Summary
Sterol regulatory element binding proteins (SREBPs) are key transcriptional regulators of lipid metabolism. To define functional differences between the three mammalian SREBPs we are using genome-wide ChIP-seq with isoform-specific antibodies and chromatin from select tissues of mice challenged with different dietary conditions that enrich for specific SREBPs. We show hepatic SREBP-2 binds preferentially to two different gene-proximal motifs. A Gene ontology analysis suggests SREBP-2 targets lipid metabolic processes as expected but apoptosis and autophagy gene categories were also enriched. We show SREBP-2 directly activates autophagy genes during cell sterol depletion, conditions known to induce both autophagy and nuclear SREBP-2 levels. Additionally, SREBP-2 knockdown during nutrient depletion decreased autophagosome formation and lipid droplet association of the autophagosome targeting protein LC3. Thus, the lipid droplet could be viewed as a third source of cellular cholesterol, which along with sterol synthesis and uptake, is also regulated by SREBP-2.
Keywords: sterol regulatory element binding protein (SREBP), ChIP-seq, Autophagy, Lipid metabolism
Introduction
The structural framework of all biological membranes is based on the unique molecular properties of lipids (van Meer et al., 2008). Lipids also have more specialized roles that have evolved over time that influence many physiologic pathways and processes that vary based on organism, tissue, developmental timing, cellular physiology, and metabolic demand (Di Paolo and De Camilli, 2006; Mann and Beachy, 2004; Toker and Cantley, 1997). Accordingly, the need for new lipid in a given cell can vary dramatically and change quickly so it is essential that a dynamic and flexible regulatory mechanism is evolving at the cellular level that is based on both need and availability.
An integrated regulatory system for mammalian lipid homeostasis operates at both transcriptional and post-transcriptional levels (Brown and Goldstein, 1980). A major part of the transcriptional control is through the sterol regulatory element binding proteins (SREBPs), which comprise a sub-family of basic helix-helix (bHLH) proteins and is conserved from simple eukaryotes to humans (Osborne and Espenshade, 2009). SREBPs are synthesized and threaded into the endoplasmic reticulum membrane where two closely spaced membrane spanning domains position the precursor molecule in a hairpin configuration. When the cellular supply of lipids is limiting, SREBPs are routed to the golgi apparatus and cleaved roughly in half by the action of two sequential acting membrane bound proteases that release the soluble amino-terminal mature SREBP transcription factor from the membrane. Mature SREBP is constitutively targeted to the nucleus where it activates genes required for synthesizing fatty acids and cholesterol (Osborne and Espenshade, 2009).
Two separate SREBPs genes, Srebf1 and Srebf2, encoding three major protein isoforms have evolved in mammals and a combination of genetics and molecular expression studies in mice and cultured cells have been used to study isoform specific roles in lipid metabolism (Horton et al., 2002). The two proteins expressed from the Srebf1 gene, SREBP-1a and SREBP-1c, have the identical DNA binding domain, and available evidence suggests they both preferentially target genes of fatty acid metabolism whereas the singular Srebf2 encoded protein preferentially activates genes of cholesterol metabolism (Horton et al., 2002). However, studies indicate there is considerable overlap in function and a few reports also suggest SREBPs may be involved in activating other physiological processes so further investigation is required to fully appreciate the broad roles of the three mammalian SREBP isoforms at the molecular level (Osborne and Espenshade, 2009).
We are complementing the over-expression and knockout investigations with genome-wide DNA binding studies to further interrogate SREBP function using a global and unbiased approach. In the current report, we have used an antibody to SREBP-2 in a chromatin immunoprecipitation-deep sequencing approach (ChIP-seq) (Johnson et al., 2007) that revealed 1800 sites of SREBP-2 binding in mouse liver chromatin. In addition to identifying genes in pathways of lipid metabolism, this analysis also identified several genes involved in autophagy as potential SREBP targets.
Autophagy provides a catabolic quality control and recycling mechanism for cells to remove damaged proteins and organelles and to recapture cellular components when nutrient building blocks are limiting (He and Klionsky, 2009). Lipid depletion has already been shown to induce autophagy (Cheng et al., 2006) and recent studies have suggested that in response to nutrient limitation cells may recycle excess cellular lipid stored in cytoplasmic droplets as cholesterol esters and triglycerides through a process termed macrolipophagy, which requires many if not all of the same proteins required for autophagy (Singh et al., 2009). We present additional studies showing that SREBP-2 activates autophagy genes during sterol depletion and that autophagosome formation and association of the key autophagosomal LC3 protein with lipid droplets in response to nutrient depletion are both significantly reduced when SREBP-2 levels are decreased. Thus, the lipid droplet could be considered a third source of cellular lipid that is also regulated by SREBPs.
Results
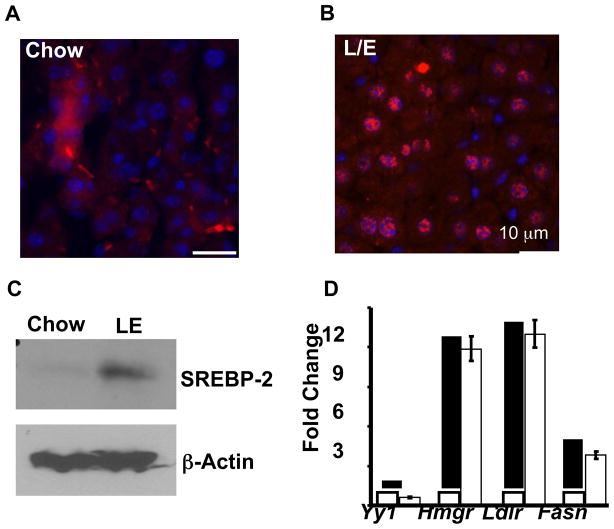
In order to evaluate SREBP-2 binding on a genome-wide scale we first prepared an antibody to a region of the mouse SREBP-2 within its amino-terminal nuclear targeted domain that bears no similarity to the corresponding region of SREBP-1 (Jeon et al., 2008). Before embarking on the ChIP-seq analysis, we first evaluated the specificity and quality of the antibody (Figure 1). Nuclear levels of hepatic SREBP-2 are significantly elevated in mice fed a chow-diet supplemented with lovastatin plus ezetimibe (LE) to inhibit cholesterol production in the liver and simultaneously limit cholesterol uptake from the diet (Bennett et al., 2008; Jeon et al., 2008). Sections cut from control and LE treated liver were stained with the SREBP-2 antibody and Figure 1A shows strong nuclear reaction with the LE sample relative to chow fed control, which shows diffuse non-nuclear staining.
