Abstract
Dendrites and axons are two major neuronal compartments with differences that are critical for neuronal functions. To learn about the differential regulation of dendritic and axonal development, we conducted a genetic screen in Drosophila and isolated the dendritic arbor reduction 1 (dar1) mutants, which display defects in dendritic but not axonal growth. The dar1 gene encodes a novel transcription regulator in the Krüppel-like factor family. Neurons lacking dar1 function have severely reduced growth of microtubule- but not F-actin-based dendritic branches. In contrast, overexpression of Dar1 dramatically increased the growth of microtubule-based dendritic branches. Our results suggest that Dar1 promotes dendrite growth in part by suppressing the expression of the microtubule-severing protein Spastin. Our study thus uncovers a novel transcriptional program for microtubule regulation that preferentially controls dendrite growth.
Introduction
Dendritic and axonal compartments have distinct morphological features that are fundamental to neuronal functions. During development, one neurite of the postmitotic neuron is specified as the axon, and then the remaining neurites are specified as dendrites. Subsequently, the developing dendrites and axons follow separate paths to form two compartments that are distinct in structure and function. The past several years have seen substantial progress in the elucidation of the molecular mechanisms underlying axon specification (Wiggin et al., 2005; Tahirovic and Bradke, 2009). However, how the dendritic and axonal compartments of a neuron diverge in their development after the postmitotic neuron is polarized remains mostly unknown.
Both the microtubule (MT) cytoskeleton and the intracellular membrane system have been proposed to be important for the differential development of dendrites and axons (Baas, 1999; Ye et al., 2007). Microtubules are oriented differently in dendrites and axons. In the axons, microtubules are oriented uniformly with their plus-ends pointing distally, whereas there are microtubules with either orientation in dendrites (Baas et al., 1988). As microtubules are essential both for transporting molecules and organelles and for extending neurites, such a differential organization between dendrites and axons is likely to have profound impact on separating dendrite and axon development.
The secretory and endocytic pathways of the intracellular membrane system also contribute to the distinction between dendrite and axon growth. The rate of endocytosis in dendrites is much higher than that in the axon (Ye et al., 2007). This leads to a greater demand of membrane supply since the vast majority of the endocytosed plasma membrane are returned to the soma (Parton et al., 1992). Indeed, when the secretory pathway function is compromised as a result of mutations in key regulators such as Sar1, the dendritic growth is preferentially reduced (Ye et al., 2007). Furthermore, dendritic but not axonal growth is altered by disruption of PKD (protein kinase D) function, which regulates membrane protein exit from trans-Golgi network (Yeaman et al., 2004), via overexpression of a dominant-negative form (Horton et al., 2005). It thus seems likely that the membrane and membrane proteins required for dendritic or axonal growth are sorted in the trans-Golgi network.
How the microtubule cytoskeleton and the intracellular membrane system are regulated to contribute to the differential development of dendrites and axons is unknown. Through a genetic screen in Drosophila, we isolated the dendritic arbor reduction 1 (dar1) complementation group that displays specific defects in dendrite development (Ye et al., 2007). The dar1 gene encodes a novel member of the Krüppel-like factor (KLF) family, which regulates gene transcription. We show that dar1 is a critical regulator of dendritic microtubule cytoskeleton. Our results also suggest that Dar1 promotes dendrite growth in part by suppressing the expression of the microtubule-severing protein Spastin. These findings lend support to the notion that dendrite and axon development are controlled by partly non-overlapping genetic programs.
Materials and Methods
Mosaic analysis with a repressible cell marker analyses.
For mosaic analysis with a repressible cell marker (MARCM) analysis of dar1 mutations, the elav-Gal4, hsFLP, UAS-mCD8::GFP; tubP-Gal80, FRT2A virgins were mated with males of dar1, FRT2A/TM6B, Tb. Eggs collected over a 2 h period were allowed to develop for another 3 h before heat shock at 38°C for 1 h. Third-instar larvae were then selected under a fluorescence stereomicroscope for green fluorescent protein (GFP) fluorescence in the peripheral nervous system (PNS), and were subsequently dissected to make fillet preparation for immunostaining. These fillets were double stained with anti-mCD8 to label mosaic clones and a pan-neuronal marker, anti-HRP, to identify neurons. Only clones in abdominal segment 4–6 (A4–6) were imaged for quantitative analyses of the dendrites and only those in A5–7 were used for measurements of axon terminals to ensure consistency.
To overexpress Knot in dar1 mutant neurons with MARCM, we crossed hsFLP; GAL421-7, UAS-mCD8::GFP; tubP-Gal80, FRT2A flies and UAS-knot; dar13010/TM6B, Tb flies.
Flip-out analysis.
Flies carrying hsFLP; ppk-GAL4, UAS-FRT-CD2stop-FRT-CD8::GFP were mated with flies carrying UAS-Dar1 or T32 (UAS-Spastin). Eggs collected for 2 h were allowed to develop for 24 h before heat shock. Third-instar larvae were then selected for single Flip-out clones, dissected to expose the ventral nerve cord, and immunostained with a chicken anti-GFP (Aves) and mouse anti-CD2. Stained samples were imaged with a Leica SP5 confocal system.
Generation of germline clones deficient for dar1.
To remove the possible maternal contribution of Dar1, we crossed hs-flp; dar13232, FRT2A/TM3, Sb virgins and w/Y; OvoD, FRT2A/TM3, Sb males. Eggs were collected for 24 h, aged for 24 more hours, and heat-shocked for 1 h at 37°C for 2–3 d to induce germline clones that are homozygous mutant for dar13232. The non-TM3, Sb virgins of the progeny (hs-flp/w; dar13232, FRT2A/OvoD, FRT2A) were then mated with dar13010, ppk-eGFP/TM3, Sb, Krüppel-GFP males. We confirmed that the OvoD, FRT2A chromosome is dominant sterile. Hence, all the progeny received maternal contribution from dar13232 germline clones. The progeny embryos were then imaged with a confocal microscope for analyses of dendrite and axon morphology. The dar1 heterozygotes can be easily distinguished by the presence of Krüppel-GFP in the balancer chromosome and the lack of ppk-eGFP. Because the progeny genotype is dar13232, FRT2A/dar13010, ppk-eGFP, they are also incapable of zygotic production of wild-type Dar1.
Generation of anti-Dar1 antibody and immunostaining of Drosophila embryos.
The EcoRI-XhoI fragment of CG12029 cDNA, which encodes 127 aa from P164 to E290, was digested from expressed sequence tag clone IP01101 and inserted into the glutathione S-transferase (GST)-expression vector pGEX4T3. This construct was then transfected into bacteria BL21 for expressing GST-fusion protein. GST-fusion protein was purified with glutathione-agarose (Sigma-Aldrich) according to the manufacturer's instructions. The GST-fusion protein was injected into guinea pigs to generate antiserum against Dar1 (Cocalico Biologicals). Immunostaining of embryos was performed following the procedure described by Patel (1994).
Generation of Dar1 transgenic fly lines.
