SUMMARY
The Bacillus subtilis genome encodes three apparent lipoyl ligase homologues: yhfJ, yqhM, and ywfL which we have renamed lplJ, lipM and lipL, respectively. We show that LplJ encodes the sole lipoyl ligase of this bacterium. Physiological and biochemical characterization of a ΔlipM strain showed that LipM is absolutely required for the endogenous lipoylation of all lipoate-dependent proteins, confirming its role as the B. subtilis octanoyltransferase. However, we also report that in contrast to E. coli, B. subtilis requires a third protein for lipoic acid assembly, LipL. B. subtilis ΔlipL strains are unable to synthesize lipoic acid despite the presence of LipM and the sulfur insertion enzyme, LipA, which should suffice for lipoic acid biosynthesis based on the E. coli model. LipM is only required for the endogenous lipoylation pathway, whereas LipL also plays a role in lipoic acid scavenging. Expression of E. coli lipB allows growth of B. subtilis ΔlipL or ΔlipM strains in the absence of supplements. In contrast, growth of an E. coli ΔlipB strain can be complemented with lipM, but not lipL. These data together with those of the companion paper (Christensen et al., 2011) provide evidence that LipM and LipL catalyze sequential reactions in a novel pathway for lipoic acid biosynthesis.
INTRODUCTION
Lipoic acid is a sulfur-containing coenzyme found in all domains of life that is required for the function of several key multienzyme complexes involved in oxidative and one-carbon metabolism. Five lipoate-dependent multienzyme complexes have been characterized. Three are closely related 2-oxoacid dehydrogenases: pyruvate dehydrogenase (PDH), 2-oxoglutarate dehydrogenase (OGDH) and branched-chain 2-oxoacid dehydrogenase (BKDH). These complexes are composed of multiple copies of each of three catalytic components referred as E1 (often produced as two proteins), E2, and E3. A fourth complex, acetoin dehydrogenase, is highly homologous to PDH and shares the three-subunit architecture of the 2-oxoacid dehydrogenases but produces acetaldehyde instead of carbon dioxide. The fifth complex, the glycine cleavage complex, has a markedly different architecture and is composed of four loosely associated proteins, called P, H (GcvH), T and L proteins (Perham, 2000). Lipoic acid is linked through an amide bond to a specific lysine residue of the lipoyl domains of the E2 and GcvH proteins where it acts as a swinging arm in transfer of covalently attached reaction intermediates among the multiple active sites of the enzyme complexes (Perham, 2000).
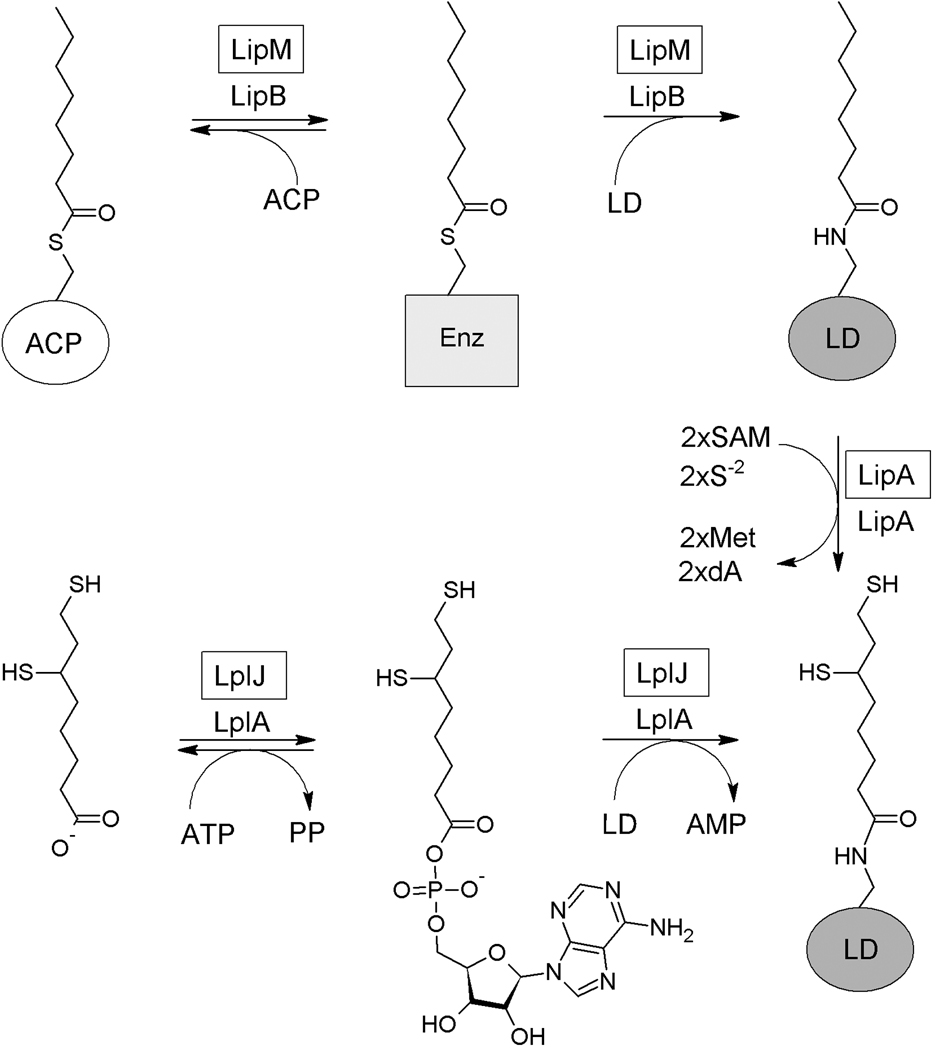
In Escherichia coli lipoylation is directly carried out by lipoyl protein ligase A (LplA) which uses exogenous lipoic acid in an ATP-dependent reaction (Morris et al., 1995, Morris et al., 1994). However, this is a scavenging rather than a biosynthetic pathway. The de novo synthesis pathway proceeds by two consecutive reactions. In the first step the octanoyl-acyl carrier protein (ACP):protein-N-octanoyltransferase (LipB) transfers endogenously produced octanoyl moieties to the target proteins (Jordan & Cronan, 2003, Morris et al., 1995, Morris et al., 1994). In the second step the octanoyl moieties are converted to lipoic acid by the S-adenosyl-L-methionine (SAM) radical enzyme, lipoyl synthase (LipA) which replaces a hydrogen atom on each of the octanoate C6 and C8 carbons atoms with a sulfur atom (Cicchillo & Booker, 2005, Zhao et al., 2003) (Fig. 1).
Fig. 1.
Current model for lipoic acid synthesis and scavenging in E. coli. E. coli uses two independent pathways for lipoic acid synthesis and scavenging. The endogenous synthesized octanoate is transferred from acyl carrier protein (ACP) to apoproteins by LipB, an octanoyl-ACP:protein-N-octanoyltransferase. The octanoylated domains then become substrates for sulfur insertion by LipA. Exogenous lipoate (or octanoate) is transferred to unmodified acceptor lipoyl domains (LD) in an ATP-dependent two-step reaction catalyzed by LplA. The dA designation denotes 5’-deoxyadenosine. The B. subtilis protein that catalyzes the same reaction as the E. coli protein is given in a box above the E. coli enzyme.
Homologues of the E. coli lipoic acid metabolism proteins are found in all domains of life and thus unraveling the pathways by which this cofactor is synthesized and transferred to lipoate-dependent proteins is of broad biological significance. In contrast to the wealth of knowledge available on lipoic acid synthesis and utilization in E. coli, the existing information about these pathways in Gram-positive bacteria is fragmentary. It has been found that Listeria monocytogenes defective in proteins homologous to the E. coli LplA enzymes are unable to scavenge lipoic acid for modification of lipoyl domains (O'Riordan et al., 2003), However, L. monocytogenes is a natural lipoate auxotroph and does not encode the enzymes necessary for lipoic acid biosynthesis. Lipoic acid synthesis and attachment to target proteins is less understood in other organisms. Despite the presence of homologues to the E. coli enzymes in fungi, plants, protists and mammals, the mechanistic details of lipoic acid synthesis still remain unclear (Fujiwara et al., 1999, Marvin et al., 2001, Thomsen-Zieger et al., 2003).
Since B. subtilis cells grown on minimal media were known to contain essential lipoate-modified proteins, this bacterium must synthesize lipoic acid and this was confirmed by demonstration of a functional lipoyl synthase (Martin et al., 2009). Moreover, the lipoate requirement of B. subtilis ΔlipA strains for growth in minimal medium was bypassed by addition of acetate and a mixture of three short-branched-chain carboxylic acids: 2-methyl butyrate, isobutyrate, and isovalerate, metabolites that yield the products of two lipoylated enzymes, PDH and BKDH (acetate utilization is mediated by acetyl-CoA synthetase, the product of the acs gene, while an unknown enzyme converts the carboxylic acids to their CoA esters in vivo). Succinate supplementation to bypass the OGDH deficiency engendered by lipoic acid starvation was not required (Martin et al., 2009).
However, despite the presence of a functional LipA, the B. subtilis genome contained no open reading frame (ORF) that resembled the E. coli LipB octanoyltransferase, an activity required for production of the substrate for LipA. Recently, an LplA homologue named LipM was identified and shown to have octanoyltransferase activity in vitro (Christensen & Cronan, 2010). We began the present study due to paucity of information on lipoic acid biosynthesis in Gram-positive bacteria. Although LipM together with LipA should be sufficient for lipoic acid biosynthesis, the only in vivo analyses reported were performed in E. coli (Martin et al., 2009; Christensen & Cronan, 2010). Moreover, the additional uncharacterized LplA homologues encoded in the genome suggested that B. subtlis lipoic acid metabolism may be more complex than in E. coli and we have found that this is the case. We have now tested the role of LipM in B. subtlis and confirmed its proposed role as octanoyltransferase in vivo. In addition to lipM, the B. subtlis genome contains two ORFs, yhfJ and ywfL, which encode proteins having significant sequence similarity to characterized lipoyl ligases. In this study we show that these genes are involved in lipoic acid metabolism (see below), so we have renamed yhfJ as lplJ and ywfL as lipL. These genes have been characterized by genetic, physiological and biochemical analyses and the resulting data demonstrate that B. subtilis synthesizes lipoic acid by a novel mechanism.
