Abstract
Oxygen consumption is a fundamental component of metabolic networks, mitochondrial function, and global carbon cycling. To date there is no method available that allows for replicate measurements on attached and unattached biological samples without compensation for extraneous oxygen leaking into the system. Here we present the Respiratory Detection System, which is compatible with virtually any biological sample. The RDS can be used to measure oxygen uptake in microliter-scale volumes with a reversibly sealed sample chamber, which contains a porphyrin-based oxygen sensor. With the RDS, one can maintain a diffusional seal for up to three hours, allowing for the direct measurement of respiratory function of samples with fast or slow metabolic rates. The ability to easily measure oxygen uptake in small volumes with small populations or dilute samples has implications in cell biology, environmental biology, and clinical diagnostics.
Keywords: Respirometry, Oxygen Consumption Rate, Reversible Diffusional Seal, Pt-Porphyrin
1. INTRODUCTION
The measurement of respiration rates can be a versatile tool for the diagnosis of cellular and metabolic state. The ability to directly determine the oxygen uptake rate of cellular samples and monitor how the rates change in response to stimuli has broad implications in furthering the understanding of a wide array of biological systems from single cells to complex ecosystems. For example, respiration rates can be indicators of subtle phenotypes and metabolic states in bacterial systems[1].
Environmentally, oxygen-linked respiration is the main sink of organic matter in nature and can be considered a fundamental component of global element cycling [2]. Differences in oxygen uptake within complex natural communities could lend insight into the utilization of energy sources in the environment as well as into primary production [2]. The availability of a simple technique that could provide direct and precise oxygen measurements in marine or freshwater samples with low cell counts would significantly improve our knowledge of aquatic ecosystem functioning at local or global scales.
In addition to applications for environmental studies, oxygen uptake rates are important for eukaryotic cell biology. Oxygen uptake rates can be directly related to mitochondrial function and have been implicated in the activation of eukaryotic cells and the progression of disease states [3–5]. The ability to predict the progression of cell death pathways would lend insight into the diagnosis, treatment, and prevention of inflammatory diseases such as infection, stroke, heart disease, diabetes, and cancer [6–8]. Potential applications for the direct measurement of respiration rates would be as a gauge of cellular health and metabolic state for stress response, toxicity studies, and the assessment of the effects on tissues from extraneous perturbation.
To date the most common method for measuring respiration rates has been electrochemical measurements via traditional Clark electrodes [1,9–11]. However, the drawbacks of electrodes include low sensitivities, signal drift, and consumption of oxygen by the electrode itself [10,11]. Other recent methods have entailed scanning electrochemical microscopy (SECM) and utilization of nanobead sensors attached to the outer membrane of cells [12,13]. However, these techniques have been employed in openly diffusible environments and only indicate oxygen flux near the cell membrane or the oxygen sensor.
Optical methods for the measurement of oxygen consumption rates (OCRs) have become both widely used and accessible in the last several years [14,15]. While palladium and ruthenium based phosphores are used for oxygen sensing, Pt-phosphores have become the most relied upon oxygen sensor dye and have been used routinely in research and industry for over twenty years [1,5,15–21]. In addition, calibration, signal acquisition, and signal processing have become routine in the utilization of phosphorescent oxygen sensors. Many of the approaches employing optical sensors are based on fabricated well arrays or on standard well plate formats commercially available [5,18,20,22–26]. In general with well and plate based systems, the sensor is deposited into the bottom of a well or positioned above the well for oxygen concentration measurements. Plate-based systems have the advantage of low costs associated with the production of mold injected plastic plates compared to the fabrication of well systems. However, the main drawback of plate-based sensing platforms is that the materials used in these techniques are themselves permeable to oxygen and require extensive characterization to separate the actual respiration rates from the leakage or diffusion of extraneous oxygen into the sample chamber [20,22,25,27–28]. In addition, difficulties with reproducible sealing of these devices present additional variability. The result is that the signal acquired for optical probes is that of flux of the oxygen near the sensor and not of a direct measurement of the OCR. Thus far, the only known method for directly measuring the OCR of cells has been an optical technique employed in a diffusionally sealed borosilicate chamber [1,18].
Herein, we describe the development of the Respiratory Detection System (RDS), which is capable of directly measuring the respiration rate of biological samples in a simple and reproducible manner. The RDS is unique in its ability to measure OCR's of dilute biological samples in a reversibly sealed sample chamber that can maintain a diffusional seal for over two hours. Commercially-available slides as well as fabricated glass well systems were tested as components of the RDS, and the latter was shown to provide rapid and reproducible measurement of OCRs for attached mammalian cell lines, low optical density bacterial cultures, and environmental samples.
2. METHODS
2.1. Cellular Samples and Growth Conditions
Methylobacterium extorquens AM1 was grown in batch culture at 28°C in minimal salts medium using either 0.3% (vol/vol) methanol or 0.4% succinate as a carbon source [29].
2.2. Lake Washington Sediment
Lake Washington sediment core samples were collected as described previously [30]. Sediment cores were transported to the laboratory on ice and stored at 9°C. The samples of the top layer of the sediment were analyzed within 24 hours after collection.
2.3. Mammalian Cell Cultures
Human lung carcinoma cells (A549) and human colon carcinoma cells (HCT-116) were gifts from the Rabinovitch Lab (Department of Pathology, University of Washington). The mouse macrophage-like cell line, RAW 267.4 was a gift from the Cookson Lab (Department of Laboratory Medicine, University of Washington).
A549 and HCT cells were cultured under identical conditions as previously described with the exception that 5% Fetal Cone III (HyClone, Logan, UT) was used [18]. RAW 267.4 cells were cultured as previously described [18].