Figure 1. Validation for SREBP-2 antibody and chromatin supplemented by chow and chow with LE.
Immunofluorescence of SREBP-2 in mouse liver from normal chow (A) or chow supplemented with lovastatin and ezetimibe (LE) for one week (B). Livers pieces were embedded in paraformaldehyde, sectioned, and stained with a rabbit anti-SREBP-2 polyclonal antibody (affinity purified), followed by fluorescein-labeled goat anti-rabbit IgG. Magnification, 400× bar shows scale for 10 μm. (C) Hepatic nuclear extracts were prepared from mice fed chow and LE diets. (D) Gene-specific ChIP analysis using chromatin from the LE group that was immuno-enriched with an antibody to SREBP-2 (Filled bars) or a control IgG (open bars). Gene specific primers for the indicated promoter regions were used in the qPCR. Data are mean ± SEM; n = 3 for 3 separate experiments.
SREBP nuclear localization results from proteolytic maturation that cleaves the ~120 kDa membrane bound precursor roughly in half. An immunoblot analysis confirmed that mature SREBP-2 was increased dramatically in the LE sample (Figure 1C). Next, chromatin from the LE sample was used in gene-specific chromatin immunoprecipitation studies with primers that flank known SREBP-2 binding sites within key target genes Hmgcr, Ldlr, and Fasn. These gene-specific ChIP reactions showed significant enrichment by the SREBP-2 antibody whereas a genomic fragment from the negative control Yy1 gene was not enriched (Figure 1D). Based on these results we were encouraged to proceed with the genome-wide binding analysis.
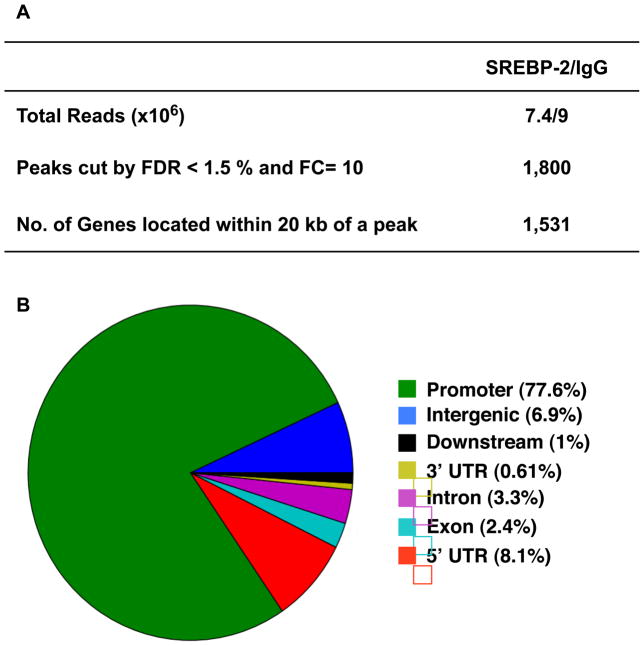
A portion of the DNA used in the confirmation ChIPs in Figure 1 was used in the ChIP-seq procedure for parallel sequencing on the Solexa Genome Analyzer II. The reads returned from the sequencer were then mapped onto the mouse genome, and putative SREBP-2 binding peaks were identified using the global identifier of target regions (GLITR) program (Tuteja et al., 2009). GLITR calculated a false discovery rate (FDR) for each peak, and we identified all binding peaks that have a FDR less than 1.5% and a fold change in peak intensity greater than 10 (relative to the control IgG). The total number of identified peaks and the number contained within 20 kb of a known gene are shown in Figure 2A. The sequencing reads were also overlayed as a track in the UCSC genome browser and the areas close to peaks associated with 4 different genes are displayed in Figure S1 as an example of the data set.
Figure 2. ChIP-seq analysis for SREBP-2 distribution on mouse liver chromatin supplemented with LE.
(A) Summary of ChIP-seq analysis by GLITR. The results were obtained using an FDR1.5% and a fold change in peak intensity of 10 relative to control IgG as cut off values. (B) Genome-wide mapping of SREBP-2 binding regions relative to RefSeq mouse genes. The ‘promoter’ and ‘downstream’ are defined as 2 kb of 5′ or 3′ flanking sequence. Intergenic region refers to all locations other than ‘promoter’, ‘5′ UTR’, ‘exon’, ‘intron’, ‘3′UTR’ or ‘downstream’. FDR: false discovery rate; FC: fold change. The total number of SREBP2 peaks was 1800 (=100%). These were distributed in the genome as follows: ‘promoter’- 1398 (= 77.6%); ‘exon’- 44 (= 2.4%); ‘downstream’- 18 (= 1.0%): ‘3utr’- 11 (= 0.61%); ‘5utr’- 146 (= 8.1%); ‘intergenic’- 124 (= 6.9%); ‘intron’- 59 (= 3.3%).
The genome-wide distribution of the peaks relative to the position of known genes is shown in Figure 2B. It is striking to note that 77% of the binding sites are localized to a region located within 2 kb of the 5′ end of known genes. By contrast, if the peaks were randomly sampled from the genome only 38 sites would be expected to be present in this region (Figure S2A), demonstrating a strong enrichment of SREBP-2 sites in proximal promoter regions (p-value < 10−6). Note that this preference of binding for proximal promoter regions is highly unusual, since most transcription factor binding sites studied by ChIP-seq tend to show more widely distributed binding throughout the entire genome (Seo et al., 2009). The gene-proximal location of the binding sites was also revealed by a plot of the distance of each peak relative to a transcription start site for a known gene (Figure S2B).
