Abstract
Objective
Pancreatic stellate cells (PSCs) are important players in pancreatic fibrosis and are major contributors to the extracellular matrix proteins observed with the stromal response characteristic of pancreatic ductal adenocarcinoma (PDAC). PSCs are also believed to secrete soluble factors that promote tumor progression, however no comprehensive analysis of the PSC proteome in either the quiescent or activated state has been reported.
Methods
Using two-dimensional tandem mass spectrometry and the RLT-PSC cell line, we present the first comprehensive study describing and comparing the quiescent and activated human PSC secreted proteomes.
Results
Very few proteins are secreted in the quiescent state. In stark contrast, activated PSCs secreted a vast array of proteins. Many of these proteins differed from those secreted by PDAC derived cell lines. Proteins associated with wound healing, proliferation, apoptosis, fibrosis and invasion were characterized. Selected proteins were verified in human tissue samples from PDAC, dysplastic pancreas, and normal pancreas using Western blot analysis and immunohistochemical staining.
Conclusions
Our study represents the first comprehensive analysis of proteins secreted by PSCs. These findings lay the foundation for characterizing PSC derived proteins involved in stroma-tumor interactions and the promotion of pancreatitis and PDAC.
Keywords: Pancreatic cancer, pancreatic stellate cells, secretome, tumor microenvironment, pancreatitis, RLT-PSC
INTRODUCTION
Since their initial description in 1998,1,2 pancreatic stellate cells (PSCs) have become increasingly recognized as important players in pancreatic fibrosis as seen in pancreatitis and pancreatic ductal adenocarcinoma (PDAC). An exuberant stromal response is a defining feature of PDAC. Indeed, cancer cells compose on average less than 40% of PDAC tumors3 and accumulating evidence supports PSCs as a major contributor to the extracellular matrix (ECM) proteins observed with this stromal response.4,5
Under normal conditions, PSCs reside in the exocrine pancreas in a quiescent state, characterized by the expression of desmin, vimentin, and glial fibrillary acidic protein (GFAP) and the ability to store vitamin A.4 PSCs become activated in vivo in response to pancreatic injury.1,2 Upon activation, PSCs proliferate, lose the ability to store vitamin A, and express α-smooth muscle actin (α-SMA) and collagen I.2,5 It has been reported that activated PSCs produce autocrine factors like PDGF, TGF-β, cytokines, and cyclooxygenase-2 to perpetuate the activated phenotype 6–10 however, no comprehensive analysis of the PSC proteome in either the quiescent or activated state has been reported.
While knowledge of the intracellular proteome is important, it is the secreted proteome, or secretome, that is of particular interest with regards to understanding intercellular interactions, for example with tumor cells. It remains difficult to characterize proteins secreted by a single cell type to the exclusion of all others in vivo. One solution to this problem is studying cell lines derived from the cell type of interest in vitro, an approach we have already used to study the secretome of PDAC cells.11
In the current study, we exploit the RLT-PSC cell line, an immortalized cell line derived from human PSCs, to characterize the secretome of both the quiescent and activated PSC. The RLT-PSC cell line can be maintained in a quiescent state when grown on a soft substrate, or in an activated state when grown on untreated culture plates.12 We report the first comparative proteomic study demonstrating a remarkable differential secretome upon PSC activation and highlighting the potential biological functions of these proteins. Expression of selected proteins are verified in human pancreatic tissue samples.
MATERIALS AND METHODS
Cell Culture
The human PDAC cell lines MiaPaCa-2 and CAPAN-2, and the immortalized human PSC line RLT-PSC12 were cultured according to standard practice using Dulbecco's Modified Eagle Medium (DMEM, Sigma, St. Louis, MO) supplemented with 10 % fetal bovine serum (Sigma) and 1 % antibiotic (penicillin/streptomycin, Invitrogen, Grand Island, NY). Secreted proteins were harvested by allowing cells to grow to 80 % confluence, washing with phosphate buffered saline (Invitrogen), and incubating the cells with serum-free media at 37 °C for 48 hr. This media was collected, passed through a 0.22 µm filter, and stored at −80 °C until used. To induce a quiescent phenotype, 80 % confluent RLT-PSCs were treated with trypsin (Invitrogen), resuspended in serum-free DMEM supplemented with 1 % antibiotic and 2.5 mM N-acetyl cysteine (Sigma), then plated at a concentration of 20,000 cells/cm2 on collagen I coated cover-slips or plates (BD Biosciences, San Jose, CA) and incubated at 37 °C for 24 hr. RLT-PSCs were plated on uncoated tissue culture plates to achieve an activated state.