Normal human bronchial epithelial (NHBE) cells were purchased from Lonza (Allendale, NJ). Cell cultures were grown in bronchial cell growth media (BEGM) with supplemental factors for NHBE cells (Lonza, Allendale, NJ).
2.4. Prototype System
Glass slides containing three concave depressions (Erie Scientific, Portsmouth NH) were employed as macrowells. The slides were cut into thirds (with a diamond tipped pen) to separate the three depressions. In order to normalize the data as oxygen consumption rate per cell, the volumes of the depressions were measured by accurately weighing the macrowell and cover slip dry and then again with the depression filled with water and sealed with the cover slip. The calculated volume and the estimated number of cells were used to convert the oxygen concentration depletion rates in the macrowell to average oxygen consumption rate per cell.
2.5. Respiratory Detection System Description
The glass chips containing the RDS-wells were fabricated in borosilicate glass as previously described resulting in square RDS-wells with dimensions of 5, 8, and 10 mm2 and a well depth of 45 μm resulting in well volumes of 1.2, 3.2 and 4.5 μl respectively (Figure 1), [18]. In addition, RDS-wells with a volume of 1.2, 3.2 and 4.5 μl had a corresponding lip area of 100, 49, and 25 mm2. The fabrication process yielded 24, 15 × 15 × 0.5 mm chips per wafer, each containing one etched square well. Lids for sealing the glass RDS-wells for experiments on biological samples were fabricated from borosilicate glass by dicing 4-inch wafers into 20 × 20 mm squares.
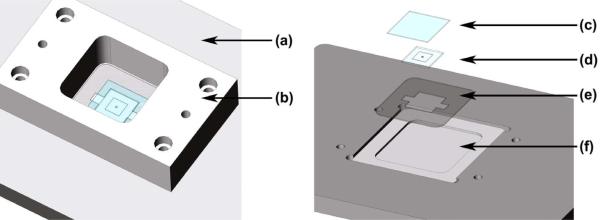
Figure 1.
Schematic of a sealed RDS-well housed in the miniature cell incubator (Molter et al. 2009). (a) Aluminum platen; (b) Delrin structure that forms the MCI-well to hold cell media; (c) Glass lid; (d) RDS-well with prophyrin sensor; (e) Chip holder; (f) Quartz viewing window.
During experiments, RDS-wells were housed within a miniature cell incubator (MCI), which consisted of a microscope platen, a quartz viewing window, and a delrin structure to form a 4 ml MCI-well (Figure 1), [18]. The MCI-well containing 2 ml growth medium was mounted directly onto the microscope stage. RDS-wells were held in place within the MCI-well with a 0.4 mm thick stainless steel chip holder. For routine experiments a quartz coverslip (VWR, West Chester, PA) was used as a lid.
2.6. Oxygen Sensor
The oxygen sensors used were 1 μm Pt phosphor carboxylate-modified microspheres (Invitrogen, Eugene, OR) rinsed in deionized water (dH2O; 1 μm beads discontinued, 40 nm beads available as product #F-20886). Approximately 0.1 μl of beads were pipetted into the RDS-wells after a 1 minute plasma etch to ensure that the surface of the well was hydrophilic. Sensor patches were cured for 10 minutes at 170°C to ensure surface adhesion by melting the microspheres. Following sensor deposition, the RDS-wells were ethanol sterilized and stored in sterile 12 well plates.
2.7. Sensor Calibration
Sensor patches were calibrated by saturating the liquid in the RDS wells with gas containing 0, 10, and 20% oxygen (5% CO2, N2 balance). The gasses were passed through 5 ml of water to humidify them and bathed over 2 ml of dH2O or the mammalian cell culture medium DMEM (Dulbecco's Modified Eagle's Medium) in the MCI at 24°C and 37°C.
2.8. Seal Tests
The lid used for seal testing the glass RDS-wells (seal test lid) was made of 5 layers of 0.5 mm thick, 20 × 20 mm borosilicate glass affixed to a piston comprised of 0.25” stainless steel drillstock with a tapered end, fabricated in-house. A thin layer of polydimethylsiloxane (PDMS) the size of the tapered tip was glued to the tip of the piston between the stainless steel and the glass as a conformal layer. The thick layers of glass ensured that no significant flexing of the piston tip would occur under pressure. All layers were glued together using medical-grade biocompatible UV-curable adhesive (Dymax Corp, Torrington, CT) and all parts were cleaned in an air plasma (Harrick Plasma Corp, Ithaca, NY) before gluing to strengthen the adhesive bond.
The diffusional seal tests were conducted in water and DMEM at 24°C by equilibrating 1ml of fluid to 0% dissolved oxygen with nitrogen, placing the seal test lid over the 4.5, 3.2, and 1.2 μl RDS-well sizes with corresponding lip areas of 100, 49, and 25 mm2 at 0 and 15 pounds psi, and turning off the gas line to allow the surrounding liquid outside of the sealed RDS-well to re-equilibrate to ambient oxygen concentrations. Sensor signal was acquired before the lid was added to ensure that the fluid was properly equilibrated and for up to 18 hours after the gas line was turned off.
2.9. Bacterial Culture and Environmental Sample Manipulation
Prior to the measurement of respiration rates, the RDS-wells were seeded with the cells of interest. For cultures of M. extorquens AM1 and Lake Washington sediment, 2 ml samples were pipetted directly into the MCI-well. Aliquots of M. extorquens AM1 cultures in logarithmic growth were diluted to OD600nm of ~0.10. The number of M. extorquens AM1 cells in the sample was calculated via the equation y = 4 × 108x − 4 × 106 such that y = colony forming units and x = OD600nm [31].