Because of the very high number of short sequence reads, Johnson et al (Johnson et al., 2007) suggested that the ChIP-seq procedure would identify binding sites very accurately. When we evaluated the sequence reads at the Hmgcr promoter relative to our previously published DNAse I footprinting analysis using recombinant SREBP and 32P labeled DNA from this region in vitro (Millinder-Vallett et al., 1996), the enrichment of the ChIP seq reads correlated very precisely with the three regions of DNA protected by recombinant SREBP in vitro (Figure S3) providing a compelling example of the congruence of the in vitro and in vivo DNA binding studies.
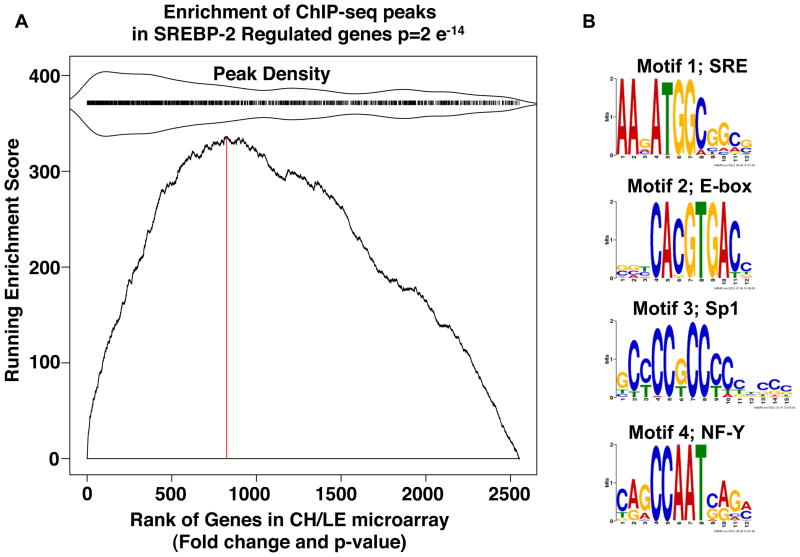
To evaluate how the genome-wide binding analysis correlates with global regulation of gene expression by SREBP-2, we also compared gene expression patterns using RNA from livers of mice fed chow supplemented with either LE where SREBP-2 nuclear levels are elevated (Figure 1) or cholesterol where nuclear levels of SREBP-2 are dramatically lower. A gene set enrichment analysis (GSEA) for this comparison is plotted in Figure 3A; the X-axis presents a rank order of all genes according to their differential expression (fold change and p value) in LE vs. cholesterol fed samples. The tick-marks above the plot indicate the presence of a ChIP-identified binding peak near the gene sampled in the expression microarray and the relative peak density within the ranked gene list is also displayed around the tick marks. The high peak density within the high differentially expressed region of the GSEA revealed a very high correlation between the two data sets (p =2e−14).
Figure 3. Analysis of SREBP-2 peaks.
(A) Gene Set Enrichment Analysis: KS plot. The gene list for the ChIP-seq peaks that were located within 20 kb of a known gene was compared for their correlation to expression of all mouse genes rank ordered for degree of differential expression in RNA isolated from livers of mice fed chow supplemented with LE versus chow supplemented with cholesterol as described in Experimental Procedures (Subramanian et al., 2005). All genes that were expressed based on the microarray (~2.5 × 104) were ranked by fold difference and p value (x-axis) and the graph plots the running enrichment score on the ordinate. Here, many of the genes from the ChIP-seq data set were located in the highly differentially expressed portion of the micorarray data set indicating the genes identified by the ChIP-seq analysis are differentially expressed in the liver of LE vs cholesterol fed mice (p-value =2×e−14). The red vertical dotted line is drawn at the microarray gene index corresponding to the maximum enrichment score (ES). Genes with a corresponding adjacent peak have a tick mark above the main plot. To indicate relative enrichment of the peak regions, peak region density is mirrored around the tick marks as indicated. (B) Web Logos for Motifs present in SREBP-2 ChIP-Seq Peaks. Four motifs were identified by running MEME on the 1800 peak sequences. Motif 1 occurred in 892 of the peaks (E-value = 1.4×10−278), motif 2 occurred in 193 of the peaks (E-value = 2.1×10−070), motif 3 occurred in 231 of the peaks (E-value = 7.8×10−047), and motif 4 occurred in 148 of the peaks (E-value = 1.4×10−031)
The motif finding program MEME (Bailey, 2002) was next applied to the sequences covered by the SREBP-2 peaks (Figure 3B). We found four motifs showing strong enrichment in the SREBP-2 peak regions (with E-values < 1.4×10−31). Interestingly, the highest scoring motif corresponds to an SREBP binding element that was defined by our previous studies using sequence alignment, in vitro mutagenesis, and chemical probing studies for recombinant SREBPs (Millinder-Vallett et al., 1996). The second highest scoring motif corresponds to an E-box that is prototypical of bHLH-LZ transcription factors. The generic E-box reads 5′-CANNTG-3′ and different bHLH proteins have a preference for the bases in the middle (Murre et al., 1994). It is interesting that the E-box identified by our global analysis 5′-CACGTG-3′ is identical to the preferred E-box identified for SREBPs by in vitro DNA site selection and mutagenesis (Kim et al., 1995). The additional over-represented motifs correspond to consensus sites for NF-Y and Sp1, two transcription factors known to function synergistically with SREBPs in transcriptional activation (Osborne, 1995).
Because the SRE motif (motif 1 above) is an atypical binding site for bHLHLZ proteins, we used site-specific manual ChIP to confirm SREBP-2 binding to several of the predicted SRE motif peaks. We designed primers to amplify the regions overlapping 12 SRE containing peaks and there was significant enrichment in all 12 with the SREBP-2 precipitated chromatin (Table 1).
Table I.