RESULTS
LipM is responsible for octanoyl transfer in vivo
B. subtilis was recently demonstrated to encode a functional lipoate synthase called LipA (Martin et al., 2009). However, BLAST searches against B. subtilis genome showed no open reading frame (ORF) that resembled the E. coli LipB octanoyltransferase, an activity required for production of the substrate of LipA. Instead, two ORFs, YhfJ and YqhM, annotated as encoding putative lipoyl ligases were present that, respectively, shared 33% and 23% identity with E. coli LplA. A third ORF, YwfL that encoded a protein of unknown function having 22% identity with YhfJ was also found. Since all three genes are involved in lipoic acid metabolism (see below), they have been renamed. In this paper we have renamed yhfJ and ywfL as lplJ and lipL, respectively, whereas yqhM was previously renamed lipM (Christensen & Cronan, 2010).
Recently, cosmids containing B. subtilis genomic fragments were isolated that complemented growth of an E. coli lipB strain and failed to complement an lplA lipA strain (Christensen & Cronan, 2010). All complementing cosmids contained the lipM gene and this gene was shown to be responsible for restoration of lipoic acid synthesis to the E. coli lipB strain. The pattern of complementation indicated that lipM encoded an octanoyltransferase and the LipM protein was shown to catalyze octanoyl transfer in vitro by the same general acyl-enzyme intermediate mechanism used by LipB (Christensen & Cronan, 2010). Based on the frequency that complementing cosmid clones were found, it was suggested that LipM might be the sole B. subtilis octanoyltransferase. However, no lipoate ligase encoding cosmids were isolated and since growth of the B. subtilis lipA strain on lipoic acid would require ligase activity, the question of cosmid bank bias was raised (Christensen & Cronan, 2010). To definitively test whether or not LipM was the sole B. subtilis octanoyltransferase we constructed strain NM57 in which lipM was replaced with a kanamycin-resistance determinant and performed its physiological and biochemical characterization.
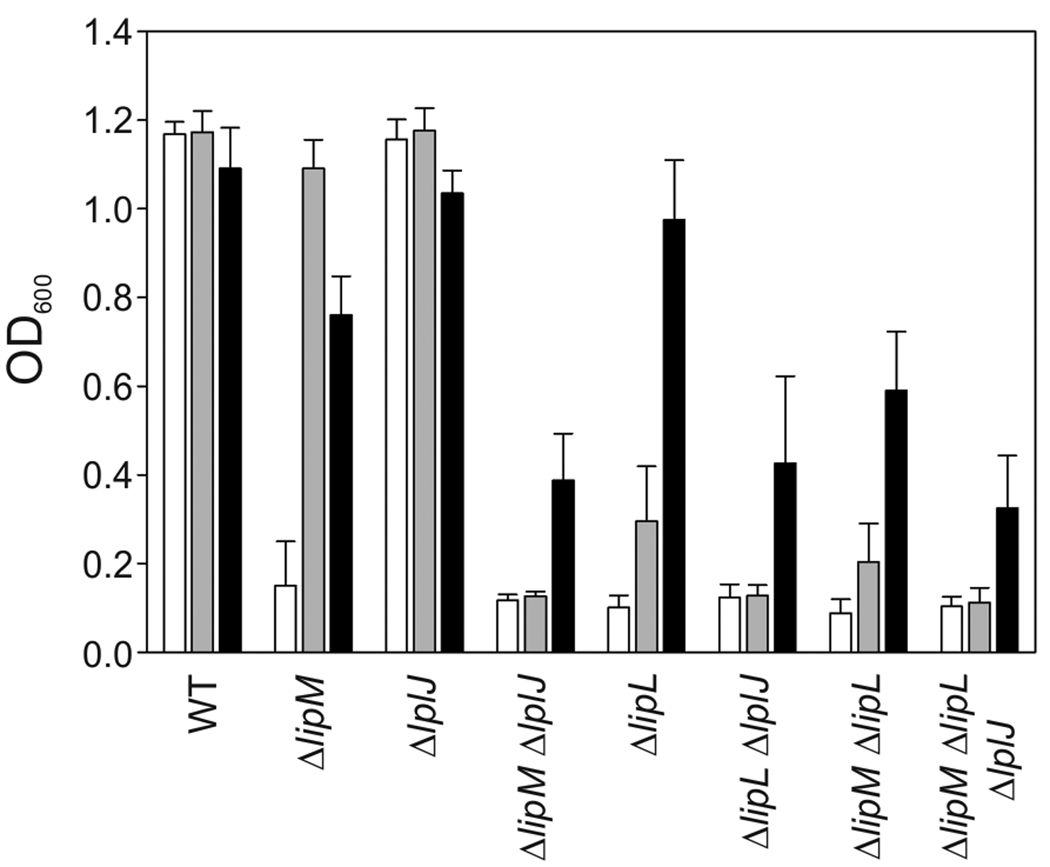
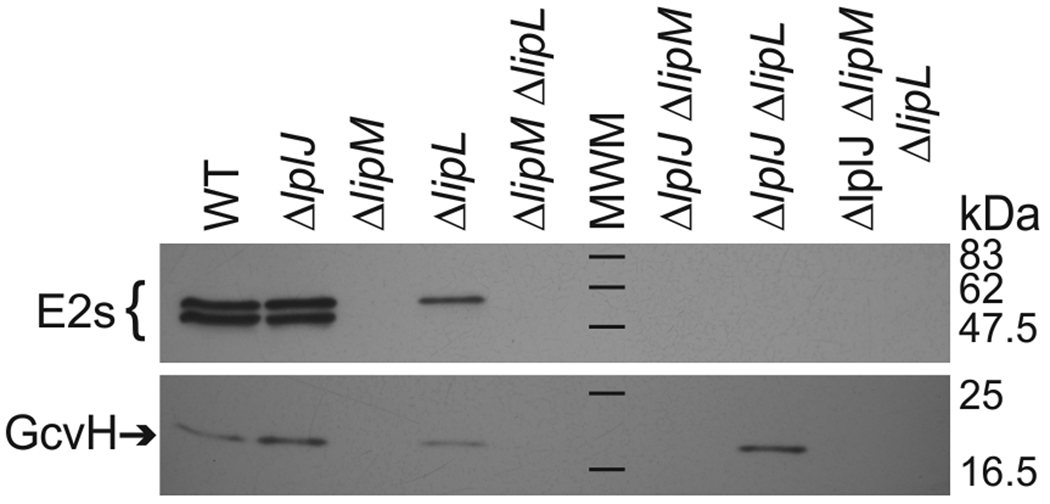
Strain NM57 (ΔlipM) was auxotrophic for lipoic acid when grown in minimal medium but grew as well as the wild type strain, JH642, in the presence of lipoic acid (Figs. 2 and 3A). As previously observed for a ΔlipA mutant strain (Martin et al., 2009) the requirement for lipoic acid could be bypassed by addition of both acetate and branched chain fatty acid (BCFA) precursors (Figs. 2 and 3A), which upon conversion to their CoA esters are the products of the two lipoylated B. subtilis enzymes PDH and BKDH required for growth in minimal medium. Cells grown in this medium were devoid of lipoylated proteins detectable by western blotting (Fig. 4). Mass spectrometry also demonstrated that lipM was required for modification of lipoyl domains by the biosynthetic pathway (see below).
Fig. 2.
Growth phenotypes of B. subtilis mutant strains The strains were grown overnight in minimal medium supplemented with acetate and branched chain fatty acid (BCFA) precursors. The cultures were centrifuged and the cells resuspended in minimal medium without supplements (white bars); minimal medium supplemented with lipoic acid (grey bars) or with acetate and BCFA precursors (black bars). The OD600 values of the cultures were measured after 17 h of growth at 37°C. The results shown are the averages of results from three independent experiments.
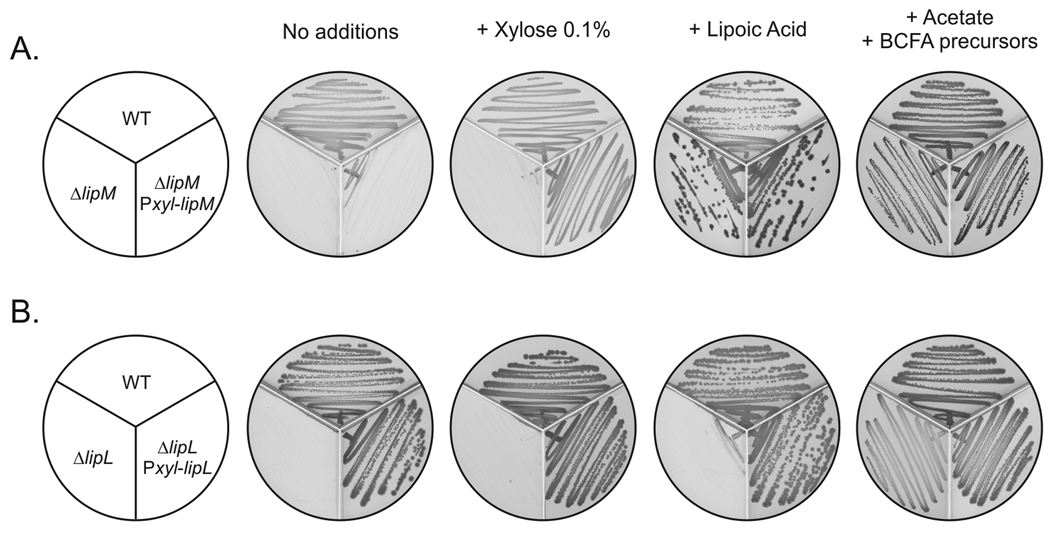
Fig. 3.
Characterization of B. subtilis strains blocked in lipoic acid (LA) synthesis. Panel A. Growth of bacterial strains JH642 (wild type), NM57 (ΔlipM) and NM08 (ΔlipM amyE::Pxyl-lipM). Panel B. Growth of wild type (JH642), NM51 (ΔlipL) and NM13 (ΔlipL amyE::Pxyl-lipL). The strains were streaked onto minimal medium-glycerol plates containing the supplements indicated above and incubated for 24 h at 37°C.
Fig. 4.
Immunoblotting analyses of mutant strains with an anti-lipoic acid antibody The strains were grown overnight in minimal medium supplemented with acetate and BCFA precursors. The cells were diluted in fresh medium of the same composition and grown for 22 h before analysis. The black bars denote the molecular weight standards (MWM).