For the mammalian cell lines, A549, HCT, and RAW 267.4 cell suspensions were centrifuged for 10 minutes at 24°C at 1200 RPM. The pellet was resuspended in fresh medium and counted using a hemacytometer with 0.1% Trypan Blue (Invitrogen, Carlsbad, CA).
2.10. Mammalian Culture Spot Seeding Method
A spot seeding method was used for all experiments with RAW 267.4 cells with the prototype system wells, initial baseline experiments with A549 cells in RDS-wells, and hypoxic experiments with A549 cells in RDS wells. Spot seeding entailed pipetting cells directly into a well containing a sensor patch, ensuring that the cell solution did not spill over onto the lip of the well. Cell suspensions of 50–150 μl RAW 267.4 cells (~200,000 cells) and 15–30 μl A549 cells (~20,000 cells) were used. The seeded prototype macrowells and RDS-wells were incubated under standard culture conditions for one hour to allow cells to attach to the glass surface. The prototype macrowells and RDS-wells, housed in a sterile 100 mm petri dish, were then immersed in 20 ml DMEM prior to the measurement of oxygen uptake rates.
Prior to the measurement of OCRs using the prototype macrowells, 350 μL of medium was added to the depression and the macrowell was sealed by the addition of a glass cover slip with the excess medium removed by pushing down on the cover slip with light force. The sealed macrowell was placed directly on a glass slide for imaging and maintained at 37°C [24]. RDS-wells spot seeded with A549 cells were manipulated for experimentation as described above.
2.11. Mammalian Culture Masked-Well Seeding Method
Prior to RDS-well seeding, Blue Tack (Semiconductor Equipment Corporation, Moorpark, CA) was applied to the surface of the sterilized chip and tape covering the well was removed leaving the tape only on the lip of the chip around the well, which will be referred to as a masked RDS-well. The masked RDS-wells were subsequently placed into one well of a 12-well plate. 100,000 A549, HCT, or NHBE cells were seeded in a 2ml volume and incubated overnight to allow cells to settle and fully attach. NHBE seeding was conducted with the addition of 0.01% fibronectin (Invitrogen, Carlsbad, CA) Prior to experiments the tape covering the lip of the RDS-well was removed and the seeded RDS-wells were manipulated as described above.
2.12. Experimental Design
Hypoxic recovery experiments were carried out by incubating A549 cultures at 1% oxygen (5% CO2, N2 balance) in a triple gas incubator (Thermo, Asheville, NC) 24 hours prior to well seeding. Once spot seeded, the wells were maintained at 1% oxygen until measurements were made. For NHBE cells, RDS-wells were masked-well seeded as described above. 24 hours prior to experiments, the 12 well plates containing seeded RDS-wells were placed within a 1% oxygen atmosphere.
For respiratory inhibition experiments, RDS-wells masked-well seeded with A549 cells were incubated in the presence of 1, 10, or 100 nM rotenone (Sigma Aldrich, St. Louis, MO) for 15 minutes. Treated RDS-wells were immediately placed into a petri dish containing 10 ml growth medium without rotenone and transferred into a MCI-well for the detection of oxygen uptake rates.
To seal an RDS-well seeded with cells, a glass lid was placed directly on top of the well and moderate pressure was applied to achieve a good surface tension seal. Measurements were conducted on a Zeiss LSM 510 META (Thornwood, NY) using a 10 × 0.3 N.A. objective at 24°C for M. extorquens AM1 cultures and Lake Washington sediment, and 37°C for mammalian cell lines. Temperature control was maintained as previously described [24]. Sensor signal acquisition was carried out from 10–60 minutes depending on conditions and cell type. All data collection and analysis was conducted as previously described [18]. For RDS-wells seeded with cells 24–48 hrs prior to experiments, the cells were stained with Calcien AM (Invitrogen, Carlsbad, CA) for 30 minutes and the entire well was imaged using the tile feature in the Zeiss LSM software (version 4.0 SP2) resulting in a 13.5 × 13.5mm image (15 × 15 tiles). Excitation was conducted with a 488nm argon laser at 1% power and detected in transmission or fluorescence detection mode.
In order to count the number cells in the RDS-wells, the tile images obtained were first converted to TIF files using the Zeiss LSM software. The constituent tiles were separated using Matlab 7.3.0.267 (Mathworks, Novi, MI) resulting in 225 tiles of 512 × 512 pixels. Each tile image was converted to its grayscale equivalent using ImageJ (http://rsbweb.nih.gov/ij/), [32], processed to remove image noise, and segmented using a modified watershed algorithm in Cell-Profiler (www.cellprofiler.org), [33], which accounted for segmentation errors due to clustered cells and over-exposure during image acquisition. The accuracy of this automated count procedure was estimated by visually inspecting the segmented cells overlaid on the original image. The maximum error thus obtained was computed to be 9.6%.
The OCRs in ppm O2 min−1 cell−1 were calculated based on the number of estimated or counted cells within the prototype macrowells or RDS-wells, the volume of the well used, and the Stern-Volmer equation [1,18]. All statistical values were calculated using standard two-sample t significance tests.
3. RESULTS
3.1. Respiratory Detection System Prototype
In order to test the concept of a small volume respiration detection system using optical sensors, a prototype system was developed. This initial prototype system consisted of phosphorescent sensor deposited into depressions of commercially available glass slides that were sealed by the application of a coverslip. Once the coverslip was applied, the cell sample in the depression consumed oxygen, and the sensor measured the rate of that decrease (the OCR). Since the oxygen concentration is monitored in real-time, the experiment can be ended by the removal of the glass lid before the dissolved oxygen concentration reaches a predetermined threshold, thereby preventing the samples from experiencing hypoxic shock. RAW267.4 cells were used to characterize the ability of the prototype system to measure OCR. The average rate per cell was found to be 1.62 +/− 0.65 fmol/min, which correlated well with previously published data (Table 1), [34]. The results verified the applicability of the prototype system to measure respiration rates. However, the depression volumes of the commercial slides were highly variable (32.6–84.2 μl), requiring measurement of each volume for OCR calculation. In order to test the concept further, a system involving inhouse fabricated wells (the RDS) was developed.