Primer pairs were designed to amplify the predicted SREBP-2 binding region from 12 genes predicted to harbor the SRE element and two separate sets of chromatin prepared from mice fed the LE diet and enriched with the SREBP-2 antibody versus control IgG were analyzed by qPCR. Hmgcr and L32 served as positive and negative controls respectively.
| Gene | Fold Change (SREBP-2/IgG) | |
|---|---|---|
| Exp1 | Exp 2 | |
| Atg4b | 3.35* | 6.28* |
| Atg4d | 1.7* | 7.28* |
| Dhps | 8.11* | 3.91* |
| Mtf2 | 2.1# | 3.29* |
| Mthfr | 9.51* | 8.69* |
| Mtmr2 | 2.46# | 2.91* |
| Ncoa3 | 5.86# | 3.8* |
| Pds5b | 3.09# | 2.25 |
| Sirt1 | 2.93# | 1.48 |
| Snx13 | 9.1* | 1.58* |
| Crk | 1.67* | 2.21# |
| Hiat1 | 9.23* | 4.86* |
| Hmgcr | 7.16* | 20.16* |
| L32 | 0.89 | 1.38 |
p< 0.005,
p<0.05
Next, we used DAVID Gene Ontology software (Dennis et al., 2003) to categorize the SREBP-2 ChIP-seq genes by apparent functional annotation. In this analysis, broad biological categories that were significantly represented were mostly metabolic processes, and more specific categories included lipid metabolic processes and cholesterol metabolism as expected (Figure S4A). Several other categories were significantly associated with the SREBP-2 ChIP-seq data set, including regulation of apoptosis and autophagy (Figure S4B). SREBPs are known to be substrates for the apoptotic Caspase 3 in vitro and SREBP cleavage in cells can be induced by signals that initiate apoptosis (Wang et al., 1996). However, there was no prior suggestion that SREBPs might be associated with autophagy.
Autophagy is a catabolic response cells utilize to adapt to limiting nutrients in order to recapture and recycle cellular building blocks as well as to degrade damaged proteins and whole organelles and to attack intracellular pathogens (Mizushima and Klionsky, 2007; Mizushima and Levine, 2010; Sumpter and Levine, 2010). SREBP-2 is key to the cells response to low levels of cellular sterols, a key class of cellular nutrients. Cellular reserves of cholesterol are stored as cholesterol-esters within cytoplasmic lipid droplets along with triglycerides (TG) and recent observations suggest that liberation of stored lipids from within these droplets may be mobilized by an autophagy related process (Singh et al., 2009). When autophagy was blocked by knockdown of a critical autophagy protein, a significant fraction of total cell TG and cholesterol accumulated within autophagosomes (Singh et al, 2009). Additionally, sterol depletion has been shown to induce autophagy directly (Cheng et al., 2006). Thus, we reasoned that the activation of SREBP-2 during autophagy might be an important part of the cellular nutrient adaptation response to low sterols.
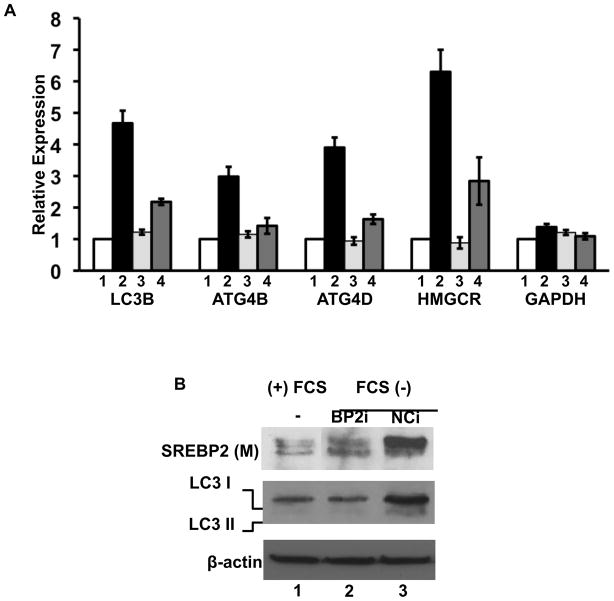
To analyze SREBP-2’s putative role in activation of genes involved in autophagy during sterol depletion, we subjected 293 cells to a standard sterol depletion protocol known to induce nuclear accumulation of SREBP-2 (Figure 4A). The autophagy genes LC3B, ATG4B, and ATG4D were predicted as SREBP-2 targets from the ChIP-seq analysis (Figure S4B) and expression of all three were induced by the sterol depletion similar to Hmgcr analyzed as a control. The induction was dependent on SREBP-2 because the inclusion of an siRNA targeting SREBP-2 dramatically reduced the level of induction of all of these genes while having no effect on expression of the control Gapdh gene.
Figure 4. SREBP-2 regulates autophagy-related gene expression.
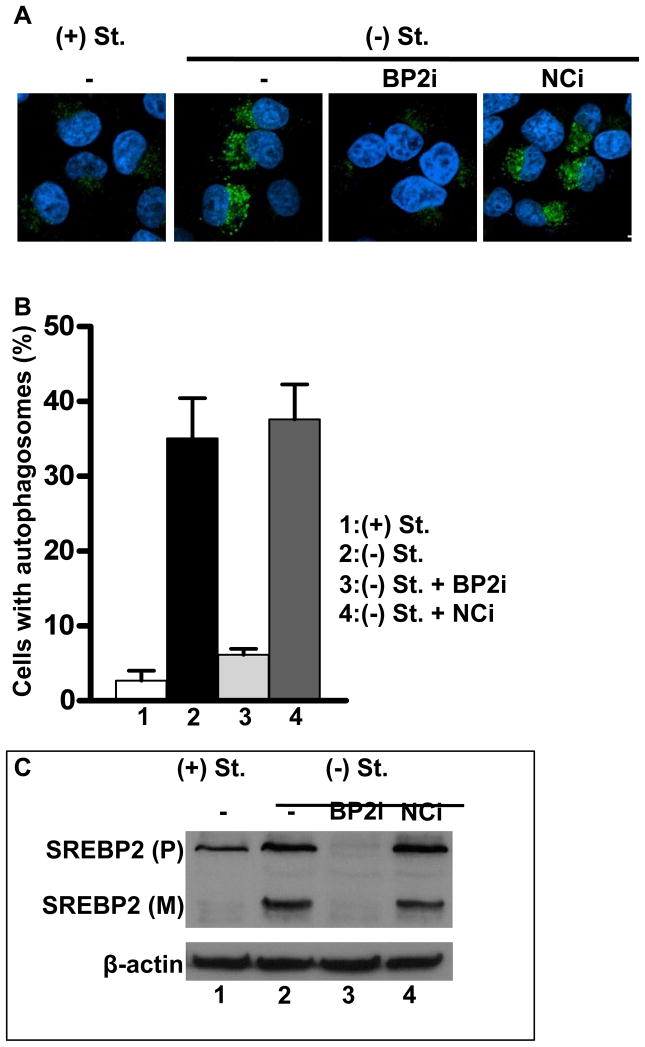
(A) 293 cells were transfected with an siRNA (10 nM) for a negative control siRNA (lanes 1 and 2) or siRNA targeting hSREBP-2 (lanes 3 and 4). Twenty-four hours after transfection, cells were cultured in 5% lipoprotein deficient media (LPDS) lanes 2 and 4 or LPDS plus sterols (12 μg/ml cholesterol and 1 μg/ml 25-hydroxycholesterol) for 24 h. and harvested for RNA or total protein (Sanchez et al., 1995). (A) mRNA levels for LC3B, ATG4B, ATG4D, HMGCR and GAPDH were analyzed by RT-qPCR and relative expression is plotted. (B) Whole cell lysates were analyzed for SREBP-2 or LC3 protein levels by immunoblotting. Data are representative of 3 separate experiments. Data are mean ± SEM; n = 3 for 3 separate experiments. HMGCR, HMG Co-A reductase; BP2i, SREBP-2 siRNA; NCi, negative control siRNA.
