely mediated by the TPR region of PP5, as no interactors with a SAINT AvgP ≥0.5 (with the exception of the bait) were detected with the TPR-mutant.
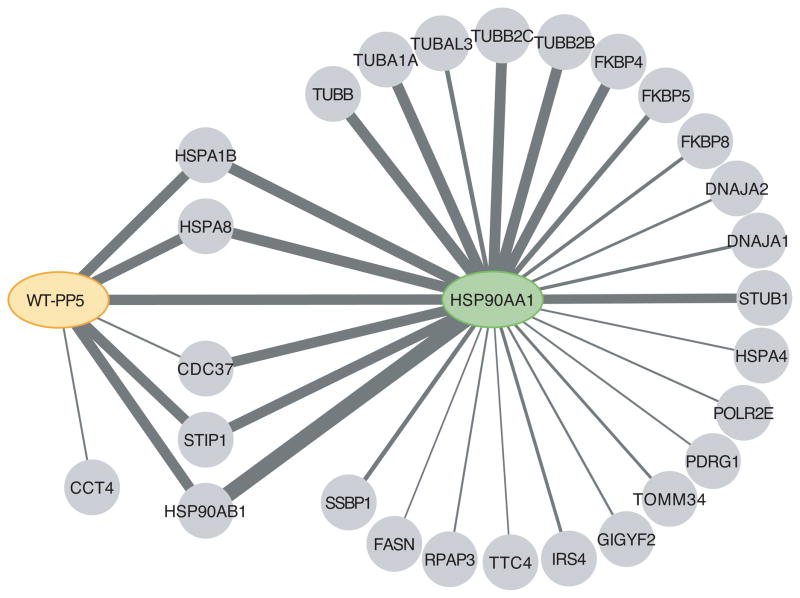
We next attempted to identify whether PP5 exhibited preferential interaction with specific Hsp90 subcomplexes or whether the apparent enrichment of STIP1 in the PP5 immunoprecipitates simply reflected the abundance of this protein in the Hsp90 complexes. To do so, we generated a stable cell line expressing a tetracycline-inducible version of Hsp90 (HSP90AA1) and analyzed four biological replicates by AP-MS and SAINT analysis. Hsp90 recovered many more statistically significant interactors with an AvgP ≥0.5 (22) as compared to PP5. Interestingly, Hsp90 was associated with a large number of Cdc37 peptides, while this co-chaperonin was detected in much smaller amounts in the PP5 immunoprecipitates. Tubulins and FK506 binding proteins were very abundant in the Hsp90 sample, yet they were very minor components of the PP5 immunoprecipitates (Figure 2; Table I, Sup Table I). Taken together, these results indicate that PP5 may preferentially interact with an Hsp90-Hsp70-STIP1 complex.
Figure 2. Cytoscape representation of the wt-PP5 and Hsp90 interaction partners.
AP-MS data is from Table I; all interaction partners have an AvgP≥0.5. The thickness of the lines is proportional to the total spectral counts for the hits in the four biological replicates purifications of the bait.
3.3 Validation of interactions
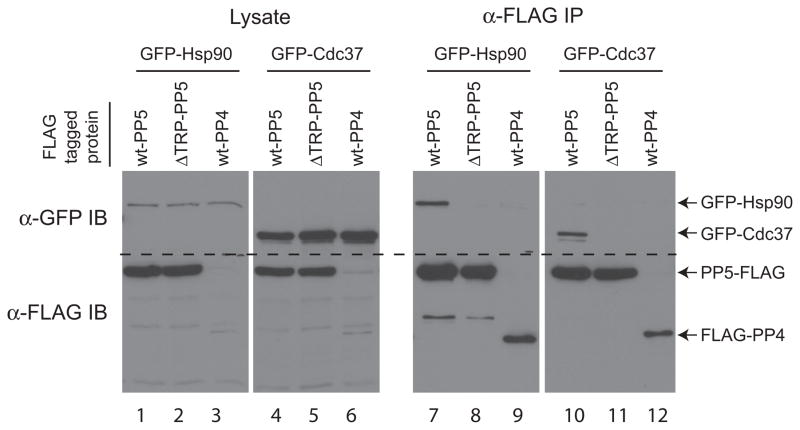
To validate the interactions detected, we first tested whether we could reproduce the interaction between wild-type PP5 and its known interactors, Hsp90 and Cdc37, in a co-immunoprecipitation/ immunoblotting assay. GFP-tagged Hsp90 or Cdc37 were co-transfected with wt-PP5-FLAG, ΔTPR-PP5-FLAG, or the related phosphatase FLAG-PP4 (used here as a negative control). FLAG-tagged proteins were precipitated using M2 agarose, and immunoblotting was performed using anti-FLAG and anti-GFP antibodies. wt-PP5-FLAG, but not ΔTPR-PP5-FLAG or FLAG-PP4, readily recovered both GFP-Hsp90 and GST-Cdc37, as expected (Figure 3; compare lane 7 to lanes 8–9, and lane 10 to lanes 11–12).