Table 1.
Compiled respiration rates measured. Data presented as mean +/− s.d.
| Cell Type | Respiration Rate (mol min−1 cell−1)1 | Range (mol min−1 cell−1)1 | Growth Condition | Seeding Method | N |
|---|---|---|---|---|---|
| Methylobacterium extorquens AM1 | 9.00 +/− 7.24 × 10−17 | 0.43 – 2.80 × 10−16 | Succinate | - | 9 |
| Methylobacterium extorquens AM1 | 1.32 +/− 0.95 × 10−16 | 0.46 – 3.71 × 10−16 | Methanol | - | 9 |
| Lake Washington Sediment | 6.20 +/− 1.83 × 10−12 | 4.11 – 7.50 × 10−12 | - | - | 3 |
| Raw267.4 (prototype system) | 1.62 +/− 0.65 m× 10−15 | 1.01 – 3.69 × 10−15 | SCC | SS | 22 |
| Human Lung Carcinoma (A549) | 4.01 +/− 1.53 × 10−15 | 2.28 – 7.24 × 10−15 | SCC | SS | 11 |
| Human Lung Carcinoma (A549) | 2.16 +/− 0.52 × 10−15 | 1.61 – 2.86 × 10−15 | 1% Atmospheric Oxygen | SS | 8 |
| Normal Human Lung Epithelium (NHBE) | 1.12 +/− 0.89 × 10−15 | 0.52 – 2.85 × 10−15 | SCC | MW | 6 |
| Normal Human Lung Epithelium (NHBE) | 2.67 +/− 0.47 × 10−15 | 1.60 – 3.2 × 10−15 | 1% Atmospheric Oxygen | MW | 8 |
| Human Lung Carcinoma (A549) | 2.06 +/− 0.36 × 10−15 | 1.39 – 3.60 × 10−15 | SCC | MW | 20 |
| Human Lung Carcinoma (A549) | 2.29 +/− 0.59 × 10−15 | 1.64 – 3.03 × 10−15 | 10 nM dose of Rotenone | MW | 7 |
| Human Lung Carcinoma (A549) | 0.75 +/− 0.55 × 10−15 | 0.25 – 1.74 × 10−15 | 100 nM dose of Rotenone | MW | 12 |
| Colon Cancer (HCT-116) | 4.16 +/− 2.14 ×× 10−15 | 1.29 – 7.73 × 10−15 | SCC | MW | 10 |
Lake Washington Sediment is shown as mol min−1 Total-Sample-Volume−1; SS = Spot seeded; MW = Masked-well seeded; SCC = Standard Culture Conditions
3.2. RDS
The RDS consists of a low volume square well etched in borosilicate glass and containing a phosphorescent porphyrin sensor used for the measurement of dissolved oxygen concentrations, (the RDS-well). As in the prototype system, a lid can be applied for a period of time to diffusionally isolate the sample in the well, and the sensor measures the OCR. Once a rate is obtained, the lid is removed. It should be noted that the microsphere immobilization protocol used in this study had no effect on sensor response. Preliminary experiments with RAW 267.4 cells demonstrated the feasibility of using the RDS to measure OCRs. Therefore, the RDS was assessed further.
3.3. Sensor Calibration and Seal Verification for the RDS
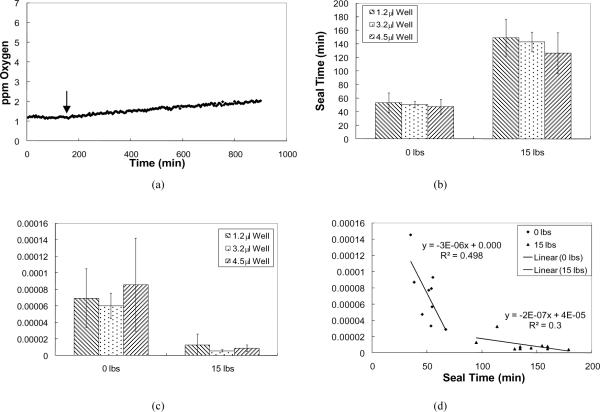
Sensor calibration was carried out as described previously, and the calibration data generated were consistent with the previous study (data not shown), [18]. Next, the RDS was tested to determine whether pressure on the lid was required to obtain an oxygen barrier for the time period expected for experimental protocols. Seal tests were conducted in triplicate on RDS-wells with lip areas of 100, 49, and 25 mm2 using 0 and 15 lbs pressure for up to 18 hours after the seal test lid was applied (Figure 2). The time for which the diffusional seal was maintained, or the seal time, ranged from 38.35 to 66.95 min for 0 lbs (force applied onto seal lid) and 94.84 to 178.88 min for 15 lbs for all RDS-well sizes combined. Figure 2(b)–(c) demonstrates that the RDS-well lip area had a negligible effect on seal times and the corresponding diffusional rate after the seal expired. However, it was found that diffusional rates of oxygen into the wells after the seal expired modestly correlated with the length of the seal time with 0 and 15 lbs force (Figure 2(d)).
Figure 2.