We also cultured 293 cells overnight in the presence and absence of serum and monitored the expression and maturation of SREBP-2 as well as the conversion of LC3 into the lipidated LC3-II form that occurs when autophagy is activated (Mizushima et al., 2010). Serum starvation induced LC3-II formation and this was also significantly reduced by the siRNA targeting SREBP-2 (Figure 4B).
The LC3-II form accumulates within the autophagosomal membrane prior to lysosomal fusion and this can be evaluated by the localization of LC3 into punctate immuno-staining foci (Mizushima et al., 2010). The LC3 staining in autophagosomes was significantly increased after serum starvation and this was severely blunted by inclusion of the SREBP-2 siRNA (Figure 5). LC3-II also accumulates within lipid droplet membranes targeted for lipoautophagy (Shibata et al., 2010; Singh et al., 2009) and our hypothesis predicts that LC3-II association with lipid droplets should also be regulated by SREBP-2 levels. When we monitored the co-localization of LC3 with lipid droplets using immunofluorescence for LC3 coupled with the fluorescent lipohilic dye bodipy to stain lipid droplets, we observed and increase in LC3 association with lipid droplets after serum starvation that was also reduced by the SREBP-2 siRNA (Figure 6A–D and S5). The degree of co-localization is unlikely to be influenced by a potential artifact resulting in red emission when the bodipy green dye stains lipid droplets (Ohsaki et al., 2010) because incubation with bodipy alone did not result in any evidence of false co-localization (Figure S5D).
Figure 5. SREBP-2 is required for autophagosome formation.
HeLa cells in 8-well chamber slides or 6-well plates were transfected with hSREBP-2 or negative control siRNA for 24h, and then cells were incubated with media containing 5% LPDS in the absence (−St) or presence (+St) of sterols (12 μg/ml cholesterol and 1 μg/ml 25-hydroxycholesterol) for 24 h. (A) Representative immunofluorescence images with LC3 antibody staining. (B) Quantitation of the percentage of cells with autophagosomes. (C) Immunoblotting with hSREBP-2 (IgG ID2) antibody. Data are mean ± SEM; n = 3 for 3 separate experiments. *p-value <0.05. Scale bar = 10 μm.
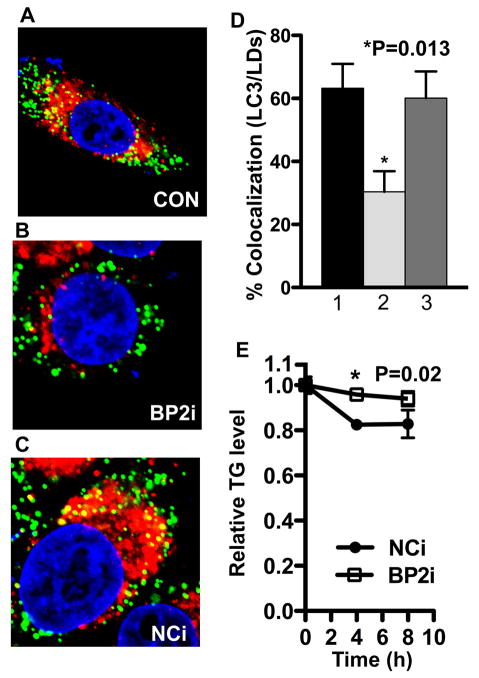
Figure 6. SREBP-2 deficiency decreases co-localization of lipid droplets (LDs) with autophagosome and triglyceride mobilization.
(A–D) HeLa cells in 8-well chamber slides were transfected with siRNA for hSREBP-2 or negative control siRNA for 24h, dishes were refed serum-depleted media with oleic acid (0.1 mM) for 24 h. (A–C): Co-localization of LDs (BODIPY 493/503, green) and autophagosome (LC3, red). (D) Quantitation of co-localization of LDs with autophagosomes. 1, control; 2, BP2i (SREBP-2 siRNA); 3, NCi (Negative control siRNA). Data are mean ± SEM; n = 3 for 3 separate experiments. *p-value <0.001. Scale bar = 2 μm. In all experiments, approximately 30 cells per sample were counted, and triplicate samples were counted per experimental condition. Additional information is provided in Experimental Procedures and additional cell images are provided in Figure S7. (E) HeLa cells were transfected with hSREBP-2 or negative control siRNA for 36h, and then cells were cultured with serum-depleted media for indicated time. Triglyceride levels were measured from total cell lysates and normalized with protein concentration. The p-value was calculated using t test (n = 3, Nci vs BP2i).
If SREBP-2 is essential for mobilization of lipid droplet contents through lipoautophagy then a knockdown of SREBP-2 during nutrient depletion conditions should delay the disappearance of cellular triglyceride levels similar to the delay observed when the autophagy pathway component ATG5 was targeted for knockdown (Singh et al., 2009). Consistent with this idea, cellular triglyceride mobilization after nutrient depletion was delayed when SREBP-2 was depleted (Figure 6E).