Cell culture immunocytochemistry
RLT-PSCs were plated on either uncoated (activated cells) or collagen I coated (quiescent cells) coverslips and incubated at 37 °C for at least 24 hr to allow attachment. Cells were formalin fixed, washed with propylene glycol (PolyScientific, Bay Shore, NY) and incubated with oil red O (0.5 % in propylene glycol, PolyScientific) for 1 hr at RT. Cells were then washed with 85 % propylene glycol, blocked with 1 % bovine serum albumin/PBS (Sigma), and labeled overnight at 4 °C with antibodies against either α-SMA (mouse, Sigma) or GFAP (rabbit, Sigma) diluted 1:1,600 and 1:600, respectively, in blocking solution. Cells were incubated with a Cy2 conjugated secondary antibody (donkey anti-mouse or -rabbit, Jackson ImmunoResearch, West Grove, PA) diluted 1:600 in blocking solution for 1 hr at RT. Finally, cells were incubated with DAPI (Sigma), diluted 1:10,000 in water, for 45 sec. Fluorescence was visualized using a Nikon (Melville, NY) E600 fluorescent microscope equipped with a CCD camera and IPLab image analysis software (BD Biosciences).
2D LC-MS/MS
Two biological replicates were analyzed for each cell line. Secreted proteins were concentrated using 3 kDa MW cutoff spin-filters (Millipore, Billerica, MA), and then precipitated using methanol-chloroform extraction. 100 µg of protein from each cell line was digested and separated by strong cation exchange (SCX) chromatography using standard methods as previously described.13 SCX fractions were subjected to reversed phase chromatography using a C18 column (0.3 × 150 mm, 4 µ, 90 Å Jupiter Proteo, Phenomenex, Torrence, CA) on a microflow high pressure liquid chromatography system (Eksigent, Dublin, CA). A chromatographic gradient increasing mobile phase B (95 % acetonitrile, 0.1 % formic acid) from 0 to 60 % over 30 min at a flow rate of 12 µl/min was used to elute peptides online to a linear ion trap mass spectrometer (LTQ, ThermoFisher, San Jose, CA) with positive ESI. Data dependent scanning was used to identify the 5 most intense precursor masses, which were then subjected to collision-induced dissociation to produce product ion spectra.
Protein identification
Raw data were submitted to Bioworks Browser version 3.2 (ThermoFisher) and batch searched through SEQUEST™ against the NCBI RefSeq human reference sequence database (version updated 03/17/09) containing 75,484 sequences consisting of 37,742 proteins and 37,742 decoy entries (1 reverse sequence for each real entry). The database was indexed using the following criteria: precursor and fragment mass tolerance of 1 amu, strict trypsin cleavage rules with up to two internal cleavage sites; differential modifications of methionine oxidation and carboxyamidomethylation on cysteine. The sequences were further processed using the Trans-Proteomic Pipeline software (version 2.8, Institute for Systems Biology, Seattle, WA) for analysis and validation of peptides and proteins using Peptide Prophet™ (version 3.0) and ProteinProphet™ (version 2.0), respectively. ProteinProphet™ results were filtered using a minimum probability score of 0.6, translating to a false discovery rate of 5 %. Proteins identified by fewer than two unique peptide sequences were discarded. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. The remaining proteins in each replicate were recombined to generate a complete list of secretome proteins for each cell line.