Summary of seal tests for RDS-wells with volumes of 1.2, 3.2, and 4.5 μl using 0 and 15 lbs of force. RDS-wells with volumes of 1.2, 3.2, and 4.5 μl correspond to lip areas of 100, 49, 25 mm2 respectively. (a) Representative data from a 4.5 μl RDS-well after a seal was initiated with 15 lbs and the diffusion rate of oxygen into the well after the seal expired (8.8 × 10−6 ppm O2 min−1 mm−2). The arrow marks the expiration of the diffusional seal after 154.7 minutes. (b) Average seal time for all well volumes using 0 and 15 lbs. (c) Average diffusional rate of oxygen into the RDS-wells after the diffusional seal expired. (d) Diffusion rates of oxygen after the seal expired versus seal times. Data displayed as mean +/− s.d.
3.4. Detection of Respiration Rates from Bacterial Cultures and Environmental Samples
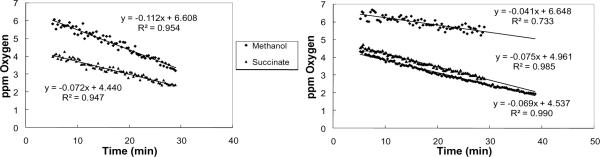
To test the use of the RDS with bacterial cultures, logarithmically growing cultures of the gram-negative bacterium Methylobacterium extorquens AM1 were diluted and tested at an OD600nm of ~0.10 during growth on succinate or methanol (Figure 3(a)). Respiration rates/cell were calculated to be 90 +/− 73 amol min−1 cell−1 and 132 +/− 95 amol min−1 cell−1 during growth on succinate and methanol respectively. The calculated rates had a range of 43 – 280 amol min−1 cell−1 for succinate growth and 46–371 amol min−1 cell−1 for methanol growth.
Figure 3.
Oxygen uptake data acquired from bacterial cultures and Lake Washington sediment. (a) Example oxygen consumption rates obtained from ~ 1.7 × 105 M. extorquens AM1 cells grown on succinate or methanol with an optical density (600 nm) of ~0.10. (b) Summary of respiration rates obtained from 3 replicates for the top aerobic layer of Lake Washington sediment.
To test the use of the RDS with environmental samples, the oxygen consumption rate from the top aerobic layer (1 cm) of Lake Washington sediment cores was measured. The minimum incubation time required for a rate calculation was 30–40 minutes, which was more than 40 times shorter compared to routine incubation approaches (Figure 3(b)), [2]. The rate of oxygen consumption in the RDS system for the sediment samples was 6.20 ± 1.83 × 10−12 mol min−1 Total-Sample-Volume−1 or 116.25 +/− 34.27 mmol O2 m−3 hr−1. The values obtained fall within the range previously reported for freshwater lakes [2].
3.5. Oxygen Consumption Rates of Mammalian Cell Lines and the Effects of Extraneous Perturbation
Experiments were carried out to demonstrate efficacy of the RDS for assessing response to perturbations with cultured mammalian cells. The respiration rates from A549, HCT-116, and NHBE cells were tested under standard tissue culture conditions and were found to be similar to literature values for the cell lines A549 [5,18] and HCT-116 [35], (Table 1).
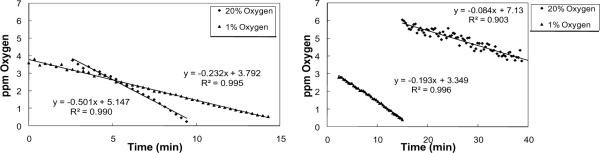
For one set of perturbation experiments, the effects of anoxic conditions on respiration rates were tested for A549 and NHBE cells grown at 20% or 1% oxygen for 24 hours prior to the measurement of respiration rates. For these experiments, A549 cells were spot seeded and NHBE cells were masked-well seeded. A549 cells grown in a 1% oxygen environment displayed an oxygen uptake rate that was half of the rate observed from control cells, matching trends in the literature (p-value < 0.005), [36], while NHBE cells displayed a two-fold increase in respiration rates compared to control cells (p-value < 0.01; Figure 4). NHBE cells were attached to the glass surface of the RDS-wells via the addition of 0.01% fibronectin to the cell seeding solution. Precoating the RDS-wells with fibronectin or adding the fibronectin to the cell solution were both found to be effective in facilitating cell attachment. However, if the lip of an RDS-well became coated with fibronectin, a diffusional seal was not possible. This problem was alleviated by applying Blue Tack Tape to the lip of an RDS-well and then removing it after seeding, which was effective in allowing both cell attachment in the wells and sealing of the wells. This process is termed masked-well seeding.
Figure 4.
Oxygen uptake data obtained from mammalian cell lines. (a) Example data from A549 cells grown 24 hours under standard tissue culture conditions and in a 1% oxygen environment. (b) Example data from NHBE cells maintained 24 hours under a 20% and 1% oxygen environment.
As a second perturbation test, masked-well seeded A549 cells were treated with 0, 1, 10, and 100 nM rotenone, a respiratory inhibitor. Concentrations greater than 100 nM were observed to completely arrest oxygen uptake rates, while a concentration of 1 nM rotenone had no effect (data not shown). Cells treated with 100 nM rotenone showed a three-fold decrease in respiration rates compared to controls (p-value < 0.0005) and cells treated with 10 nM exhibited no effect (p-value > 0.15; Figure 5).
Figure 5.
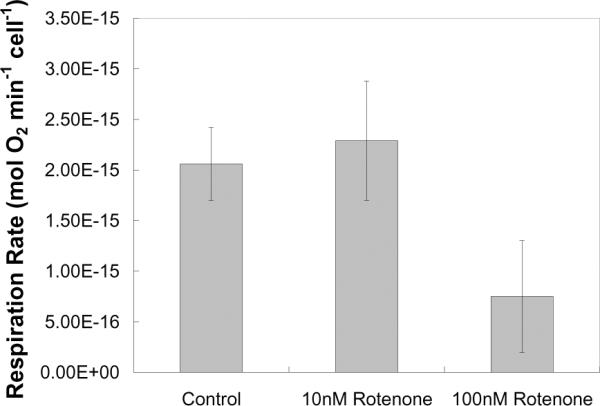
Summary of respiration rates obtained from A549 cultures treated with 10 and 100 nM rotenone. Data displayed as mean +/− s.d.