To generate UAS-Dar1 transgenic lines, CG12029 cDNA was subcloned into the transformation vector pUAST and injected into w1118 embryos, together with a DNA construct encoding the transposase Δ2-3 (Rubin and Spradling, 1982). To generate the genomic-cDNA hybrid transgene for rescuing dar1 mutants, we fused CG12029 cDNA to a 2182 bp genomic DNA (putative native promoter) upstream of the putative transcription initiation site of CG12029, and subcloned it into the transformation vector pW8.
Real-time PCR with purified Drosophila PNS neurons.
PNS neurons labeled by the marker GAL4[21-7], UAS-mCD8-GFP was dissociated from embryos and then purified by flow cytometry. The embryos were collected for 2 h and aged for an additional 14 h before dissociation. After, the chorion was removed with 50% bleach followed by extensive rinses with water. The embryos were broken apart in ice-cold Drosophila Schneider's insect medium (DSM) (Invitrogen) containing 1 U/μl RNase-Out (Invitrogen) by several gentle strokes with a Potter-Elvehjem grinder. The suspension was filtered through a 100 μm cell strainer (Falcon; BD Biosciences Discovery Labware), washed with DSM, and incubated with Liberase Blenzyme (Roche) for 30 min at 30°C. Dissociated cells were filtered through a 40 μm cell strainer. 4′,6-Diamidino-2-phenylindole (DAPI) was added and incubated for 30 min to label dead cells. GFP-positive but DAPI-negative cells are sorted by BD FACSVantage SE.
GFP-positive cells were collected for total RNA extraction with the Absolute RNA Miniprep kit (Stratagene). First-strand cDNA was synthesized from the total RNA of wild-type and dar1−/− PNS neurons with the SuperScript III First-Strand Synthesis SuperMix (Invitrogen) and used as the templates for real-time quantitative PCR. The ABsolute QPCR SYBR Green ROX mix was used for real-time PCR with the Applied Biosystems 7300 system. Gene-specific primers were included in the PCR to detect the levels of the transcripts. A pair of primers specific for Chmp1, which encodes the chromatin modifying protein 1 (Stauffer et al., 2001), was used as the control because there was no change in Chmp1 transcripts between wild-type and dar1 neurons.
Imaging, image processing, and quantification of dendrites and axons.
Live imaging of Drosophila embryos and larvae were performed with a Bio-Rad MRC-600 confocal microscope with a 40× objective lens. Staining of embryos and third-instar larval fillets was imaged with a Leica TCS SP5 confocal microscope (Leica Microsystems). The Neurolucida software (Visage Imaging) was used to quantify the length of dendrites and axons and to perform the Sholl analysis.
Results
Dar1 specifically controls dendrite development
We identified several mutants that displayed preferential defects in the dendrites but not the axons of dendritic arborization (da) sensory neurons through a genetic screen in Drosophila (Ye et al., 2007). These mutations include critical components of the early secretory pathway (Ye et al., 2007), thereby revealing a distinct sensitivity of growing dendrites to changes in the endoplasmic reticulum-to-Golgi transport. We have conducted additional analysis of another gene, dar1, and confirmed its specific function in dendritic growth.
We first took advantage of the highly selective expression of pickpocket (ppk)-eGFP transgenic marker in class IV da neurons to examine the dendrite and axon development of the dar1 mutant neurons (Grueber et al., 2003). Two alleles of the dar1 complementation group, dar13010 and dar13232, were isolated from the screen. In both dar1 mutants, major dendrites of class IV da neurons were either missing or shortened, consequently resulting in a reduced dendritic arbor (Fig. 1A). In contrast, the axonal growth of these neurons was normal (Fig. 1A).
Figure 1.

dar1 specifically controls dendrite development. A, The class IV da neurons display reduced dendritic growth but normal axonal growth in both dar13010 and dar13232 mutant embryos. Dendrites and axons were live-imaged with the class IV da neuron-specific marker ppk-eGFP. The dendritic morphology of single class IV neurons is shown in the top panels, and the axonal terminals of these neurons, located in the ventral nerve cord, are show in the bottom panels. The ladder-like structures of the axons are formed by all class IV neurons (6 in each segment). B, The class I da neurons display reduced dendritic growth in dar1 mutant embryos. Dendrites were delineated with the marker GAL42-21, UAS-mCD8-GFP. Because of the lack of a specific marker for class I da neurons, axon morphology of these neurons cannot be assessed in embryos. C, Dar1 mutant embryos without any potential maternal and zygotic contributions of Dar1 display the same dendrite and axon phenotype as zygotic dar1 mutants. The class IV da neurons were visualized with the ppk-eGFP marker. D, The dendrite phenotype of dar1 mutants is a result of defective outgrowth. Dendrite defects were visible at 16–17 h AEL shortly after the initiation of dendrite growth, the earliest time point that can be imaged with the marker GAL4109-2-80, UAS-mCD8-GFP, and became more pronounced in 18–19 h AEL.
We also examined dendrite development in class I da neurons, which have the simplest dendritic arbor among all four classes and can be visualized by using the GAL42-21 driver and found the dendrites of class I da neurons were also severely shortened (Fig. 1B). Thus, the dendrite defects in dar1 mutants are not restricted to class IV da neurons.
Similar to most types of neurons, Drosophila da neurons start to extend axons several hours before dendritic growth starts. To test the hypothesis that maternally contributed wild-type Dar1 protein is sufficient to support the axonal development but not the subsequent dendritic development, we removed the possible maternal contribution of Dar1 by generating germline clones mutant for dar1 in the mothers. We examined the embryos that are derived from such homozygous mutant germline clones and also bear two zygotic mutant alleles of dar1. These embryos with both maternal and zygotic dar1 mutations are totally devoid of Dar1 (see Materials and Methods). To visualize the class IV da neurons, we incorporated a ppk-eGFP marker into this mosaic analysis. As shown in Figure 1C, removing potential maternal Dar1 as well as zygotic Dar1 from the embryos led to the same phenotype as the zygotic mutant phenotype, that is, the dendrites were reduced but the axons were normal. These results confirm that Dar1 specifically controls dendrite but not axon development.
To determine whether the dendrite phenotype of dar1 mutants is a result of defective outgrowth or enhanced retraction of existing branches, we examined dendrite development at the earliest time point possible given the existing markers. Expressing mCD8::GFP with the pan-da neuron GAL4 driver GAL4109-2-80 allowed us to visualize dendrites as early as 16–17 h after egg laying (AEL), when da neurons normally extend a few short dendrites (Gao et al., 1999). Dendrite defects were visible at this early stage of development and became more pronounced in 18–19 h AEL (Fig. 1D). These results suggest that dar1 mutations cause reduced outgrowth of dendrites.
Dar1 functions cell-autonomously to control dendrite development
To determine whether the dendrite-specific defects in dar1 mutants were attributable to defective dar1 gene in the neurons or the surrounding tissues, we performed MARCM. This analysis allows us to generate individual neurons that are homozygous for dar1 mutation and express a transgene such as GFP in a heterozygous animal (Lee and Luo, 1999).