To confirm that the growth phenotype observed in this mutant strain was due to the absence of lipM, complementation analyses were carried out. A construct in which lipM was placed under control of a xylose-inducible promoter (PxylA) was introduced into the ΔlipM strain NM57 giving the lipM amyE::PxylA-lipM strain, NM08. Induction of lipM expression in strain NM08 allowed growth in minimal medium thereby indicating that the absence of a functional copy of lipM (rather than a polar effect on the downstream genes) was the cause of the growth phenotype of strain NM57 (Fig. 3A). Moreover, LipM could be functionally replaced by expression of E. coli lipB. The lipB gene was placed under PxylA and the construct was introduced into the ΔlipM strain NM57 to give the lipM amyE::PxylA-lipB strain, NM11. Upon induction of LipB expression strain NM11 grew in minimal medium, indicating that the LipB octanoyl transferase activity functionally replaced LipM. Similar complementation experiments were performed with a ΔlipL strain (see below). These results together with those of Christensen and Cronan (Christensen & Cronan, 2010) demonstrate that LipM functions as the sole B. subtilis octanoyltransferase.
Bacillus subtilis LplJ is a lipoate:protein ligase
The ability of exogenous lipoic acid to allow growth of both a ΔlipM strain and a lipA conditional mutant indicated that B. subtilis must encode a lipoate scavenging activity (Martin et al., 2009). The most likely candidate for this role was YhfJ (LplJ), which was annotated as a putative lipoyl ligase and has 33% identity to E. coli LplA. We first found that expression of lplJ restored E. coli lipB lplA strains to prototrophy (Fig. 5A). Since prototrophy could result from either octanoyltransfer or ligation of traces of endogenous octanoate (Hermes & Cronan, 2009), we also tested complementation of a E. coli lplA lipA strain in the presence of lipoate. Growth was also restored in this strain (Fig. 5B) and it was accompanied by activation (hence lipoate modification) of the E. coli 2-oxoacid dehydrogenases (Fig. 5C). Therefore, in complementation tests LplJ behaved like LplA. We then constructed strain NM60, a B. subtilis ΔlplJ strain. The ΔlplJ strain grew normally in minimal medium in the absence of supplements (Fig. 2) and had a wild type pattern of lipoylated proteins (Fig. 4). This behavior was expected because ΔlplA strains of E. coli lacking lipoate scavenging activity show growth defects only when the strains are additionally blocked in the lipoic acid synthetic pathway by a lipA or lipB mutation (Morris et al., 1995, Morris et al., 1994) (Fig. 1). Based on this precedent the ΔlipM ΔlplJ double mutant strain, NM65, was constructed. As expected, this strain was found to be unable to grow in minimal medium either in the presence or absence of lipoic acid (Fig. 2), indicating that LplJ is the sole B. subtilis lipoic acid salvage enzyme.
Fig. 5.
Complementation of E. coli lipoic acid auxotrophs with B. subtilis genes as described in Experimental Procedures. Panel A: Growth of the lipB lplA strain TM136 containing plasmids expressing the genes indicated on the figure. Culture absorbance at 600 nm is reported. Panel B: Growth of lipA lplA strain TM131 containing plasmids expressing genes indicated as in panel A Panel C: The 2-oxoacid dehydrogenase activities of the lipB lplA strain TM136 carrying plasmids encoding the genes indicated. The wild type strain was strain JK1. Panel D: Growth of lipB lplA fabA strain QC168 containing plasmids expressing the genes indicated. Note that the host strain was fabA to prevent possible decanoyl adduct modification of the B. subtilis proteins. Growth of the strain carrying the p(lplJ) plasmid on the unsupplemented plate is due to scavenging of the endogenous octanoic acid present in E. coli by high levels of ligase (Hermes & Cronan, 2009).
Given that proteins with significant sequence identity to E. coli LplA have been shown to catalyze octanoyl transfer, it could not be assumed that the B. subtilis lipoate salvage enzyme catalyzed a ligase reaction. Therefore, we purified the hexahistidine-tagged protein to homogeneity (Fig. 6A) and assayed it for both the overall and first partial reaction of the ligase reaction. In the absence of an acceptor domain, synthesis of both octanoyl-adenylate and lipoyl-adenylate intermediates was readily demonstrated by use of ATP labeled in the α-phosphate. Moreover, upon addition of an acceptor lipoyl domain the adenylate intermediates were hydrolyzed to AMP (Fig. 6B). Gel mobility shift assays showed that LplJ modified the E. coli lipoyl domain from PDH (E2AceF) and GcvH with either lipoate or octanoate. Western blotting with anti-lipoic acid antibody confirmed lipoylation of the acceptor proteins and demonstrated that the antibody recognized lipoylated proteins and not octanoylated proteins (Fig. 6C). Finally, modification of GcvH was confirmed by electrospray mass spectrometry of the reaction products (Fig. 6D). From these data we conclude that B. subtilis LplJ catalyzes a classical lipoate ligase reaction analogous to that of E. coli LplA.
Fig. 6.
The B. subtilis lipoate salvage enzyme is a lipoate-protein ligase. Panel A: An SDS-PAGE gel (4–20% acrylamide) containing ~2 µg of each of the indicated purified proteins is shown. The molecular weight marker standards in kilodaltons are given. Panel B: Formation of the acyl adenylate intermediates by LplJ in the first partial reaction of the ligase reaction using [α-32P]ATP incorporation assayed by thin layer chromatography. Addition of an acyl acceptor domain results in consumption of the intermediate with production of AMP. Panel C: Gel shift assay of LplJ using either the E2AceF E. coli hybrid lipoyl domain or B. subtilis GcvH as acyl acceptors. The proteins were visualized by either Coomassie R250 staining or by western blotting with anti-lipoic acid antibody (Anti-LA) as indicated. Panel D: Electrospray mass spectra of GcvH after LplJ treatment with either octanoate or lipoate as acyl donors. Panel E: Assay of purified proteins for octanoyl ligation using GcvH as the substrate acceptor domain. NE denotes the control reaction that lacked enzyme.
The unexpected requirement for LipL
A third ORF, lipL (formerly ywfL), encodes a protein annotated as of unknown function. LipL seems more divergent from authentic lipoyl ligases than LipM and has only 22% sequence identity with LplJ. Our results, together with those previously reported (Christensen & Cronan, 2010, Martin et al., 2009), indicated that B. subtilis like E. coli has an octanoyltransferase (LipM), a sulfur insertion enzyme (LipA) and a lipoate ligase (LplJ). However, unlike E. coli, these enzymes were not sufficient for either synthesis or efficient scavenging of lipoic acid as demonstrated by the phenotype of B. subtilis ΔlipL strains. Strains devoid of LipL were unable to grow in minimal medium as observed for strains lacking either lipM (Fig. 1) or lipA (Martin et al., 2009). However, in contrast to the growth phenotypes of lipM and lipA strains, addition of lipoic acid only partially restored growth of the ΔlipL strain NM51. The growth behaviour of the ΔlipL strain denoted a block in the endogenous lipoylation pathway because, like the lipA conditionally mutant strain and ΔlipM strains, the ΔlipL strain grew as well as its wild type parent in minimal medium supplemented with acetate and BCFA precursors.
The residual growth of the ΔlipL strain in minimal medium containing lipoic acid was due to LplJ activity because the ΔlipL ΔlplJ double mutant strain, NM67, failed to grow in this medium. Hence the ΔlipL strain retained some ability to transfer exogenously provided lipoate to the unmodified acceptor proteins. Expression of a functional copy of the lipL gene under the control of a xylose-dependent promoter in the non-essential amyE locus restored growth of the ΔlipL strain in minimal medium (Fig. 2B) indicating that the absence of a functional copy of lipL was the cause of the growth phenotype of strain NM51.
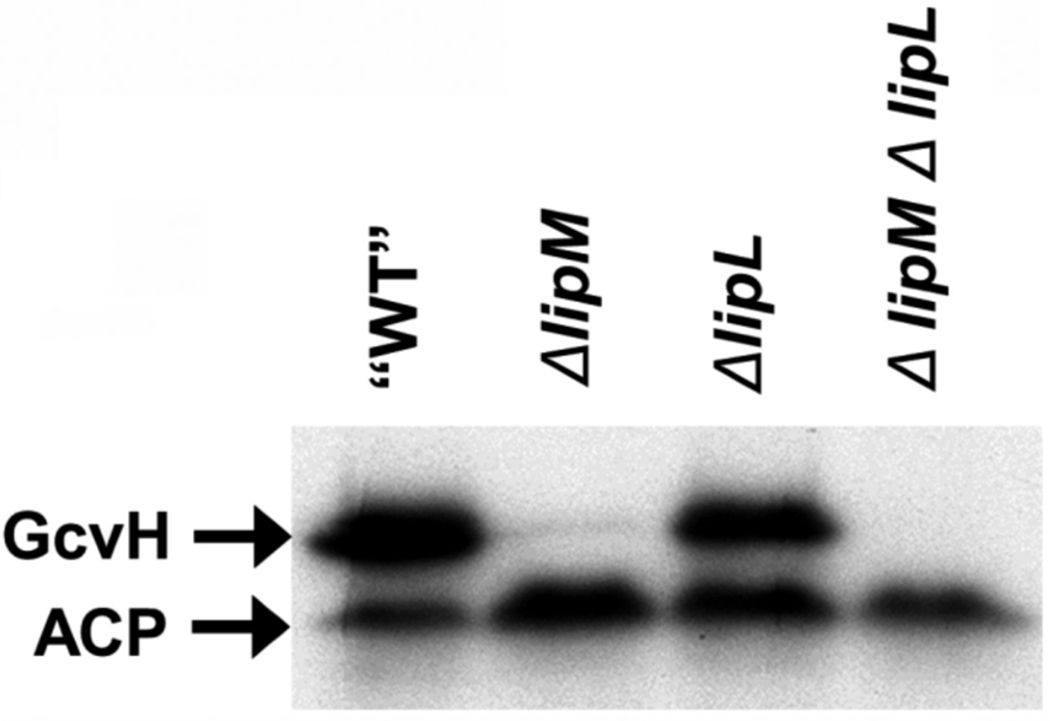
The growth phenotype of the ΔlipL strain indicated that it should have decreased levels of protein lipoylation and any residual modification should be abolished upon introduction of a ΔlplJ lesion. This was the case (Fig. 4). Immunoblot analysis of crude extracts of strain NM51 (ΔlipL) with anti-lipoate antibodies showed that GcvH was lipoylated to a level similar to that seen in the wild type strain. However, only one of the two high molecular weight bands observed in the wild type strain was detected in strain NM51 and thus the total amount of protein lipoylation of this strain was more than 3-fold less than those seen in extracts of the wild type strain (Fig. 4). Strain NM51 required both acetate and BCFA precursors for growth indicating that the BKDH and PDH E2 subunits of were nonfunctional in absence of LipL. Thus, the band observed seems likely to be the nonessential OGDH E2 subunit that may migrate together with low levels of lipoylated PdhC insufficient to overcome the acetate requirement. The putative OGDH E2 band was not present in extracts of the ΔlipL ΔlplJ strain, NM67, (Fig. 4) indicating that LplJ was responsible for the residual lipoylation seen in the absence of lipoic acid supplementation. Interestingly, in this strain the immunoblot signal of GcvH was about two-fold stronger than that of extracts from the wild type strain (Fig. 4). This observation suggests that when GcvH is lipoylated by the concerted action of LipM and LipA, lipoyl-GcvH accumulated because in the absence of LipL and LplJ the GcvH lipoyl moiety could not be transferred to other lipoyl acceptors. Another possible explanation would be that LplJ scavenges and attaches octanoate to the acceptor proteins to serve as a LipA substrate as occurs in E. coli (Hermes & Cronan, 2009). However, this does not seem to be the case in B. subtilis since no lipoylated proteins were detected in a ΔlipM strain (Fig. 4).