Figure 3. Hsp90 and Cdc37 interacts with wt-PP5, but not a TPR mutant or with the phosphatase PP4.
Immunoprecipitation on anti-FLAG (M2 agarose) beads was performed on lysate from HEK293T cells transiently co-expressing the indicated FLAG- and GFP-tagged constructs. Immune complexes were resolved by SDS-PAGE followed by transfer to nitrocellulose. Co-precipitation of GFP-tagged proteins was detected by immunoblotting (IB) for the GFP tag (top panels; the positions of the tagged proteins are indicated by arrows). The precipitated FLAG-tagged protein was detected with anti-FLAG antibodies (bottom panels). Total protein lysate (left) was analyzed in parallel to the immunoprecipitation (right) to monitor protein expression.
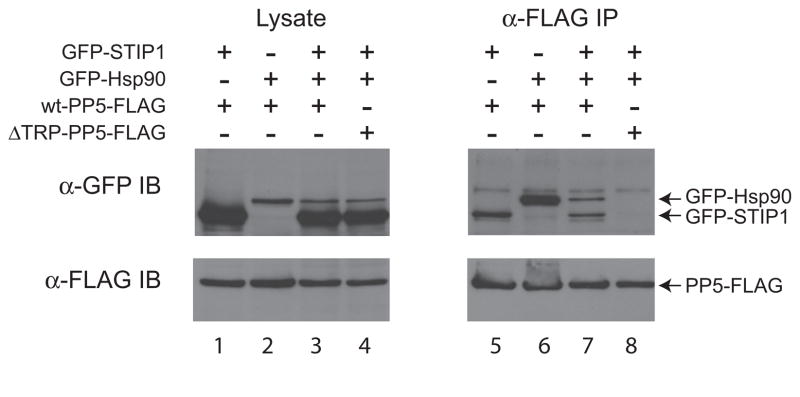
We next tested whether we could recapitulate the new interaction with STIP1 in this assay and to determine whether the interaction between STIP1 and PP5 may be affected by expression of Hsp90 (and vice versa). As shown above, GFP-Hsp90 was readily recovered with wt-PP5-FLAG (Figure 4; lane 6). GFP-STIP1 was also recovered with wt-PP5-FLAG (lane 5). Co-transfection of GFP-Hsp90 and GFP-STIP1 did not alter the amount of GFP-STIP1 recovered with wt-PP5-FLAG; however, Hsp90 recovery was markedly reduced (lane 7), indicating that STIP1 may affect association between PP5 and Hsp90. As expected, ΔTPR-PP5-FLAG was unable to interact with Hsp90 (Lane 8). In addition ΔTPR-PP5-FLAG did not interact with STIP1, confirming that the TPR domain of PP5 is needed for both interactions.
Figure 4. STIP1 interacts with wt-PP5, but not a TPR mutant.
Immunoprecipitation on anti-FLAG (M2 agarose) beads was performed on lysate from HEK293T cells transiently co-expressing the indicated FLAG- and GFP-tagged constructs. Immune complexes were resolved by SDS-PAGE followed by transfer to nitrocellulose. Co-precipitation of GFP-tagged proteins was detected by immunoblotting (IB) for the GFP tag (top panels; position of the tagged proteins are indicated by arrows). The precipitated FLAG-tagged protein was detected with anti-FLAG antibodies (bottom panels). Total protein lysate (left) was analyzed in parallel to the immunoprecipitation (right) to monitor protein expression.
4. Discussion
PP5 is expressed ubiquitously in human tissues and orthologs are highly conserved among species. Nonetheless, determining the cellular roles played by PP5 has been challenging. As described above, we have used a combination of AP-MS and statistical analysis of spectral count distribution to recapitulate the known interactions of PP5 with Hsp90 [18, 19], Cdc37[20, 21] and Hsp70 [22]. In addition, we identify STIP1 as a new interaction partner.