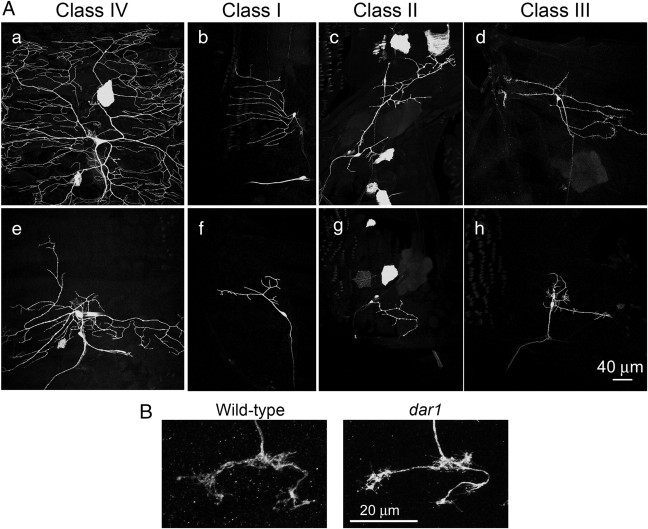
Single neurons homozygous for dar1 mutation exhibited severely reduced dendritic arbors (Fig. 2A), supporting the notion that dar1 functions cell-autonomously. MARCM analysis also allowed us to examine the functional requirement of dar1 in all four classes of da neurons. We found that, in all four classes of da neurons, dendrites were dramatically reduced by the dar1 mutation (Fig. 2A). Compared with the dendrite lengths of control da neurons, namely 1304.6 ± 120.6 μm (mean ± SEM) (n = 23) for class I (ddaD&E), 1126.2 ± 42.1 μm (n = 9) for class II (ddaB), 2582.0 ± 147.6 μm (n = 7) for class III (ddaA), and 17,788.6 ± 444.5 μm (n = 7) for class IV (ddaC), the total dendritic length in dar1 mutant neurons was dramatically reduced to 259.5 ± 147.3 μm (n = 9) for class I, 549.7 ± 32.6 μm (n = 4) for class II, 627.6 ± 162.6 μm (n = 3) for class III (excluding the dendritic filopodia), and 6498.2 ± 581.7 (n = 11) for class IV (p < 0.01 for all neuron types, t test).
Figure 2.
dar1 functions cell-autonomously to specifically control dendrite development. A, MARCM clones of da neurons homozygous for dar1 mutation. a–d, Control neurons. e–h, dar1 mutant neurons. B, Axonal growth of dar1 mutant neurons is normal. The axons of the dar1 mutant neurons not only extended normally from the body wall to the ventral nerve cord, which is ∼1700 μm away (Ye et al., 2007), but also developed terminals that are indistinguishable from those of wild-type neurons. Shown in the figure are the axonal terminals of a wild-type and a dar1 mutant class IV neuron ddaC in abdominal segment 6.
We also analyzed the axonal development of da neurons with the MARCM technique and quantified the axon terminal projections of the class IV da neurons. In contrast to dendrites, there was no significant defect in axonal growth in dar1 mutant neurons (Fig. 2B). The axon terminal length of single class IV neuron ddaC in abdominal segment 5–7 was 68.4 ± 4.2 μm (mean ± SEM) (n = 21) in wild-type and 70 ± 7.0 μm (n = 9) in dar1 mutant neurons (p = 0.84, t test).
MARCM analysis was also applied on the mushroom body neurons (Lee and Luo, 1999) to determine whether dar1 might be involved in the development of central neuron dendrites. We found no obvious defects (data not shown), consistent with our finding that Dar1 is not expressed in the CNS (see below).
Dar1 encodes a novel Krüppel-like factor
To identify the gene responsible for the dar1 phenotype, we first performed recombination mapping using the commonly used ru h th st cu sr e ca (rucuca) chromosome, which carries eight visible recessive markers on chromosome 3. For both dar13010 and dar13232 alleles, a lethal mutation between ru and h was responsible for the dendrite defects. We then conducted complementation tests between dar1 alleles and the genomic deficiencies (Df) in this region (Fig. 3A) and found that dar1 mutations lie in between CG14974 and CG10855. Results from site-specific male-recombination mapping (Chen et al., 1998) with the P-element KG07161 (Drysdale and Crosby, 2005), which is inserted distal to CG12006, also suggest dar1 is distal to CG12006 (data not shown). This mapping result was further confirmed by a recombination mapping with P-element insertions EY11305 and EY01819 (Fig. 3A).
Figure 3.
dar1 encodes a novel Krüppel-like factor. A, Genome view of the dar1 locus. Both Df(3L)GN34 and Df(3L)ΔEY01819 failed to complement lethality and dendrite defects, whereas Df(3L)R75 complemented both lethality and dendrite defects. The distal breakpoint of Df(3L)GN34 lies in between CG14974 and CG12029, and the proximal breakpoint of Df(3L)ΔEY01819 is in encore. The distal breakpoint of Df(3L)R75 lies in between CG32264 and CG10855. These results suggest that dar1 mutations lie in between CG14974 and CG10855. Recombination mapping with P-element insertions suggested that Dar1 is 0.08 cM proximal to the P-element insertion of EY11305 (in Eip63E), 0.5 cM distal to KG07161 (in CG12006), and 0.6 cM distal to EY01819 (in encore). B, The domain scheme of the protein product of CG12029. There are three zinc fingers in the C-terminal domain preceded by a NLS. dar13232, dar13010, and dar1D6 each carries a unique nonsense mutation in CG12029, which leads to truncation of the coding sequence [marked by an asterisk (*)]. C, A genomic-cDNA transgene rescues the dendrite defects of dar1 mutants. Homozygous dar1 mutant larvae carrying the rescue transgene were imaged at early third-instar stage. The bar chart presents the quantification of dendrite length in early third-instar larvae. **p < 0.01; N.S., not significant (p > 0.05, t test). Error bars indicate SEM. D, Amino acid sequence comparison of the zinc fingers of Dar1, mouse KLF5 (mKLF5), mouse KLF7 (mKLF7), and mouse (mKLF9). The numbers in the parentheses after the mouse KLFs indicate sequence similarity between each mouse KLF and Dar1. Residues identical in all four proteins are marked with yellow background. Those conserved in some of the four proteins are shown in light blue background. Residuals similar to the conserved ones are shown in pink background. Dar1 and mKLF5 also share a stretch of amino acids upstream of the zinc finger region, which is not shown here.
We sequenced the predicted exons of CG12029 and CG10862 as well as the neighboring genes CG14974, CG10858, CG32264, and CG10855, and found mutations that alter the coding regions in CG12029 only. In both dar13010 and dar13232, there was a unique nonsense mutation that generates a premature stop codon so that a fragment of only the N-terminal half of the protein can be produced (Fig. 3B). Previously, a genetic screen identified a number of lethal mutations within the 63E-64 genomic region (Harrison et al., 1995). We performed complementation tests between these mutants and dar1 alleles and found that the mutant l(3)63EdD6 failed to complement the dendrite defects and lethality of dar13010 and dar13232. These results suggest that l(3)63EdD6 is allelic to dar1, and we thus renamed this lethal mutation dar1D6. Similar to dar13010 and dar13232, dar1D6 carries a nonsense mutation, which leads to a premature stop codon after approximately two-thirds of the protein coding sequence (Fig. 3B). In addition to dar1 D6, we also found that the lethal piggyBac insertion PBac(WH)f06798 failed to complement the lethality and dendrite defects of dar1. PBac(WH)f06798 is reported to be inserted between exons 2 and 3 of CG12029 (Thibault et al., 2004), which we confirmed by inverse PCR. This again confirmed that the dar1 gene is CG12029.