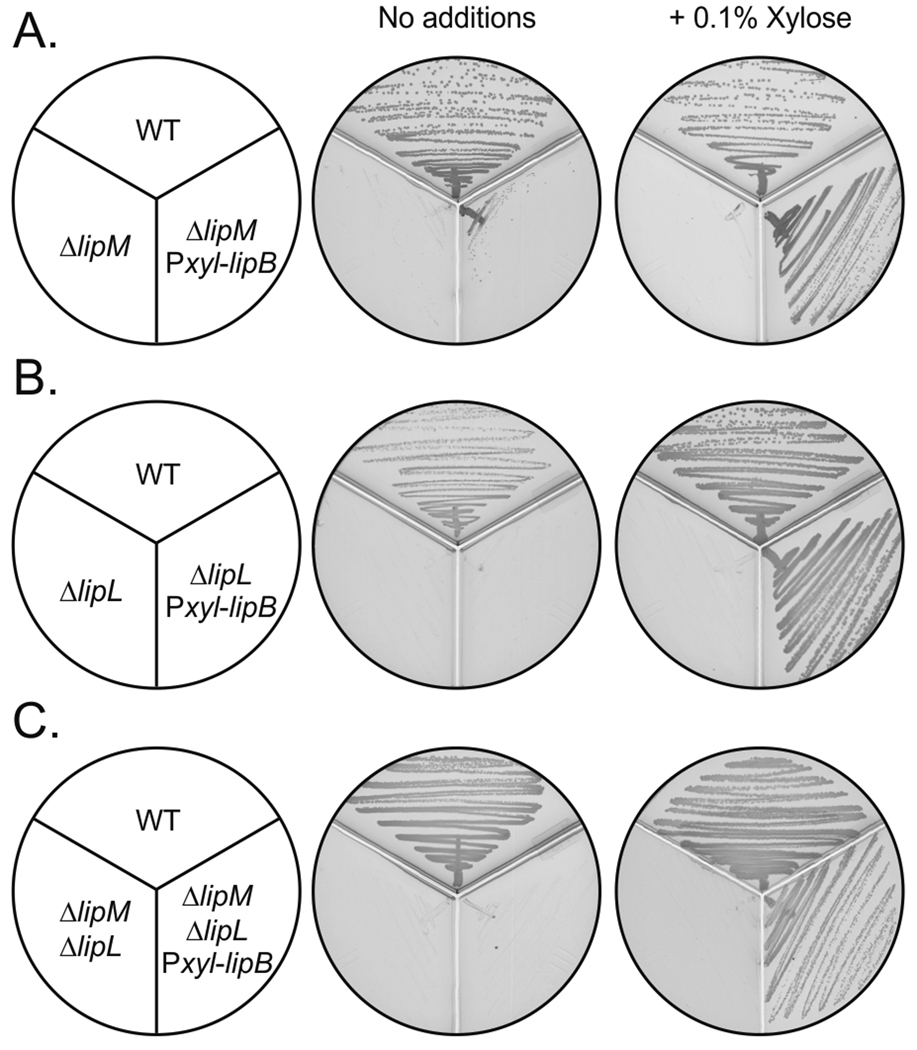
To provide further evidence for the role of LipL in protein lipoylation, complementation experiments with E. coli lipB were performed. As described above, lipB was placed under a xylose-inducible promoter (PxylA) and this construct was introduced into the chromosome of B. subtilis strain NM51 to give strain NM12 (ΔlipL amyE::Pxyl-lipB). Induction of LipB expression allowed growth not only of the ΔlipL strain NM12, but also of the ΔlipM ΔlipL double mutant strain CM28 (Fig. 7). However, when expressed in E. coli mutant strains, LipL had no detectable ability to replace either E. coli LplA or LipB (Fig. 5). A plausible explanation for this observation was that LipL might become inactive when expressed in E. coli. When B. subtilis LipM is expressed in E. coli, some molecules carry a covalently attached decanoate moiety derived from cis-3-decenoyl-ACP, an essential intermediate of unsaturated acid biosynthesis in E. coli (Christensen & Cronan, 2010) that is not present in B. subtilis. A similar inability to exclude cis-3-decenoyl-ACP was reported for Mycobacterium tuberculosis LipB (Ma et al., 2006). In the case of LipM an unmodified protein was obtained by expression of the protein in an E. coli fabA mutant which lacks the ability to make cis-3-decenoyl-ACP (Christensen et al., 2011). The inability of LipL to relieve the lipoate requirement cannot be due to formation of a covalently attached decanoate because a fabA lipB lplA strain also failed to grow (Fig. 5D).
Fig. 7.
Complementation of B. subtilis mutants deficient in lipoate synthesis by expression of E. coli LipB. B. subtilis strains JH642 (wild type), NM57 (ΔlipM), NM11 (ΔlipM amyE::Pxyl-lipB), NM51 (ΔlipL), NM12 (ΔlipL amyE::Pxyl-lipB), CM28 (ΔlipL ΔlipM) and NM14 (ΔlipL ΔlipM amyE::Pxyl-lipB) were streaked onto minimal medium-glycerol plates with or without the addition of 0.1% xylose and incubated for 24 h at 37°C.
Although LipL expression failed to complement the growth of an E. coli ΔlipB strain, lipB expression allowed growth of a B. subtilis ΔlipL strain on minimal medium. These results indicated that LipM and LipL are both required for octanoylation of E2 lipoyl domains and that the two proteins either catalyze sequential reactions or participate in octanoyl transfer as a complex. The latter explanation seems unlikely since LipM expressed in E. coli is active both in vivo and in vitro (Christensen & Cronan, 2010). Moreover, crude extracts of B. subtilis ΔlipL mutants readily catalyzed the LipM reaction; transfer of octanoate from octanoyl-ACP to GcvH (Fig. 8).
Fig. 8.
LipM acts before LipL. Autoradiogram of an SDS-PAGE gel of the products of octanoyltransfer assays from octanoyl-ACP (synthesized with AasS) to GcvH. The enzyme source were crude extracts of the indicated B. subtilis strains. All strains, including the “WT”, carried an ΔlplJ lesion to prevent possible complications by ligation of octanoate.
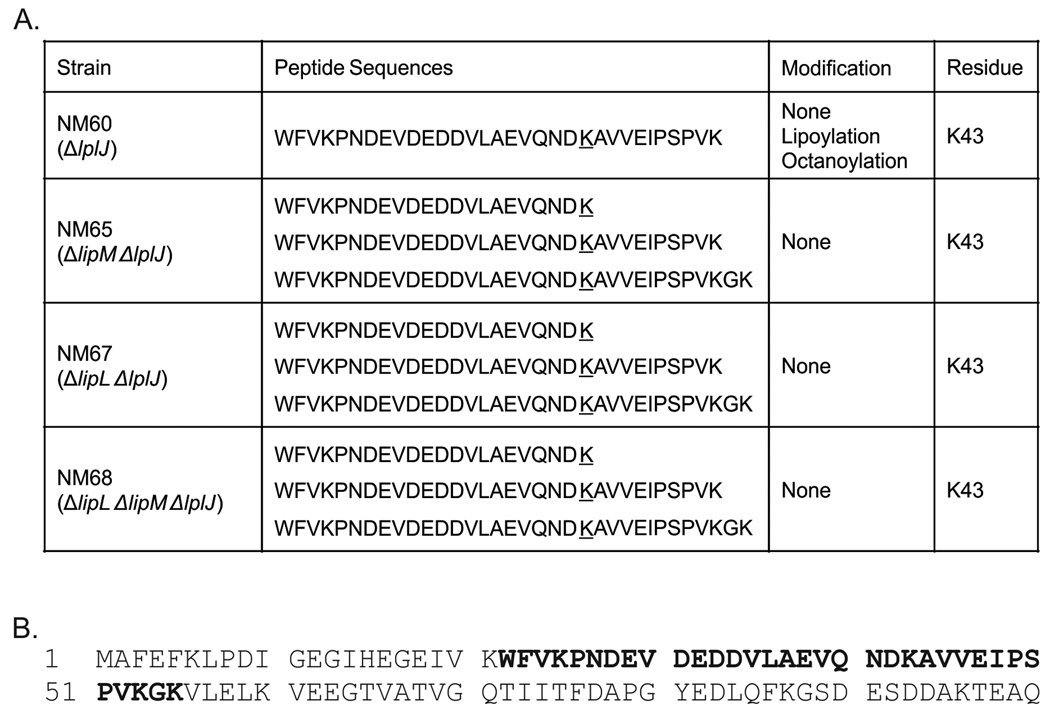
Given that lipoic acid synthesis proceeds through several sequential steps it seemed possible that intermediates attached to the lipoyl domains of the E2 subunits might accumulate in some mutant strains. Therefore we purified the PDH complexes of wild type and various mutant strains and released their lipoyl domains by limited proteolysis with the Staphylococcus aureus V8 glutamyl protease as previously described (Packman et al., 1988). The isolated domains were resolved from the other digestion products by polyacrylamide gel electrophoresis run under native conditions. The gel slices containing the domain bands were excised, crushed and subjected to in-gel trypsin digestion. The resulting peptides were analyzed by liquid chromatography-tandem mass spectrometry as described in Experimental Procedures. The PDH complexes were purified from extracts of strains NM60 (ΔlplJ); NM65 (ΔlipM ΔlplJ); NM67 (ΔlipL ΔlplJ) and NM68 (ΔlipM ΔlipL ΔlplJ) as described in Experimental Procedures (the strains all lacked LplJ in order to preclude ligation of any traces of octanoic acid or lipoic acid present in the culture medium). PDH peptides were detected with significant scores in all samples (Fig. 9) and modification was determined using an error tolerant search as described in Experimental Procedures to determine the presence or absence of posttranslational modifications of the target lysine residue (K43). K43 was found in three forms unmodified, octanoylated and lipoylated in the peptides derived from the ΔlplJ strain NM60 whereas the peptides of the other strains were unmodified (Fig. 9). These results indicate that PDH was not modified in ΔlplJ strains when either LipM or LipL was nonfunctional and that LipL is required for E2 lipoyl domain modification rather than for sulfur insertion.