Currently, the biological roles played by PP5 are a matter of considerable debate. Many studies have used siRNA or antisense oligonucleotides to suppress PP5 protein levels, and the effects of constitutive PP5 expression in trans has also been examined in both yeast and human cells. These studies suggest that changing the level of PP5 protein affects the actions of: 1) transcription factors, including p53 [23], estrogen receptors [14, 24, 25] and glucocorticoid receptors [18, 26–28]; 2) protein kinases, including ASK1 [17, 29]), eIF2alpha kinase [21] ataxia telangiectasia mutated / ATM-Rad3-related (ATM/ATR)[30, 31], IKKbeta [32], JNK [17], and Raf1 [33]; 3) and other proteins, including the G12α /G13α subunits of heterotrimeric G proteins [34] Rac [35] and Tau [36]. Interestingly, all of the proteins listed above that are influenced by altering PP5 expression are known to interact with Hsp90, mostly as clients [37]. This may indicate that the interaction of PP5 with Hsp90 acts to modulate the chaperone activity of Hsp90, a concept that has been proposed previously [20, 21].
Alternatively, Hsp90 may act to activate PP5. In vitro purified PP5 has little catalytic activity, which mutational and structural studies indicate is due to interactions between an N-terminal inhibitory domain and a novel C-terminal J-helix [11–13, 38]. In both S. cerevisiae and human cells, PP5 associates with Hsp90 via interactions between the N-terminal TPR domains of PP5 and a C-terminal “TPR-dock” in Hsp90 [12, 28]. In reconstitution studies binding to Hsp90 disrupts the auto-inhibitory conformation maintained by the interaction of the TPR-domains with the J helix and catalytic domain. Therefore, the association of PP5 with Hsp90 may “activate” PP5 by allowing substrate access to the catalytic site [13]. This model for PP5 activation is supported by studies showing that when PP5 is contained within a heterocomplex with Cdc37 and Hsp90, PP5 can efficiently dephosphorylate Cdc37 [20].
4.1 Conclusions
While further studies are needed to clarify the biological roles of PP5, essentially all of the data obtained to date is consistent with the data presented here demonstrating that the interaction of PP5 with Hsp90 represents a biologically relevant event. This physiologically relevant interaction would be missed by the application of a simple frequency filter. For example, Hsp90 is co-purified with >70% of the baits analyzed by FLAG AP-MS in our internal interaction database (n >200; [2, 39–42]). Importantly, this test case also indicates that the quantitative and statistical analysis provided by SAINT offers an efficient tool for the identification of true interaction partners, even if these partners are detected as low level background noise across multiple IPs (or even negative control data). Within the context of larger-scale experiments, the application of such approaches will be essential for the identification of meaningful interactions involving proteins often removed as likely contaminants (e.g. cytoskeletal or ribosomal proteins).
Supplementary Material
Acknowledgments
We thank Karen Colwill and Tony Pawson for the pDEST cloning vectors, Mariana Gomez for removal of the EcoRI site in pcDNA5-FLAG and Brett Larsen for advice in mass spectrometry. Supported by grants from the CCSRI to A.C.G. (20203); the NIH, to A.I.N. and A.-C.G. (R01-GM094231), and to R.E.H. (CA60750); a Canada Research Chair in Functional Proteomics to A.C.G. and the Lea Reichmann Chair in Cancer Proteomics to A.C.G. Some of the studies for this investigation (D.V.S, R.E.H.) were conducted in a facility constructed with support from Research Facilities Improvement Program Grant (C06 RR11174) from the National (USA) Center for Research Resources.
Abbreviations
- SAINT
Significance Analysis of INTeractome
- AP-MS
Affinity purification coupled to mass spectrometry
- PP5
protein phosphatase 5 (gene name PPP5C)
- TPR
tetratricopeptide repeat
- Hsp90
heat shock protein 90 (gene name HSP90AB1)
- Hsp70
heat shock protein 70 (gene name HSPA1B)
- STIP1
stress-induced phosphoprotein 1
- Cdc37
cell division cycle 37 homolog (S. cerevisiae) (gene name CDC37)
Footnotes
Conflict of interest. None of the authors have a financial or commercial conflict of interest.