Discussion
We previously reported the genome-wide binding pattern for SREBP-1 in hepatic chromatin and uncovered a preferred binding motif that was not predicted from prior studies where SREBP-1 association was evaluated one gene at a time (Seo et al., 2009). Surprisingly, over 50% of the SREBP-1 binding sites were localized to within 2 kb of the 5′ end of known genes which is much higher than by chance (Chong et al., 2010) and higher than for other transcription factors similarly analyzed to date (Seo et al., 2009). The present study revealed an even stronger 5′ proximal preference for SREBP-2 indicating this is a feature shared by the two mammalian SREBPs.
In our analysis of SREBP-1, a software platform for peak-discovery was used that did not calculate a false discovery rate along with binding site identification. We focused on 467 peaks of SREBP-1 binding that were identified simply by overrepresented sequence reads and fold-change from the background as described (Johnson et al., 2007). We independently calculated that this corresponded to very conservative false discovery rate of 0.2%. However, the GLITR program used here for SREBP-2 directly calculates a p value and FDR for each peak and when a relatively conservative FDR of 1.5% was used a total of 1800 genomic binding sites for SREBP-2 were identified.
In order to perform a comparison of the binding profiles for SREBP-1 vs SREBP-2 we first re-analyzed our SREBP-1 binding data using the GLITR program with the same criteria for peak identification as for SREBP-2. This resulted in 1597 genome-wide peaks for SREBP-1 and showed the same promoter proximal preference as before (Figure S6). When the target genes for the two different SREBPs were analyzed for overlap, 11.7 % (187/1597) of the SREBP-2 targets were contained within the SREBP-1 data set. Although the pure number of overlapping peaks between SREBP-1 and SREBP-2 are modest, their overlap is significant (p value < 10−6; if the peaks were randomly distributed, < .05% of them would overlap).
The relatively modest overlap was lower than might have been predicted based on previous promoter studies in cultured cells or from knockout and over-expression experiments in mice. In these previous studies, both SREBPs were suggested to control expression of many of the same genes however there were some quantitative differences (Horton et al., 2002; Horton et al., 2003). At least part of the reason for the low overlap may be that we used different in vivo manipulations to selectively increase the nuclear levels of each SREBP before chromatin isolation and ChIP-seq analysis; refeeding following a fast for SREBP-1 and lovastatin/ezetimibe supplementation for SREBP-2. Regardless, the results clearly demonstrate that under conditions where each protein is specifically increased in liver, they bind mostly to different sets of target sites in the liver genome.
Results from both the gene set enrichment analysis and gene ontology ranking are consistent with SREBP-2 primarily activating genes involved in sterol metabolism, which validates our approach and confirms the quality of the data set. Additional GO categories relating to apoptosis and autophagy were also significantly enriched. SREBPs were shown to be associated with apoptosis soon after their initial identification (Wang et al., 1996) but the current study provides the first information for a potential link between SREBPs and autophagy. In this regard, it is interesting that studies showed autophagy was induced upon cellular sterol depletion (Cheng et al., 2006), which is a metabolic trigger for the proteolytic maturation of SREBPs.
Direct knockdown studies (Figures 4 and 5) confirmed that SREBP-2 both activates expression of key autophagy genes and is important for the formation of autophagosomes during cellular lipid depletion. Autophagy induction occurs very rapidly and a change in gene expression is not essential for its initial induction (Mizushima et al., 2010). However, increased expression of autophagy related genes through FoxO3 is associated with skeletal muscle breakdown by autophagy in vivo (Mammucari et al., 2007). Similarly, we propose that SREBP-2 selectively activates autophagy genes during conditions where cell sterol is limited and lipophagy is required to capture the lipid stored within the intracellular lipid droplet. Consistent with this idea, we showed that a decrease in SREBP-2 reduced association of LC3 with the lipid droplet and delayed TG mobilization (Figure 6).
Sterol depletion is a selective form of nutrient depletion and many other situations that trigger autophagy are independent of cell lipid metabolism. Additionally, as mentioned above an increase in gene expression is not essential to stimulate autophagy (Mizushima et al., 2010). Thus, it is unlikely that SREBP-2 is a general autophagy activator but may selectively trigger the response when cells require new lipid. In fact, when we measured LC3-II formation in cells following treatment with either chloroquine or vinblastine which block autophagosome flux at the lysosomal degradation or autophagosome-lysosome fusion step respectively, LC3-II formation was not affected even though SREBP-2 levels were reduced (Figure S5E).
Most cells have the capacity to store at least small amounts of cholesterol and fatty acids within cytoplasmic lipid droplets as neutral cholesterol-esters and triglycerides respectively and a recent report suggests that lipid contained within the droplets may be mobilized through an autophagy-like process coined macrolipophagy (Singh et al., 2009). Putting this observation together with our Chip-seq results, it was reasonable to hypothesize that genes involved in the induction of autophagy might be activated in response to conditions where cell sterols are limiting through activation of SREBP-2 and our additional experiments support this idea. Thus, the lipid droplet could be viewed as a third source of cellular cholesterol, which similar to the de novo synthesis and LDL receptor mediated uptake pathways, is also regulated by SREBP-2.
Experimental Procedures
Animals
All animal experiments were performed in accordance with accepted standards of animal welfare and with permission of the University of California, Irvine IACUC (protocol 97–1545). Six-week-old male C57BL/6 mice were obtained from Taconic and maintained on a standard rodent chow diet (2020X, Harlan Teklad Global) for one week with a 12 h light 12 h dark cycle for acclimatization. Mice were separated into 3 groups of 6 animals per group. One group was maintained on a chow diet and the other two groups were fed either normal chow supplemented with a mixture of lovastatin (100 mg lovastatin [2.5 tablet equivalents]/100 g chow, w/w; Mylan) and ezetimibe (from MERCK/Schering-Plough Pharmaceuticals; 21 mg ezetimibe [2.1 tablet equivalents]/100 g chow, w/w) (LE) or powdered cholesterol (1%, w/w) (Ch). All mice were sacrificed by CO2 asphyxiation at 8 AM (at the end of the dark cycle).