Fig. 9.
Liquid chromatography-tandem mass spectrometric analysis of PDH lipoyl domain tryptic peptides. Panel A. The modification states of PDH residue K43 detected. Panel B. The sequence of the PDH lipoyl domain is given. The sequences of the peptides detected that contained K43 are shown in bold type. “None” denotes that no distinct peaks for modified forms were observed.
Although no novel PDH E2p-bound intermediates were present, this does not rule out a sequential mechanism in which another protein functions as an octanoyl/lipoyl carrier. Indeed the observed accumulation of lipoylated GcvH in a ΔlipL ΔlplJ strain (Fig. 4) suggested that this small protein was a good candidate for an octanoyl/lipoyl carrier.
DISCUSSION
Lipoic acid synthesis in B. subtilis is clearly more complex than in E. coli. E. coli requires only two proteins to make this cofactor, whereas in this paper we demonstrate that B. subtilis requires three proteins, LipA, LipM (an isozyme of LipB), and an unexpected protein LipL. Inactivation of any of the three genes that encode these proteins results in lipoic acid auxotrophy. The two LipA proteins are interchangeable between B. subtilis and E. coli as are LipM and LipB. In contrast, LipL has a curious behavior. Its expression in E. coli fails to complement a ΔlipB mutation whereas expression of LipB complements a B. subtilis ΔlipL strain. Therefore, LipL and LipM have distinct roles in lipoate biosynthesis. A plausible hypothesis is that LipM and LipL might form a complex to transfer an octanoyl group from octanoyl-ACP to the acceptor proteins. However this explanation seems unlikely since LipM alone complements the function of LipB in vivo (Christensen & Cronan, 2010) and the data in this and the companion paper indicates that LipM and LipL catalyze two sequential steps in octanoyl transfer with GcvH as an intermediate required for the lipoylation of most (if not all) of the other B. subtilis apoproteins.
Strains lacking LipL (ΔlipL), unlike ΔlipM strains, grow poorly in the presence of exogenously added lipoic acid (Fig. 2) indicating that LipL has a role in lipoic acid scavenging. Indeed, ΔlipL ΔlplJ strains fail to grow in the presence of exogenously supplied lipoic acid confirming that LplJ is required for the low level of lipoylation of the complexes seen in the ΔlipL strain. These data suggest that its lipoyl ligase activity may be less efficient in the absence of LipL. These results could be explained if the PDH and BKDH E2 subunits are much poorer LplJ substrates than GcvH. That is, LipL would be both a facilitator of LplJ action as well as a necessary intermediate in LipM action.
Our results clearly indicate the presence of unexpected diversity in lipoic acid metabolism among bacteria. It should be pointed out that LipL homologues can be found in pathogenic bacteria closely related to B. subtilis such as Staphyloccocus aureus and Bacillus anthracis. Thus, utilization of this pathway for protein lipoylation may be more widespread than previously appreciated. Finally, due to the involvement of lipoic acid metabolic proteins in pathogenesis, multidrug resistance and intracellular growth of pathogens, the discovery of new enzymes should provide potential new targets for antimicrobial agents. In the companion paper (Christensen et al., 2011) we report biochemical and genetic data that support the above model in which GcvH is an octanoyl/lipoyl carrier and demonstrate that LipL is a novel amidotransferase. Further work is needed to determine the importance of this new pathway in pathogenesis. The strong phenotype of a ΔlipL strains suggests LipL might be an excellent target for antimicrobials.
Experimental Procedures
Bacterial strains and growth conditions
Bacterial strains used in the present study are listed in Table 1. E. coli and B. subtilis strains were routinely grown in Luria Bertani (LB) broth (Sambrook, Fritsch and Maniatis, 1989). Spizizen salts (Spizizen, 1958), supplemented with 0.5% glucose, trace elements and 0.01% each of tryptophan and phenylalanine were used as the minimal medium for B. subtilis. Different supplements added as needed were 0.5 mM DL-α-lipoic acid, 10 mM sodium acetate and 0.1 mM each BCFA precursor (isobutyric acid, isovaleric acid and 2-methylbutiric acid). For the experiments involving gene expression under the control of the xylose-inducible promoter (PxylA), 0.5% glycerol was used as a carbon source instead of glucose. Xylose was added to 0.1% as required. Antibiotics were added to media at the following concentrations (in µg ml−1) sodium ampicillin (Amp), 100; chloramphenicol (Cm), 5; kanamycin sulfate (Km), 5 and spectinomycin sulfate (Sp), 50.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics a | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| JH642 | trpC2 pheA1 | Laboratory stock |
| NM57 | JH642 lipM:: Kmr | This study |
| NM08 | NM57 amyE::Pxyl-lipM | This study |
| NM11 | NM57 amyE::Pxyl-lipB | This study |
| NM51 | JH642 lipL:: Spr | This study |
| NM09 | JH642 amyE::Pxyl-lipB | This study |
| NM10 | JH642 amyE::Pxyl-lipL | This study |
| NM12 | NM51 amyE::Pxyl-lipB | This study |
| NM13 | NM51 amyE::Pxyl-lipL | This study |
| CM28 | NM57 lipL:: Spr | This study |
| NM14 | CM28 amyE::Pxyl-lipB | This study |
| NM60 | JH642 lplJ::Spr | This study |
| NM65 | NM60 lipM:: Kmr | This study |
| NM67 | NM60 lipL:: Spr | This study |
| NM68 | NM65 lipL:: Cmr | This study |
| E. coli | ||
| DH5α | supE44 thi-1 ΔlacU169(f80lacZΔM15) endA1 recA1 hsdR17 gyrA96 relA1 trp6 cysT329 | Laboratory stock |
| MG1655 | rph-1 | CGSCb |
| EMM99 | E. coli BL21(DE3) / pEM88 | (Martinez et al., 2010) |
| TM131 | rpsL8 lipA::Tn1000 lplA::Tn10 | (Morris et al., 1994) |
| TM136 | rpsL8 lipB::Tn1000 lplA::Tn10 | (Morris et al., 1994) |
| JK1 | rpsL8 | (Morris et al., 1994) |
| MFH120 | JC7623 lacZΔM15 φ(fabA-lacZ)1(Hyb)cat fadBA poxB::pMFH23 | (Henry & Cronan, 1992) |
| QC031 | rpsL8 lipB::Tn1000 lplA::Tn10/ pQC003 | This study |
| QC032 | rpsL8 lipB::Tn1000 lplA::Tn10/ pQC004 | (Christensen & Cronan, 2010) |
| QC035 | rpsL8 / pBAD322G | (Christensen & Cronan, 2009) |
| QC038 | rpsL8 lipA::Tn1000 lplA::Tn10/ pBAD322G | (Christensen & Cronan, 2009) |
| QC057 | rpsL8 lipB::Tn1000 lplA::Tn10/ pBAD322G | (Christensen & Cronan, 2010) |
| QC080 | rpsL8 lipB::Tn1000 lplA::Tn10/ pQC032 | This study |
| QC081 | rpsL8 lipA::Tn1000 lplA::Tn10 / pQC032 | This study |
| QC086 | rpsL8 lipB::Tn1000 lplA::Tn10/ pQC003 | This study |
| QC087 | rpsL8 lipB::Tn1000 lplA::Tn10/ pQC004 | (Christensen & Cronan, 2010) |
| QC134 | rph-1 IN(rrnD-rrnE)1 φ(fabA-lacZ)1(Hyb)cat | (Christensen & Cronan, 2010) |
| QC142 | rph-1 IN(rrnD-rrnE)1 φ(fabA-lacZ)1(Hyb)cat / pCY598, pQC015 | (Christensen & Cronan, 2010) |
| QC143 | rph-1 IN(rrnD-rrnE)1 φ(fabA-lacZ)1(Hyb)cat / pCY598,pQC033 | This study |
| QC161 | rph-1 ΔlplA::FRT ΔlipB::FRT / pQC057, pTARA | (Christensen & Cronan, 2010) |
| QC168 | rpsL lipB::Kn lplA::Tc φ(fabA- lacZ)1(Hyb)cat | This study |
| QC175 | rpsL lipB::Kn lplA::Tc φ(fabA- lacZ)1(Hyb)cat / pBAD322G | This study |
| QC176 | rpsL lipB::Kn lplA::Tc φ(fabA- lacZ)1(Hyb)cat / pQC003 | This study |
| QC177 | rpsL lipB::Kn lplA::Tc φ(fabA- lacZ)1(Hyb)cat / pQC004 | This study |
| QC178 | rpsL (SmR) lipB::Kn lplA::Tc φ(fabA-lacZ)1(Hyb)cat / pQC032 | This study |
| Plasmids | ||
| pGES40 | pBluescript (Stratagene) containing the xylR gene and PxylA promoter | (Schujman, unpublished) |
| pJM116 | Integrative vector to construct transcriptional fusions to lacZ; integrates at the amyE locus of B. subtilis; Cmr | {Dartois, 1996 #3} |
| pJM105A | Integrational vector; Cmr | (Schujman et al., 2001, Perego, 1993) |
| pJM114 | Integrational vector; Kmr | (Schujman et al., 2001, Perego, 1993) |
| pJM134 | Integrational vector; Spr | (Schujman et al., 2001, Perego, 1993) |
| pBAD322G | arabinose inducible expression vector | (Cronan, 2006) |
| pTARA | pACYC origin, arabinose inducible T7 polymerase | (Wycuff & Matthews, 2000) |
| pCY598 | RSF origin, arabinose inducible T7 polymerase | (Cronan, 2003) |
| pET101/D-TOPO | T7 promoter expression vector | Invitrogen |
| pCR2.1 | TOPO TA cloning vector | Invitrogen |
| pNM03 | pJM134 containing lipL interrupted with an spectinomycin cassette | This study |
| pNM07 | pJM114 containing lipM interrupted with a kanamycin cassette | This study |
| pNM48 | pJM134 containing lplJ interrupted with an spectinomycin cassette | This study |
| pNM47 | pJM114 containing lipL interrupted with a kanamycin cassette | This study |
| pNM53 | pJM105A containing lipL interrupted with a chloramphenicol cassette | This study |
| pNM57 | Contains xylR PxylA-lipM into BamHI site of pJM116 | This study |
| pNM58 | Contains xylR PxylA-lipL into BamHI site of pJM116 | This study |
| pNM59 | Contains xylR PxylA-lipB into BamHI site of pJM116 | This study |
| pSJ120 | E. coli LipB expression vector | (Jordan & Cronan, 2003) |
| pQC003 | LplJ expression vector | This study |
| pQC004 | LipM expression vector | (Christensen & Cronan, 2010) |
| pQC015 | N-terminal hexahistidine LipM expression vector | (Christensen & Cronan, 2010) |
| pQC032 | LipL expression vector | This study |
| pQC033 | N-terminal hexahistidine lipL expression vector | This study |
Cmr, Gmr Kmr, Spr, Ampr denote resistance to chloramphenicol, gentamicin, kanamycin, spectinomycin and ampicillin, respectively.