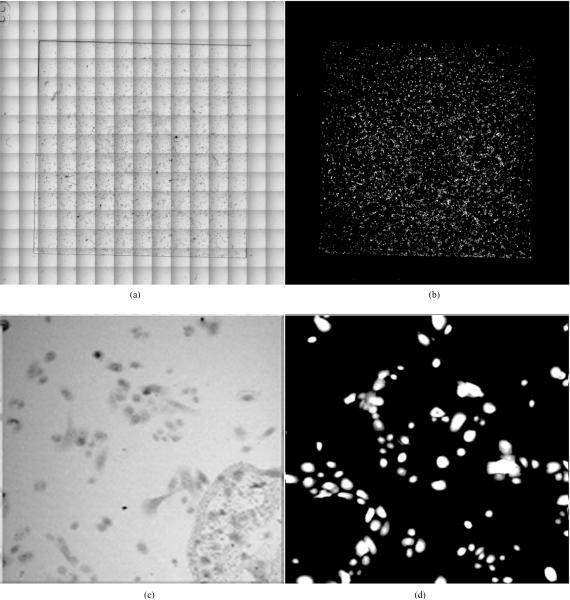
After the respiration rates from cells seeded on masked RDS-wells were measured, the cells were stained with Calcien AM and imaged (Figure 6). Since cells were seeded into the wells 24–48 hours prior to measurements, it was necessary to count the number of live cells in the wells for an accurate calculation of respiration rates per cell (see methods section). Two approaches were used for counting, hemacytometer and image analysis. Comparisons of baseline data of A549 cells spot seeded versus masked-well seeded indicate that spot seeding cells based on cell counts generated by a hemacytometer greatly underestimated the number of cells within the sample chamber when compared to directly counting the number of seeded cells via image analysis.
Figure 6.
Images of an RDS-well seeded with NHBE cells. (a) A transmission tile image in channel D. (b) Tile image of NHBE cells stained with Calcien AM. (c) Single image from a transmission tile scan showing the edge of the sensor ring. (d) Single image from the fluorescence tile scan of NHBE cells stained with Calcien AM.
4. CONCLUSION AND DISCUSSION
In this report we demonstrate the applicability of the Respiratory Detection System for directly measuring OCRs from different types of cell samples. Previous efforts have focused on the measurement of single cell respiration rates, and here we show that the RDS is a viable alternative for use with bulk populations. The RDS was demonstrated to be compatible with attached mammalian cell lines, low optical density bacterial cultures, and environmental samples. In addition, the RDS was demonstrated to maintain a diffusional seal for up to three hours to allow for OCR measurements on highly diluted or slowly respiring samples. These features and the ability to directly measure the OCR are a significant improvement compared to oxygen flux-based measurements in sample chambers made from plastics. The design parameters of the RDS integrate large glass-glass interfaces which allows for long diffusional seals. Due to the fabrication method to make RDS-wells, well dimensions can be easily changed to accommodate different sample types with the potential to measure OCR's from larger multi-cellular organisms (e.g. nematodes, zebrafish embryos). In addition to these novel characteristics, the simple design, inexpensive fabrication, and straightforward implementation of the RDS would readily allow for its integration into a preexisting plate reader format, on an optical microscope system, or as a portable device for use in the field.
Our results indicate that repeatable oxygen uptake rates could be measured with experimental times as short as ten minutes in the largest RDS-well, depending on the cellular sample. The versatility of the RDS is demonstrated by the broad spectrum of biological samples that can be tested, including mammalian cell strains that require special coatings to assist in cell attachment and proliferation.
The RDS utilizes a commercially available phosphorescent oxygen sensor to measure oxygen uptake rates. However, any optically based oxygen sensor could be employed with the RDS. Since phosphorescence-based oxygen reporters are detected by the lifetime decay of the excited phosphor [15], the RDS can be used to perform both fluorescence-based and phosphorescence-based measurements due to the difference in decay timescales, nanoseconds vs. microseconds respectively. The ability to multiplex oxygen sensing with additional fluorescence reporters allows for a wide array of fluorescence-based reporters to be used simultaneously, which could be imaged on a microscope, detected with a plate reader, or with a basic filter-based light emitting diode/photo multiplier tube optical train [5,9].
The RDS is a device that can be used to directly measure the respiration rate of bulk samples in a reversible diffusionally sealed sample chamber. The simplicity of the device and the ability to use microliter-scale cell samples makes it a logical alternative or enhancement to other established methods for measuring oxygen. By integrating the RDS design with existing systems, OCR measurements would directly reflect oxygen consumption rather than the flux of oxygen near the sensor. These characteristics of the RDS would allow the system to be employed as a portable device, a stand-alone bench top system, a microscope mounted device, or it could be integrated into a standard plate reader format. The latter approach would allow taking advantage of the absorbance, fluorescence, and luminescence detection capabilities of microplate readers to multiplex a wide array of assays with the detection of oxygen uptake rates [37].