Chromatin preparation for ChIP assay and ChIP-seq
Chromatin preparation for ChIP assays and ChIP Seq using mouse livers were performed as previously described (Bennett et al., 2008; Seo et al., 2009). For gene specific ChIP, qPCR for SREBP-2 binding to specific gene promoters was analyzed in triplicate with a standard dilution curve of the input DNA performed in parallel and enrichment was measured by cyber green incorporation using an I-cycler (Bio-Rad). Analysis was performed by the standard curve method and values were normalized to a non-target control region from the ribosomal L32 gene. The qPCR oligonucleotide pairs for the mouse promoters were as previously described (Seo et al., 2009) or are included in Figure S7.
To prepare chromatin for the ChIP-seq, after isolating the ChIP-enriched DNA, gene-specific enrichment for known SREBP-2 target promoters relative to IgG control chromatin was verified. 20 ng of ChIP enriched DNA or control DNA was sent to Ambry Genetics (Aliso Viejo, CA) for high throughput DNA sequencing. The samples were blunt ended and adapters were ligated to the ends, according to the library preparation protocol from Illumina. Then DNA fragments with 200 ± 25bp in length were selected for the construction of the ChIP-seq DNA library. After size selection, all the resulting ChIPed DNA fragments were amplified and sequenced simultaneously using Solexa/Illumina Genome Analyzer.
For qPCR validation of Motif 1 Target sites, twelve randomly selected peaks were chosen for validation by gene specific ChIP. Chromatin from the LE samples was analyzed with primers designed to flank the peaks and to produce a PCR product of ~200 bases. The fold change is the fold increase for the signal from SREBP-2 antibody enriched chromatin relative to a control reaction where a control IgG fraction was used in place of the SREBP-2 antibody.
Preprocessing ChIP-Seq data
The ChIP-seq dataset was analyzed to determine peaks which contain binding sites of SREBP-2 to its target genes. Short reads of 39-bp were produced from Solexa/Illumina Genome Analyzer, and mapped to a reference genome by Ambry Genetics using ELAND, allowing one mismatch. Short sequence reads that mapped to simple and complex repeats or that were not unique by chance were removed from the analysis. We converted these files to BED files using the ChIP-Seq mini 2.0.1 suite developed by the Wold lab at California Institute of Technology (Johnson et al., 2007). The BED files were used as input to downstream processing, as well as visualization in the UCSC Genome Browser (http://genome.ucsc.edu/index.html).
Finding peaks and distance from peak to transcription start sites (TSSs)
To determine where the SREBP-2 bound to the genome, we looked for areas where there were significantly more enriched reads mapped in the ChIP sample than in the IgG. This was accomplished using GLITR (Tuteja et al., 2009). For each peak, the distance from the peak to the nearest transcription start site was determined, and plotted. The TSSs were taken from a RefSeq file obtained from NCBI. The background was determined by placing peaks at random locations on the genome and by determining distances to TSS.
Motif analysis
DNA sequences were retrieved using Galaxy (http://main.g2.bx.psu.edu) and used for motif search using MEME (Bailey, 2002). MEME represents motifs as position-dependent letter-probability matrices (PWM), which was used to determine score cutoff represented as a z-score. Sampling the genome hundreds of thousands of times, we determined a background score distribution and used a z-score threshold of 4.27 when searching for instances of the MEME motifs.
Annotation of genes and gene ontology (GO) analysis
All SREBP-2 binding sites were assigned to nearest genes based on the Mus musculus NCBI genome assembly (mm9; July 2007). GO analysis of SREBP-2 target genes was conducted by using the NIH Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) (Dennis et al., 2003). This analysis was used to classify the nearest gene list into functionally related gene groups by using ‘PANTHER Biological Process’ term.
Kolmogorov-Smirnov analysis
The obtained ChIP-seq data was compared with expression microarray data by using a Kolmogorov-Smirnov (KS) plot, a modified method of gene set enrichment analysis (GSEA) (Subramanian et al., 2005). The KS plot tests the null hypothesis that the ranks of the genes identified by ChIP-seq is uniformly distributed throughout the SREBP-2 expression microarray. KS plot was obtained by calculating the running sum statistics for our ChIP-seq gene set to observe enrichment in the ranked gene list from the expression microarray data.
Cell culture and RNA interference
293 and HeLa cells are were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) and antibiotics in an atmosphere of 5% CO2 at 37°C. The siRNA targeting human SREBP-2 (sc-36559) and siRNA control (sc-37007) were purchased from Santa Cruz Biotechnology. The cells were transfected for 24 h with 10 nM of each siRNA by using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions, and dishes and chamber slides were washed and refed with media with or without serum for 24 h. To block autophagic flux, HeLa cells were incubated with complete media containing 50 μM of chloroquine (Sigma-Aldrich) for 6 h.
RNA isolation, RT-qPCR, and gene expression profiling
Total RNA was isolated from mouse liver and 293 cells with Trizol (Invitrogen) and QIAGEN RNAeasy isolation kits (Qiagen) and used for RT-qPCR with a iQ5 Real-Time PCR Detection System (Bio-Rad). Primer sequences used in this study were shown in Figure S7. mRNA levels were normalized for expression of mouse ribosomal protein L32 and human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA as control and calculated by the comparative threshold cycle method. Gene expression profiling was carried out using the Mouse Gene 1.0 OST (Affymetrix) by hybridizing RNA from LE and Ch livers in triplicate at the Sanford-Burnham Genomics Core facility in Lake Nona Fl. Differential expression was then assessed using the Partek Genomics Suite (Partek Inc.) and cyberT (Long et al., 2001).
Immunoblotting
Polyclonal rabbit anti-SREBP-2 antibody against mouse SREBP-2 was generated as previously described (Jeon et al., 2008). Hepatic nuclear proteins and whole-cell lysates were subjected to SDS-PAGE (10 or 12 %), followed by transfer to a polyvinylidene difluoride membranes. The membranes are incubated with anti-SREBP-2 antiserum for mouse, anti-SREBP-2 (ID2) for human, anti-LC3B (Cell Signaling), anti-YY1 (Santa Cruz Biotechnology), and anti-β-actin (Sigma-Aldrich) as primary antibodies. After incubation with horseradish peroxidase-conjugated secondary antibodies, blots were developed using the SuperSignal West Femto Maximum Sensitivity Substrate from Thermo Scientific Inc.