CGSC denotes the Coli Genetic Stock Center.
Genetic techniques
E. coli competent cells were transformed with supercoiled plasmid DNA using the calcium chloride procedure (Sambrook & Russell, 2001). Transformation of B. subtilis was carried out by the method of Dubnau and Davidoff-Abelson (Dubnau and Davidoff-Abelson, 1971). The amy phenotype was assayed with colonies grown for 48 h in LB starch plates by flooding the plates with 1% I2-KI solution (Sekiguchi et al., 1975). Under these conditions amy positive colonies produced a clear halo, whereas Δamy colonies gave no halo.
Plasmids and strains construction
In all cases DNA fragments were obtained by PCR using the oligonucleotides described in Table 2. Chromosomal DNA from wild type B. subtilis was used as a template. Sequencing corroborated the identity and correct sequence of all the cloned fragments.
TABLE 2.
Oligonucleotide primers
| Primer name | Sequence (5’-3’)a |
|---|---|
| I | ggagctcGTTGTAAATCTCAGTGCAGCC |
| II | CTTctAgaGCCTGAGTCTATAAACCGCCA |
| III | AgaGctcGACATACAAACGAGAATGAGC |
| IV | TTTCccggGCATCAGTAAATCAATC |
| V | gGagCtcGATGCTGATATAGAACAGTTTCA |
| VI | ATTctagaGCGGATCATTGATATTTTGATTG |
| VII | AAACgtcGacCGCTTCAGATGAATG |
| VIII | CTTcTcgAgGGGCATCATCTTCTTC |
| IX | CCGaAgcttTGAGCGGAATGCAAAG |
| X | GTgGtAccCCACCAGGAGTTTCGCG |
| XI | TGGGgtCgacACGAAAGAGGATTTC |
| XII | CAGCcTcgaGCTTGGCCACATAATA |
| XIII | AGTTgTCGacCAATAAGCCTAACATGAAAGGG |
| XIV | AGGgtAccgGatcCTTGAGATAAAAAATGCATG |
| XVII | CTATgtcgACGGTAAGGAAGGTCTTAAAATGCA |
| XVIII | CGGGTaccggatCCTGTTTACCGCTTATAATTC |
| XL | GTGTcgacGAATggaggCTTCATATGTATCAGG |
| XLI | AGgtacCGgatcCAAGCTTAAGCGGTAATATATTCG |
| XLVI | TGAAAgcTtTGGTATCTTCTTATTGTAGTGCTG |
| XLVII | CTTggtaccggATCCTGTTTTAGTCTTCTTGTG |
| L | AGTGGaTccAAGAGCATGGGAAAG |
| LI | TTgtCgACTCGTGTTCTCCTGAGTAA |
| LII | GAGtcGACACAAGAAGACTAAAACAG |
| LIII | AAAGActGCAGTGAAATTCACCGCCA |
| Q003 | GAGACATGTTATTTATAGACAATCAAAATATCAATG |
| Q004 | TATAAGCTTCTCCTGCCTCCATTATTT |
| Q021 | CACCATGCATCATCATCATCATCATATGTTATTTATAGACAATCAAAATATCAATG |
| Q036 | GAGCAATTGACCATGCATCATCATCATCATCATATGTTATTTATAGACAATCAAAATATCAATG |
| Q039 | CACCATGGCAAACCAACCGATTGA |
| Q040 | CGTTCACCCAAATACCTTTGCA |
| Q043 | CACCATGCATCATCATCATCATCATATGGCAAACCAACCGATTGA |
Lowercase letters indicate variations with respect to the wild-type B. subtilis sequence. Restrictions sites are underlined
A strain with a deletion of the yqhM (lipM) gene was constructed by gene replacement with a kanamycin resistance determinant, through a double crossover event. For this purpose a 571 bp fragment, corresponding to the 5’ end of lipM and the upstream gene was PCR amplified with primers I and II (Table 2) and cloned into SacI and XbaI sites of plasmid pJM114 (Perego, 1993). A 525 bp fragment containing the 3’ end of lipM and part of the downstream gene was PCR amplified with primers VII and VIII (Table 2) and cloned into SacI and XbaI sites of the previously generated plasmid. The plasmid obtained pNM07, was linearized with ScaI and used to transform strain JH642. Transformants were selected for kanamycin resistance. The resulting strain was named NM57.
An yhfJ (lplJ) knock out mutant was constructed as follows. A 520 bp fragment of the 5’ end of lplJ plus upstream sequences was PCR amplified using oligonucleotides V and VI (Table 2) and cloned into the XbaI site of vector pJM134 (M. Perego, unpublished) orientation of this insert was checked by restriction enzyme digestion with EcoRI and PstI. Afterwards, a 516 bp fragment of the 3’ end of lplJ and downstream region was PCR amplified using oligonucleotides XI and XII (Table 2) and inserted into the SalI and XhoI sites of that plasmid to give pNM48. Strain JH642 was transformed with pNM48 linearized with PvuII, yielding strain NM60.
To construct a strain containing a deletion mutation in ywfL (lipL) gene a 552 bp fragment of the 5’end of lipL plus upstream gene sequences was PCR amplified with oligonucleotides III and IV (Table 2) and inserted between the SacI and SmaI sites of plasmid pJM134 (M. Perego, personal communication). Then, a 581 bp fragment containing the 3’ end of lipL and part of the downstream gene was PCR amplified using oligonucleotides IX and X (Table 2) and inserted between the HindIII and KpnI sites of the previously generated plasmid to render plasmid pNM03. Plasmid pNM03 was linearized with ScaI and used to transform strain JH642. The resulting strain was named NM51. It should be noted that essentially the entire coding sequences of the lipM, lipL and lplJ genes were removed in construction of the deletion strains.
Strain CM28 was constructed by transformation of strain NM57 with plasmid pNM03 previously linearized with ScaI. Plasmid pNM07 was linearized with ScaI and was used to transform strain NM60, rendering strain NM65. To construct strain NM67 the spectinomycin resistant cassette from pNM03 was replaced by a kanamycin resistance cassette from vector pJM114 (Perego, 1993), yielding plasmid pNM47. This plasmid was linearized by digestion with ScaI and use to transform strain NM60, to give strain NM67. Strain NM68 was constructed as follows. A replacement of the spectinomycin resistant cassette from pNM03 with a chloramphenicol resistant cassette from vector pJM105A (Perego, 1993) was performed. The resulting plasmid, pNM53, was linearized by digestion with ScaI and used to transform strain NM65. For strains NM65, NM67 and NM68 selection of transformants was carried out in LB supplemented with acetate and BCFA precursors.
To test complementation of strain NM57 with a wild type copy of the lipM gene, a 882 bp fragment containing lipM with its ribosome binding site was PCR amplified with oligonucleotides XVII and XVIII (Table 2) and the product inserted between the SalI and KpnI sites of pGES40 (Schujman, unpublished). This plasmid was digested with BamHI to obtain a fragment containing xylR PxylA-lipM, which was cloned into pJM116 previously digested with BamHI, yielding plasmid pNM57. Strain NM57 was transformed with this plasmid; transformants were screened for kanamycin and chloramphenicol resistance and amyE phenotype. The resulting strain was named NM08.
Strain NM51 was complemented with a wild type copy of lipL gene as follows. A 952 bp fragment containing lipL with its ribosome-binding site was PCR amplified with oligonucleotides XIII and XIV (Table 2) and cloned into SalI and KpnI sites of pGES40 (Schujman, unpublished). This plasmid was digested with BamHI to obtain a fragment containing xylR PxylA-lipL, which was cloned into pJM116 previously digested with BamHI, yielding plasmid pNM58. Strain JH642 was transform with plasmid pNM58 to yield strain NM10 which was further transformed with plasmid pNM03 to give strain NM13.
To express E. coli lipB in B. subtilis plasmid pNM59 was constructed as follows. A 682 bp fragment containing the lipB gene was PCR amplified from plasmid pYFJ29 (Zhao et al., 2005) with oligonucleotides XL and XLI (Table 2) and inserted between the SalI and KpnI sites of pGES40 (Schujman, unpublished). This plasmid was digested with BamHI to obtain a fragment containing xylR PxylA-lipB, which was cloned into pJM116 previously digested with BamHI, yielding plasmid pNM59. Strain JH642 and NM57 were transformed with plasmid pNM59 to yield strains NM09 and NM11. Strains NM09 and NM11 were then transformed with pNM03 to yield strain NM12 and NM14, respectively.