To determine whether the dendrite defects caused by dar1 mutations could be rescued by expressing wild-type CG12029, we generated a transgenic line that carries the CG12029 cDNA fused to a 2182 bp genomic DNA upstream of the putative transcription initiation site of CG12029. This genomic-cDNA hybrid transgene rescued the dendrite defects in both class IV (Fig. 3C) and class I (data not shown) neurons mutant for dar1, further confirming that CG12029 is the dar1 gene.
Sequence analysis with the basic local alignment search tool suggests that CG12029 encodes a novel Krüppel-like factor in Drosophila. The hallmark of KLFs is the presence of three zinc fingers in the C-terminal domain after a nuclear localization signal (NLS). The sequences before the NLS are highly variable among family members. Dar1 contains all three zinc fingers and an N-terminal fragment that displays little homology to any other protein in fly or mammalian genome (Fig. 3D). The closest mammalian homologs of Dar1, based on the zinc finger alignments, are transcription factor KLF5 and 7. Dar1 and KLF5 also share a stretch of amino acids before the zinc finger region, including a PY motif that could bind E3 ubiquitin ligase and is responsible for ubiquitin-mediated degradation of KLF5 (Chen et al., 2005). The NLS and zinc fingers are absent in the dar13010, dar13232, and dar1D6 alleles because of their premature stop codons.
We then generated antibodies against Dar1 and performed immunostaining in embryo to study its expression pattern. Dar1 was highly enriched in the PNS and absent in the CNS (Fig. 4A). Interestingly, Dar1 was present in neurons that elaborate more than one primary dendrite, including da neurons, bd neurons, and the dmd1 neuron, but undetectable in neurons with single dendrites, such as external sensory organ (es) neurons and chodotonal neurons (Fig. 4B). In addition to PNS, Dar1 immunoreactivity was also observed at low levels in non-neuronal cells at the segmental borders and part of the hindgut (data not shown). In all the cells that express Dar1, Dar1 was localized to the nucleus, consistent with the role of KLFs as transcriptional regulators.
Figure 4.
Localization of Dar1 protein in wild-type and dar1 mutant embryos. Stage 13 embryos were stained with antibodies against Dar1 (green) and neuron-specific transcription factor Elav (magenta). A, Immunostaining of a wild-type embryo showing that Dar1 protein is present in the PNS but absent in the CNS. B, Immunostaining of the dorsal cluster of the PNS. Dar1 is present in the nuclei of neurons extending more than one dendrite (the da neurons, dbd neuron, and dmd1 neuron) but is absent in neurons having a single dendrite (es neurons). C, Cytoplasmic rather than nuclear Dar1 immunoreactivity in dar1D6 mutant embryos carrying a premature stop codon preceding the coding sequences for the NLS and zinc fingers. The immunosignal in each neuron occupies a larger area than that of the nuclear staining in wild-type animals.
The antigen used for raising the antibody is a GST-fusion protein of the distinctive N-terminal fragment of Dar1. In the dar13010, dar13232, and dar1D6 mutant embryos, this fragment might still be present. Consistent with this, immunostaining revealed that the Dar1 antigen became cytoplasmic because of the loss of the NLS (Fig. 4C). We subsequently generated another allele, dar1sy, by random mutagenesis with diepoxybutane and selecting for its failure to complement the dar13010 allele. This mutation turned out to be a deletion that removed CG12029 in addition to CG14947. There was no detectable immunostaining in the embryos of this allele (data not shown), confirming the specificity of the antibody.
Dar1 preferentially regulates microtubules
The da neurons in Drosophila PNS offer a wealth of features for analyzing different aspects of dendrite development, including different compositions of microtubule and actin cytoskeleton in different types of dendritic branches. The class III da neurons have characteristic dendritic filopodia (also termed “dendritic spikes”) (Grueber et al., 2002), which are distributed along major dendrites and are usually straight and devoid of additional branching. These dendritic spikes are enriched with filamentous actin (F-actin) and are essentially free of microtubules (Andersen et al., 2005; Jinushi-Nakao et al., 2007). In contrast, the major dendrites of the same neurons are enriched with microtubules and contain F-actin at a level much lower than that in the dendritic spikes. The separation of F-actin- and microtubule-based dendritic branches allowed us to investigate the effect of dar1 mutations on these two types of cytoskeleton.
Although the major dendrites were dramatically reduced by ∼75% in the class III da neurons mutant for the dar1 gene (Fig. 2A), the density of dendritic spikes was much less affected. The densities of dendritic spikes were 1.9 ± 0.4 (n = 8) and 1.3 ± 0.4 (n = 6) per 10 μm of major dendrite (mean ± SD) in wild-type and dar1 mutant class III da neurons (p = 0.04, t test), respectively. This raised the possibility that dar1 preferentially regulates microtubule cytoskeleton. To test this hypothesis, we overexpressed Rac1 in class I da neurons mutant for dar1. Rac1 is a small GTPase that regulates actin cytoskeleton. Overexpression of Rac1 (OE Rac1) in many neuron types, including the da neurons (Lee et al., 2003; Emoto et al., 2004), leads to hyperbranching of dendrites. The dar1 mutations did not block the branching activity of Rac1 (Fig. 5A,B), suggesting that Dar1 is not required for regulating actin-cytoskeleton.
Figure 5.

dar1 is not required for forming F-actin-enriched dendritic filopodia. A, Representative images showing that dar1 mutations do not block filopodium formation induced by overexpressing Rac1. GAL42-21 was used to drive the expression of mCD8-GFP and Rac1 in class I da neurons. B, Quantification of terminal branch number per 100 μm of major dendrites. Note that class I neurons in dar1 mutant embryos display increased number of terminal branches that are transient as they disappear in larval stages and are not visible in MARCM clones. *p < 0.05, t test. Error bars indicate SEM. C, Overexpression of Dar1 results in overgrowth of dendrites. Representative images of control and Dar1-overexpressing (“OE Dar1”) class IV da neuron. GAL44-77 was used to express mCD8-GFP and Dar1 in class IV da neurons. Images were collected from early third-instar larvae. D, Sholl analysis of the dendritic morphology of control and Dar1-overexpressing class IV da neurons. Diamond, Control neurons; dot, Dar1-overexpressing neurons.