The respiration rate of a cell can be used as a gauge of metabolic state and cellular health. Thus, the RDS has a broad range of possible applications, especially in cases in which the sample size is limited. The ability to maintain a diffusional seal allows for greater sensitivity when measuring subtle changes in OCRs and when the sample exhibits a slow respiration rate such as dilute bacterial cultures or environmental samples. This sensitivity could be beneficial in the study of the effects of experimental drugs on target cells as an assay in a commercial environment or as a clinical screen of fertilized embryos to be used for in vitro fertilization [16]. This same concept is relevant in gauging the effects of stimuli on cells in the investigation of stress response and toxicity studies. Lastly, the compatibility of the RDS with attached cells and an optically based oxygen sensor allows for fast replicate measurements without the need for stirring, as compared to detection methods utilizing openly diffusible sample chambers. These characteristics impart significant improvements to existing methods for the measurement of respiration rates from biological samples.
Acknowledgments
This work was supported by an NIH National Human Genome Research Institute (NHGRI) Centers of Excellence in Genomic Sciences grant (P50 HG 002360) and the U. S. Department of Energy (DE-PS02-07ER07-14).
REFERENCES
- [1].Strovas TJ, Dragavon JM, Hankins TJ, Callis JB, Burgess LW, Lidstrom ME. Measurement of respiration rates of Methylobacterium extorquens AM1 cultures by use of a phosphorescence-based sensor. Applied Environmental Microbiology. 2006;72:1692–1695. doi: 10.1128/AEM.72.2.1692-1695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bidanda B. Respiration in aquatic ecosystems. In: Del Giorgio PA, Williams P.J. le B., editors. Journal of Plankton Research. Oxford University Press; New York: 2005. [Google Scholar]
- [3].Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infection and Immunity. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Weissig V, Boddapati SV, Jabr L, D'Souza GG. Mitochondria-specific nanotechnology. Nano-medicine. 2007;2:275–285. doi: 10.2217/17435889.2.3.275. [DOI] [PubMed] [Google Scholar]
- [5].Wu M, Neilson A, Swift AL, Moran R, Tamagnine J, Parslow D, Armistead S, Lemire K, Orrell J, Teich J, Chomicz S, Ferrick DA. Multi-parameter metabolic analysis reveals a close link between attenuated mitochondrial bioenergetic function and enhanced glycolysis dependency in human tumor cells. American Journal of Physiology: Cellular Physiology. 2007;292:125–136. doi: 10.1152/ajpcell.00247.2006. [DOI] [PubMed] [Google Scholar]
- [6].Bell EL, Chandel NS. Mitochondrial oxygen sensing: Regulation of hypoxia-inducible factor by mitochondrial generated reactive oxygen species. Essays in Biochemistry. 2007;43:17–27. doi: 10.1042/BSE0430017. [DOI] [PubMed] [Google Scholar]
- [7].Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cellular Microbiology. 2007;9:2562–2570. doi: 10.1111/j.1462-5822.2007.01036.x. [DOI] [PubMed] [Google Scholar]
- [8].Maley CC. Multistage carcinogenesis in Barrett's esophagus. Cancer Letters. 2007;245:22–32. doi: 10.1016/j.canlet.2006.03.018. [DOI] [PubMed] [Google Scholar]
- [9].Eklund SE, Thompson RG, Snider RM, Carney CK, Wright DW, Wikswo J, Cliffel DE. Metabolic discrimination of select list agent by monitoring cellular responses in a multianalyte microphysiometer. Sensors. 2009;9:2117–2133. doi: 10.3390/s90302117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ramamoorthy R, Dutta PK, Akbar SA. Oxygen sensors: Materials, methods, designs, and applications. Journal of Material Science. 2003;38:4271–4282. [Google Scholar]
- [11].Renger G, Hanssum B. Oxygen detection in biological systems. Photosynthesis Research. 2009;102:487–498. doi: 10.1007/s11120-009-9434-2. [DOI] [PubMed] [Google Scholar]
- [12].Kuang Y, Walt DR. Detecting oxygen consumption in the proximity of Saccharomyces cerevisiae cells using self-assembled fluorescent nanosensors. Biotechnology Bioengineering. 2007;96:318–325. doi: 10.1002/bit.21092. [DOI] [PubMed] [Google Scholar]
- [13].Saito T, Wu CC, Shiku H, Yasukawa T, Yokoo M, Ito-Sasaki T, Abe H, Hoshi H, Matsue T. Oxygen consumption of cell suspension in a poly (dimethylsiloxane) (PDMS) microchannel estimated by scanning electrochemical microscop. Analyst. 2006;131:1006–1011. doi: 10.1039/b600080k. [DOI] [PubMed] [Google Scholar]
- [14].Warkentin M, Freese HM, Karsten U, Schumann R. New and fast method to quantify respiration rates of bacterial and plankton communities in freshwater ecosystems by using optical oxygen sensor spots. Applied Environmental Microbiology. 2007;73:6722–6729. doi: 10.1128/AEM.00405-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Papkovsky DB, O'Riordan TC. Emerging applications of phosphorescent metalloporphyrins. Journal of Fluorescence. 2007;15:569–584. doi: 10.1007/s10895-005-2830-x. [DOI] [PubMed] [Google Scholar]
- [16].Abe H. A non-invasive and sensitive method for measuring cellular respiration with a scanning electrochemical microscopy to evaluate embryo quality. Journal of Mammalian Ova Research. 2007;24:70–78. [Google Scholar]