For E. coli complementation analyses, coding sequences were amplified from genomic DNA by PCR and inserted into pBAD322G (Cronan, 2006). The lplJ coding sequence was amplified with primers Q003 and Q004 and the product was ligated to the vector NcoI and HindIII sites to give pQC003. The lipL coding sequence was amplified with primers Q039 and Q040 and inserted into pCR2.1 using the TA cloning kit (Invitrogen) and then inserted into the NcoI and HindIII sites of pBAD322G to give pQC032. These manipulations placed these genes under the control of an arabinose inducible promoter.
For purification of the protein products of the genes, they were amplified with primers that added a sequence encoding an N-terminal hexahistidine tag. These PCR products were first inserted into pET101 using the TOPO Cloning Kit (Invitrogen). The lplJ coding sequence was amplified using primers Q021 and Q004 to give pQC014, whereas the lipL coding sequence was amplified using primers Q0043 and Q0040 to give pQC033. These manipulations placed these genes under control of a T7 polymerase-dependent promoter (Studier & Moffatt, 1986).
Growth curves of B. subtilis cultures
Strains were grown overnight on liquid minimal medium supplemented with acetate and BCFA precursors. Cells were washed once with minimal medium and used to inoculate fresh media at an OD600 of 0.1–0.15. Cells were grown using a Bioscreen C with 300 µL per well with continuous and strong shaking. Growth (OD600) was measured every h.
Complementation of E. coli lipoate auxotrophs
B. subtilis genes were expressed in E. coli from plasmids with an arabinose inducible promoter as described (Christensen & Cronan, 2010). Complementation of the lipB lplA strain TM136 was tested in M9 minimal medium containing 0.2% arabinose, 0.1% Vitamin-Assay Casamino Acids, and gentamycin. Complementation of the lipA lplA strain TM131 was tested using the same medium containing 5 µg/ml sodium lipoate. Growth was measured by absorbance (OD600) in a Beckman DU600 spectrophotometer.
The activities of the lipoate dependent dehydrogenases were assayed using the continuous spectrophotometric assay previously described (Christensen & Cronan, 2009). Briefly, derivatives of TM136 carrying various plasmids were subcultured to an OD600 of 0.1 in LB with 0.2% arabinose, 5 mM acetate and 5 mM succinate pH 7.0. Cells were harvested in late exponential phase at an OD600 of 0.7. The cells were lysed by two passages through a French pressure cell and protein was quantified using the Bradford assay reagent (Bio-Rad) with bovine gamma globulin (Pierce) as the standard. PDH and OGDH activities were measured spectrophotometrically at 366 nm following the reduction of acetyl-pyridine adenine dinucleotide.
Strain QC168 was used to test the complementation properties of B. subtilis genes in a strain deficient in unsaturated fatty acid biosynthesis. Cultures were grown on M9 minimal agar with 0.4% glycerol, 0.1% Vitamin-Assay Casamino Acids, 5 mM acetate, 5 mM succinate, 0.5 mM sodium oleate, and 0.1% Tergitol NP-40 and then restreaked on the same medium lacking acetate and succinate to test for complementation.
Protein purifications
Purification of GcvH
Hexahistidine-tagged B subtilis GcvH was heterologously expressed in E. coli and purified by nickel affinity and anion exchange chromatographic steps as previously described (Christensen & Cronan, 2010). GcvH was verified to be in the unmodified form lacking the N-terminal methionine residue by electrospray mass spectrometry as previously described (Christensen & Cronan, 2010) and was quantified by absorbance at 280 nm using a calculated extinction coefficient of 16,960 M−1 cm−1.
Purification and Modification of B. subtilis AcpP
The native acyl carrier protein of B. subtilis was purified by nickel affinity and ion exchange chromatographic steps, as previously described. Holo and octanoyl forms of ACP were also prepared as previously described (Christensen & Cronan, 2010).
Purification of Lipoate Metabolism Enzymes
LipB was purified by nickel affinity and anion exchange as described (Jordan & Cronan, 2003). LipB was quantified by absorbance at 280 nm using an extinction coefficient of 22,920 M−1 cm−1. LplJ was purified from strain QC103 by nickel affinity and anion exchange as described for T. acidophilum LplA (Christensen & Cronan, 2009). LplJ was quantified by absorbance at 280 nm using an extinction coefficient of 34,380 M−1 cm−1. LipM and LipL were also purified from the E. coli fabA strains QC142 and QC143, respectively, as described for LipM and analyzed by MALDI MS as previously described (Christensen & Cronan, 2010). LipL was also purified from strain QC083 grown in the absence of triclosan. LipM and LipL were quantified by absorbance at 280 nm using extinction coefficients of 45,380 and 25,900 M−1 cm−1, respectively. The purified proteins were concentrated with Vivaspin centrifuge concentrators (GE Healthcare) and flash frozen for storage as above except that the buffer contained 100 mM sodium chloride.
Lipoate ligation Assay
Lipoate ligase activity was assayed by observing a mobility shift upon modification by native gel electrophoresis as originally described by Miles and Guest (Miles & Guest, 1987). Assays contained 100 mM sodium phosphate (pH 7.0), 5 mM DTT, 1 mM sodium lipoate, 1 mM magnesium chloride, 1 mM ATP, 20 µM lipoyl domain and 1 µM LplJ. For assay of octanoylation by various enzymes, sodium octanoate was substituted for sodium lipoate and 10 µM enzyme was used. The assays were incubated at 37°C for 1 h and 10 µl of the assays were subjected to native Tris-glycine gel electrophoresis using a 20% acrylamide gel for four h at 100 V. The proteins were visualized by staining with Coomassie Blue R-250 (Sambrook & Russell, 2001) or by western blotting with anti-lipoic acid antibody (Calbiochem) and anti-rabbit IgG HRP conjugate (Roche). Western blotting was carried out using standard methods with 5% dehydrated milk (Carnation) for blocking and antibody incubation steps (Ausubel, 1987).
Acyl-adenylate intermediate formation was assayed by thin layer chromatography and autoradiography as described for BirA (Xu & Beckett, 1997). The reactions contained 100 mM sodium phosphate (pH 7.0), 5 mM TCEP, 0.1 µM α-32P ATP (6000 Ci/mmol), 1 µM MgCl2, 0.1 mM sodium lipoate or octanoate, and 10 µM LplJ. GcvH was also added to 50 µM where indicated. Reactions were incubated at 37°C for 30 min before spotting onto a cellulose TLC plate.
Assay of octanoyl-[Acyl Carrier Protein]:Protein N-octanoyltransfer
For assays using B. subtilis extracts, 100 µg of extract protein was added instead of enzyme. Cultures were grown to an OD600 of 0.6, pelleted by centrifugation, resuspended 1:100 of the culture volume in 100 mM sodium phosphate buffer (pH 7.0), and sonicated for 10 min using a Misonix cup-horn sonicator cooled with circulating 50% polyethylene glycol. Extract were cleared by centrifugation and quantified using the Bradford assay (Bio-Rad) with bovine gamma globulin as a standard. For assay of transfer from octanoyl-ACP to both LipM and LipL, 50 µM sodium [1-14C]octanoate and 10 µM enzyme were used in the absence of lipoyl domain. The reaction was analyzed using a modification of the method of Laskey and Mills (Laskey & Mills, 1975) in which 10 µl of the reactions were subjected to SDS-PAGE on a 4–20% gradient gel which was soaked in Amplify fluorographic reagent (GE Healthcare), dried, and exposed to preflashed Biomax XAR film (Kodak) at −70°C for 24 h.
Immunoblotting analyses
B. subtilis wild type and mutants strains were grown overnight in minimal medium supplemented with acetate and BCFA precursors at 37°C. Cells were resuspended in fresh media of the same composition and cultured at 37°C. A 1-ml aliquot of each culture was harvested after 22 hours of growth. The samples were centrifuged and the pellets were washed with buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl). They were resuspended in 180 µl of lysis buffer (50 mM Tris-HCl [pH 8.0], PMSF 10 µM) per OD600 unit. Resuspended cells were disrupted by incubation with lysozyme (100 µg/ml) for 15 min at 37°C followed by 5 min of boiling in the presence of loading buffer. Each sample was fractionated by sodium dodecyl sulfate-gel electrophoresis in a 12% acrylamide gel. Proteins were electroeluted to a nitrocellulose membrane and detected using anti-lipoate rabbit antibody and a secondary anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Bio-Rad). The bands were visualized by use of the ECL Plus Western Blotting Detection System (GE). The blots were scanned and the intensity of the bands was quantified by ImageQuant 5.2.
Purification of the B. subtilis PDH complex
Cultures were grown at 37°C in LB supplemented with 0.5% glucose, 10 mM sodium acetate, 0.1 mM of each BCFA precursor and the appropriate antibiotic until an OD600 of 1–1.5 when cells were harvested. Purification was carried out similarly as previously described by (Hodgson et al., 1983). Briefly, cell pastes were suspended in 50 mM sodium phosphate buffer, (pH 7.0) containing 5 mM EDTA, Complete EDTA free protease inhibitor cocktail (Roche), lysozyme (6 mg/ml) (Sigma) and DNAse I (5 µg/ml) (Sigma) and stirred for 2 h. Cells were lysed by three passages through a French pressure cell at ~20,000 psi. Lysates were cleared by centrifugation at 44,000 × g for 30 min. The cleared lysate was treated with 32 µg/ml ribonuclease and incubated at 15°C for 70 min to degrade ribosomes. The samples were centrifuged for 30 min. at 44,000 × g. The solution of approximately 50 mg of protein/ml was layered on top of a sucrose step gradient (12.5% w/v and 70% w/w). Centrifugation was performed for 3 h at 180,00 × g. The brown protein band at the interface of the 12.5% and 70% sucrose layers was drawn off. Sucrose was removed by overnight dialysis against 50 mM sodium phosphate buffer (pH 7.0) containing 5 mM EDTA and 0.15 mM phenylmethanesulphonylfluoride. The samples were concentrated using Vivaspin concentrators (Sartorius), loaded onto a column of Sephacryl HR-500 26/16 (GE) and eluted with 50 mM sodium phosphate buffer containing 5 mM EDTA (pH 7.0). Fractions containing pure complex as judged by SDS PAGE were pooled and dialyzed overnight against 50 mM sodium phosphate buffer, 15% glycerol and 1 mM tris(2-carboxyethyl)phosphine (Sigma). Samples were then concentrated using Vivaspin concentrators (Sartorius), flash frozen in dry ice and ethanol, and stored at −80°C.