To further test whether Dar1 is capable of promoting MT growth, we generated transgenic lines to express Dar1 in da neurons. Overexpressing Dar1 (OE Dar1) with the driver GAL44-77 caused dramatic overgrowth of dendrites in the class IV da neurons in early third-instar larvae (Fig. 5C). Control neurons had 350.2 ± 14.0 (mean ± SEM; n = 6) branch points, excluding the dendrites around the segmental borders where dendrites of different neurons intermingle. In contrast, neurons overexpressing Dar1 had 930.7 ± 17.3 (n = 6) branch points (p < 0.01, t test). Similarly, the total dendritic length of control neurons, excluding the dendrites around the segmental borders, was 9159.0 ± 217.0 μm (n = 6), whereas that of neurons overexpressing Dar1 was 18,044.0 ± 278.8 μm (n = 6) (p < 0.01, t test). These results suggest that Dar1 is not only necessary but also sufficient for promoting dendritic growth. Sholl analysis showed that the increase in branch number and length took place throughout the dendritic fields rather than being restricted to particular regions (Fig. 5D). No change in the size of dendritic fields or dendritic tiling was observed.
We also used the Flip-out technique (Jiang and Struhl, 1995) to study the effects of overexpressing Dar1 on class IV da neuron axons. In contrast to its effects on dendrites, overexpression of Dar1 did not significantly affect axon growth. The axon terminal length of wild-type class IV neurons was 68.4 ± 4.2 μm (mean ± SEM) (n = 21), and that of class IV neuron overexpressing Dar1 was 77.7 ± 5.7 μm (n = 26) in dar1 mutant neurons (p = 0.21, t test).
Overexpressing Dar1 in class I da neurons also dramatically increased dendrite growth (data not shown). Strikingly, when Dar1 was overexpressed in the class III da neurons, we observed an increased number of long curvy terminal branches (Fig. 6) that are reminiscent of microtubule-containing branches.
Figure 6.

Dar1 promotes microtubule-based dendritic growth. F-actin marker GMA (green channel in left column) and microtubule marker tubulin-GFP (green channel in middle and right columns) were expressed in the class III neuron ddaA together with the membrane marker mCD8-mRFP (red channel in all three columns) for dendritic morphology by the driver GAL419-12. The blue arrowheads point to several examples of the dendritic spikes, which are enriched with GMA (F-actin) but lack tubulin-GFP. Overexpression of dar1 leads to elongated branches that contain tubulin-GFP.
To directly visualize microtubule and F-actin in the class III da neurons overexpressing Dar1, we made use of tubulin-GFP and the F-actin marker GMA. GMA is a chimeric protein with the actin binding domain of Drosophila moesin fused to the C terminus of GFP (Dutta et al., 2002) and is enriched at the dendritic spikes of class III da neurons (Andersen et al., 2005; Jinushi-Nakao et al., 2007). Overexpressing Dar1 in class III da neurons led to the presence of MT in the ectopic terminal branches (Fig. 6). No enrichment of F-actin was observed in these branches (data not shown), suggesting overexpression of Dar1 promoted the growth of microtubule-containing major dendrites but not the F-actin-enriched filopodia.
Together, these results suggest that dar1 preferentially regulates microtubule cytoskeleton to mediate its specific control of dendrite growth.
Dar1 restricts the expression of the microtubule-severing protein Spastin
The microtubule-severing protein Spastin has been found to regulate dendrite development in Drosophila da neurons (Jinushi-Nakao et al., 2007). Because Dar1 preferentially regulates microtubule-based dendritic growth, we asked whether Dar1 could regulate Spastin expression. Currently, no antibody against Drosophila Spastin provides sufficient sensitivity to detect endogenous Spastin protein level. We therefore determined the levels of Spastin transcripts in dar1 mutant neurons by quantitative real-time PCR (qPCR). The qPCR technique was preferred to other techniques, such as in situ hybridization, for both consistency in quantification and sensitivity. Total RNA was extracted from purified da neurons of dar1 mutant embryos as well as from those of wild-type embryos. Real-time PCR was used to compare the amounts of Spastin transcripts between wild-type and dar1 mutant da neurons. The levels of Spastin transcripts were significantly elevated in dar1 mutant neurons. Transcripts from dar1 mutant neurons on average took 1.44 less PCR cycles to reach the threshold cycle (Ct) for amplifying Spastin than those from wild-type neurons (n = 4; p < 0.05, t test), suggesting a 3.2-fold increase in Spastin transcript level in dar1 mutant neurons. In contrast, there is no difference in the Ct for amplifying the chromatin modifying protein 1 (Chmp1) (Stauffer et al., 2001) between wild-type and dar1 mutant neurons.
Consistent with the reduced dendrite phenotype in dar1 mutants, upregulation of Spastin with the EP insertion T32 (SpaT32), which is known to cause a modest overexpression (Sherwood et al., 2004), led to a dramatic reduction in both total dendritic length and branch number (Fig. 7A). Whereas the total dendritic length in wild-type class IV neurons was 17,789 ± 444.5 μm (mean ± SEM) (n = 7), it was reduced to 10,582 ± 580.3 μm (n = 4) in neurons overexpressing SpaT32 (p < 0.01, t test). Similarly, the number of branch points was reduced from 767.9 ± 22.8 in wild-type (n = 7) to 403.3 ± 43.5 in SpaT32-overexpressing neurons (p < 0.01, t test).
Figure 7.

Upregulation of the microtubule-severing protein Spastin in dar1 mutant neurons may be responsible for the dendrite defects. A, Representative images showing that upregulation of Spastin by overexpressing SpaT32 reduced both total dendritic length and branch numbers in class IV da neurons. B, Dar1 is essential for the transcription factor Knot, which promotes Spastin expression, to promote dendritic growth in class I da neurons. The MARCM technique was used to overexpress Knot in single class I da neurons. Representative images are shown. Overexpressing Knot causes dendritic overgrowth in wild-type class I da neurons (left), and dar1 mutations abolish the effects of Knot overexpression (right). C, Quantification of the total dendritic length in class I neurons overexpressing Knot, those defective of dar1, and those overexpressing Knot in dar1 mutant background. Error bars indicate SEM. *p < 0.01, t test. D, A model that explains how the Dar1–Spastin pathway differentiates dendrite and axon growth. Dar1 suppresses Spastin expression, either directly or indirectly, in addition to positively control the expression of an inhibitory mechanism of microtubule severing (e.g., MAPs or microtubule posttranslational modification) specifically in the axon. Spastin is capable of severing microtubules in both dendrites and axons. Whether the microtubule severing by Spastin promotes or inhibits dendrite/axon growth depends on the expression levels of Spastin.
These results together suggest that Dar1 restricts the expression of Spastin either directly or through a transcription repressor, allowing dendrite growth. Different from dar1 mutants, Spastin upregulation by expressing SpaT32 also resulted in a mild yet significant reduction in axonal growth from 68.4 ± 4.2 μm (n = 21) in wild-type class IV neurons to 55.2 ± 2.1 μm (n = 17) in SpaT32-expressing neurons (p < 0.05, t test).