References
- 1.Chen GI, Gingras AC. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 2.Goudreault M, D’Ambrosio LM, Kean MJ, Mullin MJ, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breitkreutz A, Choi H, Sharom JR, Boucher L, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigaut G, Shevchenko A, Rutz B, Wilm M, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 5.Ho Y, Gruhler A, Heilbut A, Bader GD, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 6.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 7.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol Biosyst. 2007;3:354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 8.Sardiu ME, Cai Y, Jin J, Swanson SK, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi H, Larsen B, Lin Z-Y, Breitkreutz A, et al. SAINT: probabilistic scoring of affinity purification–mass spectrometry data. Nat Mehods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swingle MR, Honkanen RE, Ciszak EM. Structural basis for the catalytic activity of human serine/threonine protein phosphatase-5. J Biol Chem. 2004;279:33992–33999. doi: 10.1074/jbc.M402855200. [DOI] [PubMed] [Google Scholar]
- 12.Cliff MJ, Harris R, Barford D, Ladbury JE, Williams MA. Conformational diversity in the TPR domain-mediated interaction of protein phosphatase 5 with Hsp90. Structure. 2006;14:415–426. doi: 10.1016/j.str.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Roe SM, Cliff MJ, Williams MA, et al. Molecular basis for TPR domain- mediated regulation of protein phosphatase 5. Embo J. 2005;24:1–10. doi: 10.1038/sj.emboj.7600496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Golden T, Aragon IV, Rutland B, Tucker JA, et al. Elevated levels of Ser/Thr protein phosphatase 5 (PP5) in human breast cancer. Biochim Biophys Acta. 2008;1782:259–270. doi: 10.1016/j.bbadis.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingras AC, Caballero M, Zarske M, Sanchez A, et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Zhang J, Larsen B, Stark C, et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol. 28:1015–1017. doi: 10.1038/nbt1010-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou G, Golden T, Aragon IV, Honkanen RE. Ser/Thr protein phosphatase 5 inactivates hypoxia-induced activation of an apoptosis signal-regulating kinase 1/MKK-4/JNK signaling cascade. J Biol Chem. 2004;279:46595–46605. doi: 10.1074/jbc.M408320200. [DOI] [PubMed] [Google Scholar]
- 18.Silverstein AM, Galigniana MD, Chen MS, Owens-Grillo JK, et al. Protein phosphatase 5 is a major component of glucocorticoid receptor.hsp90 complexes with properties of an FK506-binding immunophilin. J Biol Chem. 1997;272:16224–16230. doi: 10.1074/jbc.272.26.16224. [DOI] [PubMed] [Google Scholar]
- 19.Russell LC, Whitt SR, Chen MS, Chinkers M. Identification of conserved residues required for the binding of a tetratricopeptide repeat domain to heat shock protein 90. J Biol Chem. 1999;274:20060–20063. doi: 10.1074/jbc.274.29.20060. [DOI] [PubMed] [Google Scholar]
- 20.Vaughan CK, Mollapour M, Smith JR, Truman A, et al. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Mol Cell. 2008;31:886–895. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao J, Hartson SD, Matts RL. Evidence that protein phosphatase 5 functions to negatively modulate the maturation of the Hsp90-dependent heme-regulated eIF2alpha kinase. Biochemistry. 2002;41:6770–6779. doi: 10.1021/bi025737a. [DOI] [PubMed] [Google Scholar]
- 22.Zeke T, Morrice N, Vazquez-Martin C, Cohen PT. Human protein phosphatase 5 dissociates from heat-shock proteins and is proteolytically activated in response to arachidonic acid and the microtubule-depolymerizing drug nocodazole. Biochem J. 2005;385:45–56. doi: 10.1042/BJ20040690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Urban G, Golden T, Aragon IV, Cowsert L, et al. Identification of a functional link for the p53 tumor suppressor protein in dexamethasone-induced growth suppression. J Biol Chem. 2003;278:9747–9753. doi: 10.1074/jbc.M210993200. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda K, Ogawa S, Tsukui T, Horie-Inoue K, et al. Protein phosphatase 5 is a negative regulator of estrogen receptor-mediated transcription. Mol Endocrinol. 2004;18:1131–1143. doi: 10.1210/me.2003-0308. [DOI] [PubMed] [Google Scholar]