PDH activity was assayed in the preparation purified from strain NM60 extracts to confirm that the purified complex was indeed the PDH complex.
Preparation of the lipoyl domain of B. subtilis pyruvate dehydrogenase complex
B. subtilis pyruvate dehydrogenase complex (1 ml; 5–20 mg/ml) in 50 mM sodium phosphate buffer (pH 7.0) was digested at 30°C with Staphylococcus aureus V8 protease (Wako) (1% w/w) for 100 min at which time a second addition of protease (1% w/w) was made. Following a total digestion time of 180 min phenylmethanesulphonylfluoride was added to a final concentration of 1 mM to block further digestion. After digestion the samples were centrifuged for 30 min at 14,000 rpm in a bench top centrifuge and supernatants analyzed by 20% native polyacrylamide-gel electrophoresis. The peptides were sliced from the gel and submitted to the UIUC Mass Spectrometry Lab for LC-MS/MS analysis.
Liquid Chromatography Tandem Mass Spectrometry (LC-MS/MS) Analysis
Trypsin digestion and LC-MS/MS analysis was performed by the University of Illinois Biotechnology Center Protein Sciences Facility as follows. Gel slices were crushed and rinsed with water. 25 µl of trypsin (12.5 µg/ml) in 25 mM ammonium bicarbonate was added and the samples were digested in a CEM Discover microwave reactor for 15 min at 55°C and 50 watts. The gel pieces were extracted with 100 µl of 50% acetonitrile and 5% formic acid for 10 minutes with sonication. The extracts were dried down in a vacuum centrifuge and resuspended in 13 µl of 5% acetonitrile and 0.1% formic acid. Analysis by LC-MS was carried out on a Waters Q-Tof API-US Quad-ToF mass spectrometer with a nanoAcquity UPLC system. The columns used were Waters nanoAcquity UPLC (75 m × 150 mm 3 µm Atlantis dC18) and Atlantis dC18 5 µm Nanoease trap columns. A 60 min linear gradient of 1% to 60% acetonitrile in 0.1% formic acid was used to elute the peptides from the columns. MS/MS data were collected using the Data Directed Analysis method in MassLynx to fragment the top four ions in each survey scan. ProteinLynx (Waters) was used to process the mass spectral data into peak list files for analysis by Mascot (Matrix Science). Database searches were performed against the NCBI non-redundant database with taxonomy restrictions.
ACKNOWLEDGEMENTS
This investigation was supported by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Agencia Nacional de Promoción Científica y Tecnológica (FONCYT), Fundación Josefina Prats and the National Institutes of Health under Ruth L. Kirschstein National Research Service Award 5 T32 GM070421 from the National Institute of General Medical Sciences and research grant AI15650 from the National Institute of Allergy and Infectious Disease. N. Martin is a Fellow of CONICET and M. C. Mansilla and D. de Mendoza are Career Investigators of the same institution. D. de Mendoza is an International Research Scholar of the Howard Hughes Medical Institute. We thank Dr. M. Perego for research materials.
REFERENCES
- Ausubel FB, Kingston R, Moore RE, Seidman DD, Smith JA JD, Struhl K. Current protocols in molecular biology. 1987;2:10.18.11–10.18.16. [Google Scholar]
- Christensen Q, Martin N, Mansilla M, de Mendoza D, Cronan J. A novel transamidase required for lipoic acid cofactor assembly in Bacillus subtilis. Mol. Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07598.x. Companion paper. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen QH, Cronan JE. The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J. Biol. Chem. 2009;284:21317–21326. doi: 10.1074/jbc.M109.015016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen QH, Cronan JE. Lipoic acid synthesis: A new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry. 2010 doi: 10.1021/bi101215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicchillo RM, Booker SJ. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: Both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J. Am. Chem. Soc. 2005;127:2860–2861. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
- Cronan JE. Cosmid-based system for transient expression and absolute off-to-on transcriptional control of Escherichia coli genes. J Bacteriol. 2003;185:6522–6529. doi: 10.1128/JB.185.22.6522-6529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE. A family of arabinose-inducible Escherichia coli expression vectors having pBR322 copy control. Plasmid. 2006;55:152–157. doi: 10.1016/j.plasmid.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Suzuki M, Okumachi Y, Okamura-Ikeda K, Fujiwara T, Takahashi E, Motokawa Y. Molecular cloning, structural characterization and chromosomal localization of human lipoyltransferase gene. Eur J Biochem. 1999;260:761–767. doi: 10.1046/j.1432-1327.1999.00204.x. [DOI] [PubMed] [Google Scholar]
- Henry MF, Cronan JE. A new mechanism of transcriptional regulation: Release of an activator triggered by small molecule binding. Cell. 1992;70:671–679. doi: 10.1016/0092-8674(92)90435-f. [DOI] [PubMed] [Google Scholar]
- Hermes F, Cronan J. Scavenging of cytosolic octanoic acid by mutant LplA lipoate ligases allows growth of Escherichia coli strains lacking the LipB octanoyltransferase of lipoic acid synthesis. J Bacteriol. 2009;191:6796. doi: 10.1128/JB.00798-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson JA, Lowe PN, Perham RN. Wild-type and mutant forms of the pyruvate dehydrogenase multienzyme complex from Bacillus subtilis. Biochem J. 1983;211:463–472. doi: 10.1042/bj2110463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SW, Cronan JE., Jr The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein Transferase. J. Bacteriol. 2003;185:1582–1589. doi: 10.1128/JB.185.5.1582-1589.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey RA, Mills AD. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhao X, Eddine AN, Geerlof A, Li X, Cronan JE, Kaufmann SHE, Wilmanns M. The Mycobacterium tuberculosis LipB enzyme functions as a cysteine/lysine dyad acyltransferase. Proc Natl Acad Sci U S A. 2006;103:8662–8667. doi: 10.1073/pnas.0510436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Lombardia E, Altabe SG, de Mendoza D, Mansilla MC. A lipA (yutB) mutant, encoding lipoic acid synthase, provides insight into the interplay between branched-chain and unsaturated fatty acid biosynthesis in Bacillus subtilis. J Bacteriol. 2009;191:7447–7455. doi: 10.1128/JB.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MA, Zaballa ME, Schaeffer F, Bellinzoni M, Albanesi D, Schujman GE, Vila AJ, Alzari PM, de Mendoza D. A novel role of malonyl-ACP in lipid homeostasis. Biochemistry. 2010;49:3161–3167. doi: 10.1021/bi100136n. [DOI] [PubMed] [Google Scholar]
- Marvin ME, Williams PH, Cashmore AM. The isolation and characterisation of a Saccharomyces cerevisiae gene (LIP2) involved in the attachment of lipoic acid groups to mitochondrial enzymes. FEMS Microbiol Lett. 2001;199:131–136. doi: 10.1111/j.1574-6968.2001.tb10663.x. [DOI] [PubMed] [Google Scholar]
- Miles JS, Guest JR. Subgenes expressing single lipoyl domains of the pyruvate dehydrogenase complex of Escherichia coli. Biochem J. 1987;245:869–874. doi: 10.1042/bj2450869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris T, Reed K, Cronan J. Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol. 1995;177:1–10. doi: 10.1128/jb.177.1.1-10.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris TW, Reed KE, Cronan JE., Jr Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J Biol Chem. 1994;269:16091–16100. [PubMed] [Google Scholar]
- O'Riordan M, Moors MA, Portnoy DA. Listeria intracellular growth and virulence require host-derived lipoic acid. Science. 2003;302:462–464. doi: 10.1126/science.1088170. [DOI] [PubMed] [Google Scholar]
- Packman LC, Borges A, Perham RN. Amino acid sequence analysis of the lipoyl and peripheral subunit-binding domains in the lipoate acetyltransferase component of the pyruvate dehydrogenase complex from Bacillus stearothermophilus. Biochem J. 1988;252:79–86. doi: 10.1042/bj2520079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein JAHAL, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington DC: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. CSHL Press; 2001. [Google Scholar]
- Schujman GE, Choi KH, Altabe S, Rock CO, de Mendoza D. Response of Bacillus subtilis to cerulenin and acquisition of resistance. J Bacteriol. 2001;183:3032–3040. doi: 10.1128/JB.183.10.3032-3040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Takada N, Okada H. Genes affecting the productivity of alpha-amylase in Bacillus subtilis Marburg. J Bacteriol. 1975;121:688–694. doi: 10.1128/jb.121.2.688-694.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc Natl Acad Sci U S A. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Thomsen-Zieger N, Schachtner J, Seeber F. Apicomplexan parasites contain a single lipoic acid synthase located in the plastid. FEBS Lett. 2003;547:80–86. doi: 10.1016/s0014-5793(03)00673-2. [DOI] [PubMed] [Google Scholar]
- Wycuff DR, Matthews KS. Generation of an araC-araBAD promoter-regulated T7 expression system. Anal Biochem. 2000;277:67–73. doi: 10.1006/abio.1999.4385. [DOI] [PubMed] [Google Scholar]
- Xu Y, Beckett D. Biotinyl-5'-adenylate synthesis catalyzed by Escherichia coli repressor of biotin biosynthesis. Meth Enzymol. 1997;279:405–421. doi: 10.1016/s0076-6879(97)79045-1. [DOI] [PubMed] [Google Scholar]
- Zhao X, Miller JR, Cronan JE. The reaction of LipB, the octanoyl-[acyl carrier protein]:protein N-octanoyltransferase of lipoic acid synthesis, proceeds through an acyl-enzyme intermediate. Biochemistry. 2005;44:16737–16746. doi: 10.1021/bi051865y. [DOI] [PubMed] [Google Scholar]
- Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol. 2003;10:1293–1302. doi: 10.1016/j.chembiol.2003.11.016. [DOI] [PubMed] [Google Scholar]