The Collier/Olf1/EBF family transcription factor Knot has been proposed to regulate Spastin expression and promotes microtubule-based dendrite growth in class IV da neurons (Jinushi-Nakao et al., 2007). We asked whether Dar1 and Knot genetically interact to control dendrite development. Overexpressing Knot (OE Knot) in class I da neurons, which have the simpler dendritic arbors, leads to an increase in dendrite growth (Hattori et al., 2007; Jinushi-Nakao et al., 2007). We expressed Knot in single class I da neurons using the MARCM technique and observed significant increase in total dendrite length (Fig. 7B). In contrast, when Knot was expressed in neurons with dar1 mutation, the dendrites were severely reduced (Fig. 7B). To test whether Knot regulates Dar1 expression, we examined Dar1 protein levels by immunostaining in class I neurons and found no difference between control neurons and neurons overexpressing Knot. The mean intensity of Dar1 immunofluorescence was 88.1 ± 5.7 (arbitrary unit; n = 12) in control neurons and 80.6 ± 5.7 (n = 12) in neurons overexpressing Knot (p = 0.36, t test). Therefore, it is unlikely that Knot controls class IV dendrite development by regulating Dar1 expression.
Discussion
In this study, we show that a novel molecule in the KLF family of transcriptional regulators, Dar1, regulates the microtubule cytoskeleton during the differential development of dendrites and axons. Dar1 is specifically required for dendritic growth. Neurons lacking dar1 function exhibit reduced growth of microtubule-based dendritic branches. However, overexpression of Dar1 in neurons increased the growth of microtubule-based dendritic branches. Dar1 suppresses the expression of the microtubule-severing protein Spastin, either directly or indirectly. Upregulation of Spastin expression leads to a dendrite phenotype similar to that observed in dar1 mutant neurons.
Transcriptional programs that differentiate dendrite and axon development
Substantial progress has been made in the past several years on the specific regulation of axon growth. The anaphase-promoting complex (APC) and its activator Cdh1 form a multiunit complex of ubiquitin ligase in the nucleus of cerebellar granule cells to specifically restrict axon growth (Konishi et al., 2004). This function of Cdh1-APC requires the transcriptional repressor SnoN, which is a target of the ubiquitin-dependent degradation mediated by Cdh1-APC (Stegmüller et al., 2006). SnoN in turn requires a scaffold protein Ccd1 to promote axon growth (Ikeuchi et al., 2009). The TGFβ-regulated signaling protein Smad2 plays an important role in regulating the Cdh1-APC-SnoN (Stegmüller et al., 2008).
Neural activities can specifically induce dendrite growth by activating transcription factors. In cerebellar granule cells, knockdown of calcium/calmodulin-dependent protein kinase IIα (CaMKIIα) or the transcription factor NeuroD results in reduced growth of dendrites but not axons (Gaudillière et al., 2004). CaMKIIα phosphorylates NeuroD in response to neural activities. Moreover, synaptic activity-induced dendritic growth can be blocked by reducing NeuroD level, suggesting that CaMKII and NeuroD mediate the dendritic growth promoted by synaptic activity.
In this study, we have identified a novel transcriptional program that is specifically involved in the development of dendrites. Whereas whether Dar1 plays a role in activity-dependent dendritic growth remains to be determined, Dar1 is likely to be part of the activity-independent transcription program that controls dendrite growth in early development before the exposure to sensory inputs or the establishment of neuronal circuits, because the dendrite defects were observed soon after the initiation of dendritic growth in embryo (Gao et al., 1999) (Fig. 1D). The fact that dar1 is both necessary and sufficient for dendrite growth suggests that it may determine multiple aspects required for dendrite development. Indeed, although dar1 is specifically involved in dendrite growth and is likely a regulator of Spastin expression, Spastin is involved in both dendrite and axon development. This raises the possibility that Dar1 also controls the expression of other molecules which suppresses Spastin function in the axon. It will be interesting to identify such mechanisms by systematically identifying downstream molecules of Dar1.
The microtubule-severing protein Spastin in dendrite and axon development
Microtubule severing by Katanin and Spastin is important for axon and dendrite development (Yu et al., 1994; Jinushi-Nakao et al., 2007; Lee et al., 2009), possibly by keeping microtubules sufficiently short to be efficiently transported into developing neurites (Ahmad et al., 1999) or by creating more free ends of microtubules to interact with other proteins in developing processes. The Spastin gene, which encodes a microtubule-severing protein, is mutated at high frequency in autosomal dominant hereditary spastic paraplegia (Hazan et al., 1999). RNA interference (RNAi)-mediated knockdown of Spastin leads to reduced axon length in cultured hippocampal neurons from rats (Yu et al., 2008) and reduced dendrite growth in Drosophila da neurons in vivo (Jinushi-Nakao et al., 2007). Overexpression of Spastin in cultured hippocampal neurons has no effect in total axon length although results in increased branch numbers (Yu et al., 2008). In Drosophila neuromuscular junction, loss-of-function mutant of spastin exhibits an increase in synaptic bouton number but a reduction in bouton size (Sherwood et al., 2004). In the present studies, we found that overexpressing Spastin dramatically reduced dendrite growth in Drosophila da neurons. The fact that both RNAi knockdown and overexpression of Spastin lead to reduced dendrite growth is consistent with the notion that proper Spastin levels are important for neurite growth (Riano et al., 2009). We also observed a slight reduction in axonal length in da neurons on overexpressing Spastin. Our in vivo technique does not provide enough resolution to determine whether there is an increase in the number of fine branches of the axons (Yu et al., 2008). It is possible that PNS (this study) and CNS neurons (Sherwood et al., 2004) require distinct levels of Spastin for proper axon development. Alternatively, there might be a difference between mammalian and fly neurons in their requirement of Spastin for axon development.
The different phenotypes in axon development between dar1 mutants and Spastin overexpression may be explained by additional factors regulated by Dar1 (Fig. 7D), which inhibits Spastin functions in the axon. It is known that microtubule-associated proteins (MAPs), such as Tau, can shield microtubules from being severed by the severing proteins (Yu et al., 2008). In addition, microtubule-severing abilities of the severing proteins are also influenced by posttranslational modifications of tubulin (Riano et al., 2009). It is possible that Dar1 also regulates MAPs and/or posttranslational modification of tubulin in concert with Spastin expression.
Little is known about how the expression of Spastin is controlled. Our results suggest that Dar1, either directly or indirectly, suppresses the transcription of Spastin. As proper Spastin levels are important for neurite growth (Riano et al., 2009), it is likely that Dar1 is required for maintaining proper levels of Spastin in neurons for dendrite growth. In Drosophila, the Collier/Olf1/EBF (COE) family transcription factor Knot, which is expressed in the class IV (Hattori et al., 2007; Jinushi-Nakao et al., 2007; Crozatier and Vincent, 2008) but not other classes of da neurons, has been proposed to positively regulate Spastin expression as overexpressing Knot increases Spastin transcription (Jinushi-Nakao et al., 2007). Ectopic expression of Knot in the class I da neurons, which normally have the simplest dendritic arbors among the four classes of da neurons, leads to dendrite overgrowth (Hattori et al., 2007; Jinushi-Nakao et al., 2007). However, overexpressing Spastin in class I neurons did not result in dendrite overgrowth that resembles that caused by Knot overexpression (data not shown). This raises the possibility that, in the class I neurons, Knot induces the expression of other factors together with Spastin to cause dendrite overgrowth. Our results showed that the effects of Knot overexpression in class I neurons requires Dar1. Knot overexpression might require factors that are positively regulated by Dar1 to cause dendrite overgrowth. Alternatively, the absence of Dar1 function in that scenario probably results in Spastin levels that are too high to allow dendrite growth.