- [17].Borisov SM, Nuss G, Klimant I. Red light-ex- citable oxygen sensing materials based on platinum(II) and palladium(II) benzoporphyrins. Analytical Chemistry. 2008;80:9435–9442. doi: 10.1021/ac801521v. [DOI] [PubMed] [Google Scholar]
- [18].Molter TW, McQuaide SC, Suchorolski MT, Strovas TJ, Burgess LW, Meldrum DR, Lidstrom ME. A microwell array device capable of measuring single-cell-oxygen consumption rates. IEEE Sensors and Actuators B. 2009;135:678–686. doi: 10.1016/j.snb.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].O'Donovan C, Twomey E, Alderman J, Moore T, Papkovsky D. Development of a respirometric biochip for embryo assessment. Lab on a Chip. 2006;6:1438–1444. doi: 10.1039/b607622j. [DOI] [PubMed] [Google Scholar]
- [20].O'Mahony FC, O'Donovan C, Hynes J, Moore T, Davenport J, Papkovsky DB. Optical oxygen microrespirometry as a platform for environmental toxicology and animal model studies. Environmental Science and Technology. 2005;39:5010–5014. doi: 10.1021/es048279x. [DOI] [PubMed] [Google Scholar]
- [21].Wilson DF, Vanderkooi JM, Green TJ, Maniara G, Defeo SP, Bloomgarden DC. A versatile and sensitive method for measuring oxygen. Advances in Experimental Medical Biology. 1987;215:71–77. doi: 10.1007/978-1-4684-7433-6_9. [DOI] [PubMed] [Google Scholar]
- [22].Alderman J, Hynes J, Floyd SM, Krüger J, O'Conner R, Papkovsky DB. A low-volume platform for cell-respirometric screening based on quenched- luminescence oxygen sensing. Biosensors and Bioelectronics. 2004;19:1529–1535. doi: 10.1016/j.bios.2003.12.008. [DOI] [PubMed] [Google Scholar]
- [23].Birmele M, Roberts M, Garland J. Characterization of methods for determining sterilization efficacy and reuse efficiency of oxygen biosensor multiwell plates. Journal of Microbiological Methods. 2006;67:619–623. doi: 10.1016/j.mimet.2006.06.003. [DOI] [PubMed] [Google Scholar]
- [24].Dragavon J, Molter T, Young C, Strovas T, McQuaide S, Holl M, Zhang M, Cookson B, Jen A, Lidstrom M, Burgess L. A cellular isolation system for real-time single-cell oxygen consumption monitoring. Journal of the Royal Society Interface. 2008;5(S2):S151–S159. doi: 10.1098/rsif.2008.0106.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Will Y, Hynes J, Ogurtsov VI, Papkovsky DB. Analysis of mitochondrial function using phosphorescent oxygen-sensitive probes. Nature Protocols. 2007;1:2563–2572. doi: 10.1038/nprot.2006.351. [DOI] [PubMed] [Google Scholar]
- [26].Wodnicka M, Guarino RD, Hemperly JJ, Timmins MR, Stitt D, Pitner JB. Novel fluorescent technology platform for high throughput cytotoxicity and proliferation assays. Journal of Biomolecular Screening. 2000;5:141–152. doi: 10.1177/108705710000500306. [DOI] [PubMed] [Google Scholar]
- [27].Arain S, Weiss S, Heinzle E, John GT, Krause C, Klimant I. Gas sensing in microplates with optodes: Influence of oxygen exchange between sample, air, and plate material. Biotechnology and Bioengineering. 2005;90:271–280. doi: 10.1002/bit.20348. [DOI] [PubMed] [Google Scholar]
- [28].Gerencser AA, Neilson A, Choi SW, Edman U, Yadava N, Oh RJ, Ferrick DA, Nicholls DG, Brand MD. Quantitative microplate-based respirometry with correction for oxygen diffusion. Analytical Chemistry. 2009;81:6868–6878. doi: 10.1021/ac900881z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Strovas TJ, Sauter LM, Guo X, Lidstrom ME. Cell-to-cell heterogeneity in growth rate and gene expression in Methylobacterium extorquens AM1. Journal of Bacteriology. 2007;189:7127–7133. doi: 10.1128/JB.00746-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kalyuzhnaya MG, Bowerman S, Nercessian O, Lidstrom ME, Chistoserdova L. Highly divergent genes for methanopterin-linked C1 transfer reactions in Lake Washington, assessed via metagenomic analysis and mRNA detection. Applied Environmental Microbiology. 2005;71:8846–8854. doi: 10.1128/AEM.71.12.8846-8854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Guo X. Ph.D. dissertation. University of Washington, Department of Chemical Engineering; 2006. Intergrative study of metabolism of Methylobacterium extorquens AM1 from a metabolomic perspective. [Google Scholar]
- [32].Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- [33].Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nalwaya N, Deen WM. Nitric oxide, oxygen, and superoxide formation and consumption in macrophage cultures. Chemical Research in Toxicology. 2005;18:486–93. doi: 10.1021/tx049879c. [DOI] [PubMed] [Google Scholar]
- [35].Leschelle X, Goubern M, Andriemihaja M, Blottiere HM, Coupland E, Gonzalez-Barroso MD, Petite C, Pagniez A, Chaumentet C, Mignotte B, Bouillaud F, Blachier F. Adaptative metabolic response of human colonic epithelial cells to the adverse effects of the luminal compound sulfide. Biochimica et Biophysica Acta. 2005;1725:201–212. doi: 10.1016/j.bbagen.2005.06.002. [DOI] [PubMed] [Google Scholar]
- [36].Heerlein K, Schulze A, Hotz L, Bartsch P, Mairbaurl H. Hypoxia decreases cellular ATP demand and inhibits mitochondrial respiration of A549 cells. American Journal of Respiratory Cell and Molecular Biology. 2005;32:44–51. doi: 10.1165/rcmb.2004-0202OC. [DOI] [PubMed] [Google Scholar]
- [37].Lidstrom ME, Meldrum DR. Life-on-a-chip. Nature Reviews Microbiology. 2003;1:158–164. doi: 10.1038/nrmicro755. [DOI] [PubMed] [Google Scholar]