Immunofluorescnce and Immunohistochemistry
Sections of paraffin-embedded livers were dewaxed in xylene and rehydrated, and antigen retrieval was performed with 10 mM citrate buffer. Sections were blocked for 30 minutes in 1% BSA, 0.02% Triton X-100, and 10% normal goat serum (NGS). Serial sections were incubated with SREBP-2 antibody (1:500), and then incubated with Cy5-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories Inc.). The sections are mounted by Vectashield with DAPI (Vector Laboratories). Images from serial sections were acquired by an inverted microscope Axioskop using the AxioVision camera and software (Zeiss) (Jeon et al., 2008). For immunocytochemistry, HeLa cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, blocked with 10% NGS, and incubated with anti-LC3B primary antibody followed by Alexa fluor 488-conjugated goat anti-rabbit secondary antibody before mounting. LDs were stained by incubating cells with BODIPY 493/503 (Invitrogen) for 30 min, fixed and processed for immunofluorescence as described above. Images from cells were acquired with a Laser Scanning Confocal microscope (A1R VAAS with ×60/numerical aperture 1.49 oil-immersion lens, Nikon) along with NIS-Elements AR software (Nikon). In all experiments, approximately 30 cells per sample were counted, and triplicate samples were counted per experimental condition. Quantification for autophagy was performed in deconvoluted and threshold limited images using the ‘Analyze Particle’ function in ImageJ software (NIH). For quantification of colocalizations, the ImageJ JACoP plug-in (Bolte and Cordelieres, 2006) was used to calculate the percentage of colocalization from Manders’ overlapping coefficients (fraction of red overlapping green) (Manders et al., 1992).
Data sets
Data sets including genome-mapped sequencing reads as well as micorarray expression values are provided at http://cbcl.ics.uci.edu/CompBioLab/index.php/Supplemental_Data.
Statistics
The data are presented as mean ± SEM. Differences between the means of the individual groups were assessed by 1-way ANOVA with Dunnet’s multiple comparison test; differences were considered significant at p-value < 0.05. The statistical software package Prism 5.0 (GraphPad Software) was used for these analyses.
Supplementary Material
Acknowledgments
We thank the Functional Genomics and Imaging Core facilities at Sanford Burnham Medical Research Institute at Lake Nona for help with the Affymetrix microarray and confocal imaging analysis respectively. This work was supported in part by NIH grant HL48044 to TO and NSF grant DBI-0846218 to XX. JB and HC were supported by NIH/NLM bioinformatics training grant T1507443. The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey TL. Curr Protoc Bioinformatics. Unit 2. Chapter 2. 2002. Discovering novel sequence motifs with MEME; p. 4. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Seo YK, Datta S, Shin DJ, Osborne TF. Selective Binding of SREBP isoforms and Co-Regulatory Proteins to Promoters for Lipid Metabolic Genes in Liver. The Journal of biological chemistry. 2008;283:15628–15637. doi: 10.1074/jbc.M800391200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21:505–517. [PubMed] [Google Scholar]
- Cheng J, Ohsaki Y, Tauchi-Sato K, Fujita A, Fujimoto T. Cholesterol depletion induces autophagy. Biochemical and biophysical research communications. 2006;351:246–252. doi: 10.1016/j.bbrc.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Chong HK, Infante AM, Seo YK, Jeon TI, Zhang Y, Edwards PA, Xie X, Osborne TF. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TI, Zhu B, Larson JL, Osborne TF. SREBP-2 regulates gut peptide secretion through intestinal bitter taste receptor signaling in mice. J Clin Invest. 2008;118:3693–3700. doi: 10.1172/JCI36461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science (New York, NY. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kim JB, Spotts GD, Halvorsen Y-D, Shih H-M, Ellenberger T, Towle HC, Spiegelman BM. Dual DNA binding specificity of ADD1/SREBP-1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. The Journal of biological chemistry. 2001;276:19937–19944. doi: 10.1074/jbc.M010192200. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S, Sandri M. FoxO3 controls autophagy in skeletal muscle in vivo. Cell metabolism. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mann RK, Beachy PA. Novel lipid modifications of secreted protein signals. Annu Rev Biochem. 2004;73:891–923. doi: 10.1146/annurev.biochem.73.011303.073933. [DOI] [PubMed] [Google Scholar]
- Millinder-Vallett S, Sanchez HB, Rosenfeld JM, Osborne TF. A direct role for sterol regulatory element binding protein in activation of 3-hydroxy-3-methylglutaryl coenzyme A reductase gene. J Biol Chem. 1996;271:12247–12253. doi: 10.1074/jbc.271.21.12247. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nature cell biology. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochimica et Biophysica Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Shinohara Y, Suzuki M, Fujimoto TA. pitfall in using BODIPY dyes to label lipid droplets for fluorescence microscopy. Histochem Cell Biol. 2010;133:477–480. doi: 10.1007/s00418-010-0678-x. [DOI] [PubMed] [Google Scholar]
- Osborne TF. Transcriptional control mechanisms in the regulation of cholesterol balance. Crit Rev Euk Gene Exp. 1995;5:317–335. doi: 10.1615/critreveukargeneexpr.v5.i3-4.50. [DOI] [PubMed] [Google Scholar]
- Osborne TF, Espenshade PJ. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it’s been. Genes & development. 2009;23:2578–2591. doi: 10.1101/gad.1854309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez HB, Yieh L, Osborne TF. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. The Journal of biological chemistry. 1995;270:1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- Seo YK, Chong HK, Infante AM, Im SS, Xie X, Osborne TF. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata M, Yoshimura K, Tamura H, Ueno T, Nishimura T, Inoue T, Sasaki M, Koike M, Arai H, Kominami E, Uchiyama Y. LC3, a microtubule-associated protein1A/B light chain3, is involved in cytoplasmic lipid droplet formation. Biochemical and biophysical research communications. 2010;393:274–279. doi: 10.1016/j.bbrc.2010.01.121. [DOI] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumpter R, Jr, Levine B. Autophagy and innate immunity: triggering, targeting and tuning. Semin Cell Dev Biol. 2010;21:699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A, Cantley LC. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zelenski NG, Yang J, Sakai J, Brown MS, Goldstein JL. Cleavage of sterol regulatory element binding proteins (SREBPs) by CPP32 during apoptosis. EMBO J. 1996;15:1012–1020. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.