Krüppel-like factors and their roles in neural development
The KLF family of transcriptional regulators has been implicated in a variety of biological processes. At least 17 members of the KLF family have been identified in mammals to date (Pearson et al., 2008). In contrast, the Drosophila genome encodes only three KLFs—Luna and two predicted molecules, CG12029 and CG9895. Luna has been reported to be a potential fly homolog of KLF6 and 7 (De Graeve et al., 2003). RNAi and overexpression studies found that luna is required for embryonic development and cell differentiation in adult eyes; no luna mutant is currently available for additional genetic analysis.
Several KLFs are known to be involved in neurite development. KLF7 is required for dendrite and axon development. In the CNS of KLF7-null mice, both dendrite growth and axon growth are reduced (Laub et al., 2005). In the PNS of these mice, TrkA expression is reduced in the DRG neurons (Lei et al., 2005). Consequently, NGF-dependent nociceptive neurons undergo increased apoptosis. In trigeminal ganglion neurons, KLF7 cooperates with the POU homeodomain protein Brn3a to control TrkA expression (Lei et al., 2006). KLF9 also serves important functions in the nervous system. Its expression is strongly induced by thyroid hormone (Denver et al., 1999) and stress (Bonett et al., 2009). Furthermore, it is required for neurite growth (Cayrou et al., 2002). It remains to be determined whether KLF9 differentially regulates dendrite or axon development. KLF4 has recently been identified as a suppressor of neurite growth in mammalian CNS neurons (Moore et al., 2009). Overexpression of KLF4 reduces both dendritic and axonal length. Consistently, axon length is increased both in cultured neurons and in injured optic nerve of KLF4 knock-out mice.
The closest mammalian homolog of Dar1 is KLF5. Overexpression of KLF5 in cultured retinal ganglion cells leads to a modest reduction of neurite length (Moore et al., 2009); the functions of KLF5 in neuronal development have not been explored using loss-of-function studies or in vivo studies. KLF5 has been reported to be expressed in many tissues, including the brain (Yanagi et al., 2008), and is downregulated in schizophrenia patients. KLF5 homozygous mutant mice die before embryonic day 8.5 and the heterozygous mutant mice have defects in angiogenesis, cardiovascular remodeling, gut development, and adipogenesis (Shindo et al., 2002). In light of the involvement of Dar1 in Drosophila dendrite development, it will be interesting to examine these KLF5 mutant mice with respect to neural development.
It is unknown whether any of the mammalian KLFs function specifically in either dendrite or axon development. Whereas the simplest scenario of such evolutionary conservation is that one of the mammalian KLFs is the ortholog of Dar1, it is equally possible that multiple KLFs coordinate their activities (as proposed by Moore et al., 2009) to perform the equivalent of Dar1 function.
In summary, we have identified a novel transcription factor and demonstrated its requirement for dendritic but not axonal growth in Drosophila. Future studies that elucidate the regulatory mechanisms of Dar1 and its downstream effector genes will shed light on the genetic program that differentiates dendrite and axon development.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants R00MH080599 and R01MH091186 and The Whitehall Foundation (B.Y.), and NIH Grants R37NS40929 and R01NS47200 (Y.N.J.). B.Y. is a Pew Scholar in Biomedical Sciences. Y.N.J. and L.Y.J. are investigators of the Howard Hughes Medical Institute. We thank Drs. Tadashi Uemura and Michèle Crozatier for UAS-knot flies, Dr. Daniel Kiehart for UAS-GMA flies, Drs. Yang Xiang, Hsiu-Hsiang Lee, and Ulrike Heberlein for GAL19-12 flies, Dr. Nina Tang Sherwood for UAS-Spastin and T32 flies, and Laura Essex for technical assistance. We also thank Drs. Melih Acar, Qing Li, Jiandie Lin, and Di Ma for advice on cell sorting and real-time PCR, Dr. Zhe Han for advice on cell dissociation from fly embryos, and Drs. Wesley Grueber, Ye Zhang, Rebecca Yang, and Jill Wildonger for discussion during the studies. Dr. Yonghua Wang generated the genomic-cDNA hybrid construct used for rescuing dar1 mutants.
The authors declare no competing financial interests.
References
- Ahmad FJ, Yu W, McNally FJ, Baas PW. An essential role for katanin in severing microtubules in the neuron. J Cell Biol. 1999;145:305–315. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen R, Li Y, Resseguie M, Brenman JE. Calcium/calmodulin-dependent protein kinase II alters structural plasticity and cytoskeletal dynamics in Drosophila. J Neurosci. 2005;25:8878–8888. doi: 10.1523/JNEUROSCI.2005-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW. Microtubules and neuronal polarity: lessons from mitosis. Neuron. 1999;22:23–31. doi: 10.1016/s0896-6273(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Baas PW, Deitch JS, Black MM, Banker GA. Polarity orientation of microtubules in hippocampal neurons: uniformity in the axon and nonuniformity in the dendrite. Proc Natl Acad Sci U S A. 1988;85:8335–8339. doi: 10.1073/pnas.85.21.8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonett RM, Hu F, Bagamasbad P, Denver RJ. Stressor and glucocorticoid-dependent induction of the immediate early gene Krüppel-like factor 9: implications for neural development and plasticity. Endocrinology. 2009;150:1757–1765. doi: 10.1210/en.2008-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Denver RJ, Puymirat J. Suppression of the basic transcription element-binding protein in brain neuronal cultures inhibits thyroid hormone-induced neurite branching. Endocrinology. 2002;143:2242–2249. doi: 10.1210/endo.143.6.8856. [DOI] [PubMed] [Google Scholar]
- Chen B, Chu T, Harms E, Gergen JP, Strickland S. Mapping of Drosophila mutations using site-specific male recombination. Genetics. 1998;149:157–163. doi: 10.1093/genetics/149.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT. Human Krüppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280:41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Vincent A. Control of multidendritic neuron differentiation in Drosophila: the role of Collier. Dev Biol. 2008;315:232–242. doi: 10.1016/j.ydbio.2007.12.030. [DOI] [PubMed] [Google Scholar]
- De Graeve F, Smaldone S, Laub F, Mlodzik M, Bhat M, Ramirez F. Identification of the Drosophila progenitor of mammalian Krüppel-like factors 6 and 7 and a determinant of fly development. Gene. 2003;314:55–62. doi: 10.1016/s0378-1119(03)00720-0. [DOI] [PubMed] [Google Scholar]
- Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. evidence for a role in neurite outgrowth. J Biol Chem. 1999;274:23128–23134. doi: 10.1074/jbc.274.33.23128. [DOI] [PubMed] [Google Scholar]