- 25.Golden T, Aragon IV, Zhou G, Cooper SR, et al. Constitutive over expression of serine/threonine protein phosphatase 5 (PP5) augments estrogen-dependent tumor growth in mice. Cancer Lett. 2004;215:95–100. doi: 10.1016/j.canlet.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Leung DY, Nordeen SK, Goleva E. Estrogen inhibits glucocorticoid action via protein phosphatase 5 (PP5)-mediated glucocorticoid receptor dephosphorylation. J Biol Chem. 2009;284:24542–24552. doi: 10.1074/jbc.M109.021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rossie S, Jayachandran H, Meisel RL. Cellular co-localization of protein phosphatase 5 and glucocorticoid receptors in rat brain. Brain Res. 2006;1111:1–11. doi: 10.1016/j.brainres.2006.06.106. [DOI] [PubMed] [Google Scholar]
- 28.Chen MS, Silverstein AM, Pratt WB, Chinkers M. The tetratricopeptide repeat domain of protein phosphatase 5 mediates binding to glucocorticoid receptor heterocomplexes and acts as a dominant negative mutant. J Biol Chem. 1996;271:32315–32320. doi: 10.1074/jbc.271.50.32315. [DOI] [PubMed] [Google Scholar]
- 29.Morita K, Saitoh M, Tobiume K, Matsuura H, et al. Negative feedback regulation of ASK1 by protein phosphatase 5 (PP5) in response to oxidative stress. Embo J. 2001;20:6028–6036. doi: 10.1093/emboj/20.21.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Bao S, Furumai R, Kucera KS, et al. Protein phosphatase 5 is required for ATR-mediated checkpoint activation. Mol Cell Biol. 2005;25:9910–9919. doi: 10.1128/MCB.25.22.9910-9919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali A, Zhang J, Bao S, Liu I, et al. Requirement of protein phosphatase 5 in DNA- damage-induced ATM activation. Genes Dev. 2004;18:249–254. doi: 10.1101/gad.1176004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang CW, Liu WK, Chiang CW, Chou CK. Phosphorylation-dependent association of the G4-1/G5PR regulatory subunit with IKKbeta negatively modulates NF-kappaB activation through recruitment of protein phosphatase 5. Biochem J. doi: 10.1042/BJ20100247. [DOI] [PubMed] [Google Scholar]
- 33.von Kriegsheim A, Pitt A, Grindlay GJ, Kolch W, Dhillon AS. Regulation of the Raf-MEK-ERK pathway by protein phosphatase 5. Nat Cell Biol. 2006;8:1011–1016. doi: 10.1038/ncb1465. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi Y, Katoh H, Mori K, Negishi M. Galpha(12) and Galpha(13) interact with Ser/Thr protein phosphatase type 5 and stimulate its phosphatase activity. Curr Biol. 2002;12:1353–1358. doi: 10.1016/s0960-9822(02)01034-5. [DOI] [PubMed] [Google Scholar]
- 35.Gentile S, Darden T, Erxleben C, Romeo C, et al. Rac GTPase signaling through the PP5 protein phosphatase. Proc Natl Acad Sci U S A. 2006;103:5202–5206. doi: 10.1073/pnas.0600080103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong CX, Liu F, Wu G, Rossie S, et al. Dephosphorylation of microtubule- associated protein tau by protein phosphatase 5. J Neurochem. 2004;88:298–310. doi: 10.1111/j.1471-4159.2004.02147.x. [DOI] [PubMed] [Google Scholar]
- 37.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 38.Kang H, Sayner SL, Gross KL, Russell LC, Chinkers M. Identification of amino acids in the tetratricopeptide repeat and C-terminal domains of protein phosphatase 5 involved in autoinhibition and lipid activation. Biochemistry. 2001;40:10485–10490. doi: 10.1021/bi010999i. [DOI] [PubMed] [Google Scholar]
- 39.O’Donnell L, Panier S, Wildenhain J, Tkach JM, et al. The MMS22L-TONSL Complex Mediates Recovery from Replication Stress and Homologous Recombination. Mol Cell. 40:619–631. doi: 10.1016/j.molcel.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawo S, Bashkurov M, Mullin M, Ferreria MG, et al. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 41.Chen GI, Tisayakorn S, Jorgensen C, D’Ambrosio LM, et al. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem. 2008;283:29273–29284. doi: 10.1074/jbc.M803443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakada S, Tai I, Panier S, Al-Hakim A, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 466:941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.