- Drysdale RA, Crosby MA FlyBase Consortium. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Bloor JW, Ruiz-Gomez M, VijayRaghavan K, Kiehart DP. Real-time imaging of morphogenetic movements in Drosophila using Gal4-UAS-driven expression of GFP fused to the actin-binding domain of moesin. Genesis. 2002;34:146–151. doi: 10.1002/gene.10113. [DOI] [PubMed] [Google Scholar]
- Emoto K, He Y, Ye B, Grueber WB, Adler PN, Jan LY, Jan YN. Control of dendritic branching and tiling by the Tricornered-kinase/Furry signaling pathway in Drosophila sensory neurons. Cell. 2004;119:245–256. doi: 10.1016/j.cell.2004.09.036. [DOI] [PubMed] [Google Scholar]
- Gao FB, Brenman JE, Jan LY, Jan YN. Genes regulating dendritic outgrowth, branching, and routing in Drosophila. Genes Dev. 1999;13:2549–2561. doi: 10.1101/gad.13.19.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudillière B, Konishi Y, de la Iglesia N, Yao G, Bonni A. A CaMKII-NeuroD signaling pathway specifies dendritic morphogenesis. Neuron. 2004;41:229–241. doi: 10.1016/s0896-6273(03)00841-9. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol. 2003;13:618–626. doi: 10.1016/s0960-9822(03)00207-0. [DOI] [PubMed] [Google Scholar]
- Harrison SD, Solomon N, Rubin GM. A genetic analysis of the 63E–64A genomic region of Drosophila melanogaster: identification of mutations in a replication factor C subunit. Genetics. 1995;139:1701–1709. doi: 10.1093/genetics/139.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y, Sugimura K, Uemura T. Selective expression of Knot/Collier, a transcriptional regulator of the EBF/Olf-1 family, endows the Drosophila sensory system with neuronal class-specific elaborated dendritic patterns. Genes Cells. 2007;12:1011–1022. doi: 10.1111/j.1365-2443.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- Hazan J, Fonknechten N, Mavel D, Paternotte C, Samson D, Artiguenave F, Davoine CS, Cruaud C, Dürr A, Wincker P, Brottier P, Cattolico L, Barbe V, Burgunder JM, Prud'homme JF, Brice A, Fontaine B, Heilig B, Weissenbach J. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- Horton AC, Rácz B, Monson EE, Lin AL, Weinberg RJ, Ehlers MD. Polarized secretory trafficking directs cargo for asymmetric dendrite growth and morphogenesis. Neuron. 2005;48:757–771. doi: 10.1016/j.neuron.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Stegmüller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A. A SnoN-Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci. 2009;29:4312–4321. doi: 10.1523/JNEUROSCI.0126-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80:563–572. doi: 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and Cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmüller J, Matsuda T, Bonni S, Bonni A. Cdh1-APC controls axonal growth and patterning in the mammalian brain. Science. 2004;303:1026–1030. doi: 10.1126/science.1093712. [DOI] [PubMed] [Google Scholar]
- Laub F, Lei L, Sumiyoshi H, Kajimura D, Dragomir C, Smaldone S, Puche AC, Petros TJ, Mason C, Parada LF, Ramirez F. Transcription factor KLF7 is important for neuronal morphogenesis in selected regions of the nervous system. Mol Cell Biol. 2005;25:5699–5711. doi: 10.1128/MCB.25.13.5699-5711.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Lee HH, Jan LY, Jan YN. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc Natl Acad Sci U S A. 2009;106:6363–6368. doi: 10.1073/pnas.0902051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lei L, Laub F, Lush M, Romero M, Zhou J, Luikart B, Klesse L, Ramirez F, Parada LF. The zinc finger transcription factor Klf7 is required for TrkA gene expression and development of nociceptive sensory neurons. Genes Dev. 2005;19:1354–1364. doi: 10.1101/gad.1227705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Zhou J, Lin L, Parada LF. Brn3a and Klf7 cooperate to control TrkA expression in sensory neurons. Dev Biol. 2006;300:758–769. doi: 10.1016/j.ydbio.2006.08.062. [DOI] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL. KLF family members regulate intrinsic axon regeneration ability. Science. 2009;326:298–301. doi: 10.1126/science.1175737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Simons K, Dotti CG. Axonal and dendritic endocytic pathways in cultured neurons. J Cell Biol. 1992;119:123–137. doi: 10.1083/jcb.119.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH. Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Riano E, Martignoni M, Mancuso G, Cartelli D, Crippa F, Toldo I, Siciliano G, Di Bella D, Taroni F, Bassi MT, Cappelletti G, Rugarli EI. Pleiotropic effects of spastin on neurite growth depending on expression levels. J Neurochem. 2009;108:1277–1288. doi: 10.1111/j.1471-4159.2009.05875.x. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sherwood NT, Sun Q, Xue M, Zhang B, Zinn K. Drosophila Spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2004;2:e429. doi: 10.1371/journal.pbio.0020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, et al. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–863. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J Cell Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- Stegmüller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Stegmüller J, Huynh MA, Yuan Z, Konishi Y, Bonni A. TGFβ-Smad2 signaling regulates the Cdh1-APC/SnoN pathway of axonal morphogenesis. J Neurosci. 2008;28:1961–1969. doi: 10.1523/JNEUROSCI.3061-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S, Bradke F. Neuronal polarity. Cold Spring Harb Perspect Biol. 2009;1:a001644. doi: 10.1101/cshperspect.a001644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Wiggin GR, Fawcett JP, Pawson T. Polarity proteins in axon specification and synaptogenesis. Dev Cell. 2005;8:803–816. doi: 10.1016/j.devcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Yanagi M, Hashimoto T, Kitamura N, Fukutake M, Komure O, Nishiguchi N, Kawamata T, Maeda K, Shirakawa O. Expression of Krüppel-like factor 5 gene in human brain and association of the gene with the susceptibility to schizophrenia. Schizophr Res. 2008;100:291–301. doi: 10.1016/j.schres.2007.11.042. [DOI] [PubMed] [Google Scholar]
- Ye B, Zhang Y, Song W, Younger SH, Jan LY, Jan YN. Growing dendrites and axons differ in their reliance on the secretory pathway. Cell. 2007;130:717–729. doi: 10.1016/j.cell.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman C, Ayala MI, Wright JR, Bard F, Bossard C, Ang A, Maeda Y, Seufferlein T, Mellman I, Nelson WJ, Malhotra V. Protein kinase D regulates basolateral membrane protein exit from trans-Golgi network. Nat Cell Biol. 2004;6:106–112. doi: 10.1038/ncb1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Ahmad FJ, Baas PW. Microtubule fragmentation and partitioning in the axon during collateral branch formation. J Neurosci. 1994;14:5872–5884. doi: 10.1523/JNEUROSCI.14-10-05872.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Qiang L, Solowska JM, Karabay A, Korulu S, Baas PW. The microtubule-severing proteins spastin and katanin participate differently in the formation of axonal branches. Mol Biol Cell. 2008;19:1485–1498. doi: 10.1091/mbc.E07-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]