Bioinformatics
Pathway analysis was performed on proteins using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database 14 as a reference using methods previously described.11 Gene ontology (GO) analysis was also conducted using the functional annotation tool available through the DAVID Bioinformatics Database.15,16
Western blot analysis
The following primary polyclonal rabbit anti-human antibodies were obtained: pyruvate dehydrogenase kinase (PKM2) (GenWay, San Diego, CA), urokinase plasminogen activator preproprotein (PLAU) (Abgent, San Diego, CA) and transforming growth factor, beta-induced, 68kDa (TGFBI) (Proteintech Group, Inc., Chicago, IL). Primary goat anti-human serpin peptidase inhibitor, clade I (pancpin), member 2 (SERPINI2) antibody was obtained (Everest, Oxfordshire, UK). The following mouse monoclonal antibodies were obtained: ubiquitin carboxyl-terminal esterase L1 (UCHL1), matrix metallopeptidase 2 (MMP2), plasminogen activator inhibitor-1 (SERPINE1), non-metastatic cells 1, protein (NM23A) (NME1), ezrin (EZR), annexin A2 (ANXA2), and galectin-3-binding protein (LGALS3BP) (Santa Cruz Biotechnology, Santa Cruz, CA). Alexa Fluor 680 conjugated secondary donkey anti-mouse, anti-rabbit, and anti-goat antibodies were obtained (Invitrogen). Cells were lysed in either 25 % sample loading buffer (LDS, Invitrogen) or 6 M urea/2 M thiourea containing HALT™ Protease Inhibitor Cocktail (Pierce, Rockford, IL). Secreted proteins were concentrated using 3 kDa MW cutoff spin filters, precipitated using methanol-chloroform, then resuspended in 6 M urea/2M thiourea. Protein concentration of reconstituted whole cell lysates and secreted proteins was determined according to the Bradford method. 8 µg of protein was loaded per well on NuPAGE® 4–12 % Bis-Tris gels (Invitrogen) then separated in MOPS buffer (Invitrogen). Proteins were transferred to a PVDF membrane (Millipore, Billerica, MA), blocked with 5 % powdered milk in TBST (Teknova, Hollister, CA), then incubated with primary antibody overnight at 4 °C at a dilution of 1:200. After washing with TBST, membranes were incubated with secondary antibody for 30 min at RT at a dilution of 1:10,000 in the dark, then washed again with TBST. Fluorescence was measured used the Odyssey® Imaging System (LI-COR Biosciences, Lincoln, NE).
Tissue immunohistochemistry
The following primary polyclonal rabbit anti-human antibodies were obtained: PKM2 (GenWay, San Diego, CA), and TGFBI (Proteintech Group, Inc., Chicago, IL). Primary goat anti-human SERPINI2 antibody was obtained (Everest, Oxfordshire, UK). The following mouse monoclonal antibodies were obtained: MMP2 (Calbiochem, San Diego, CA), NME1, EZR, ANXA2, and LGALS3BP (Santa Cruz Biotechnology, Santa Cruz, CA), α-SMA, vimentin, CD31, and UCHL1 (Dako, Glostrup, Denmark). Biotinylated secondary donkey anti-mouse, -rabbit, or -goat antibodies were obtained (Vector, Burlingame, CA). Formalin-fixed paraffin embedded human pancreatic tissue was obtained from patients treated at the Hospital of the University of Pennsylvania through Institutional Review Board approved protocols. In total, 5 tissue samples of normal human pancreas, 5 tissue samples of human PDAC, and 2 tissue samples of PDAC metastasis to lymph node were analyzed. 4 µm serial sections were dewaxed, slides were microwaved in 10 mM citric acid pH 6 solution to achieve antigen retrieval, then endogenous peroxidases were quenched using 30 % hydrogen peroxide solution. Slides were incubated with successive solutions of avidin and biotin blocking solutions (Vector) and StartingBlock blocking buffer (Pierce, Rockford, IL) for 15 min each at RT and primary antibody overnight at 4 °C. Slides were incubated with biotin conjugated secondary antibody followed by the VECTSTAIN® HRP-ABC reagent (Vector, Burlingame, CA), each for 30 min at 37 °C. After developing the signal with diaminobenzadine (Vector), slides were washed with water and counterstained with hematoxylin. Images were captured using a Nikon (Melville, NY) E600 fluorescent microscope equipped with a CCD camera and IPLab image analysis software (BD Biosciences).
RESULTS
RLT-PSCs recapitulate primary human PSCs
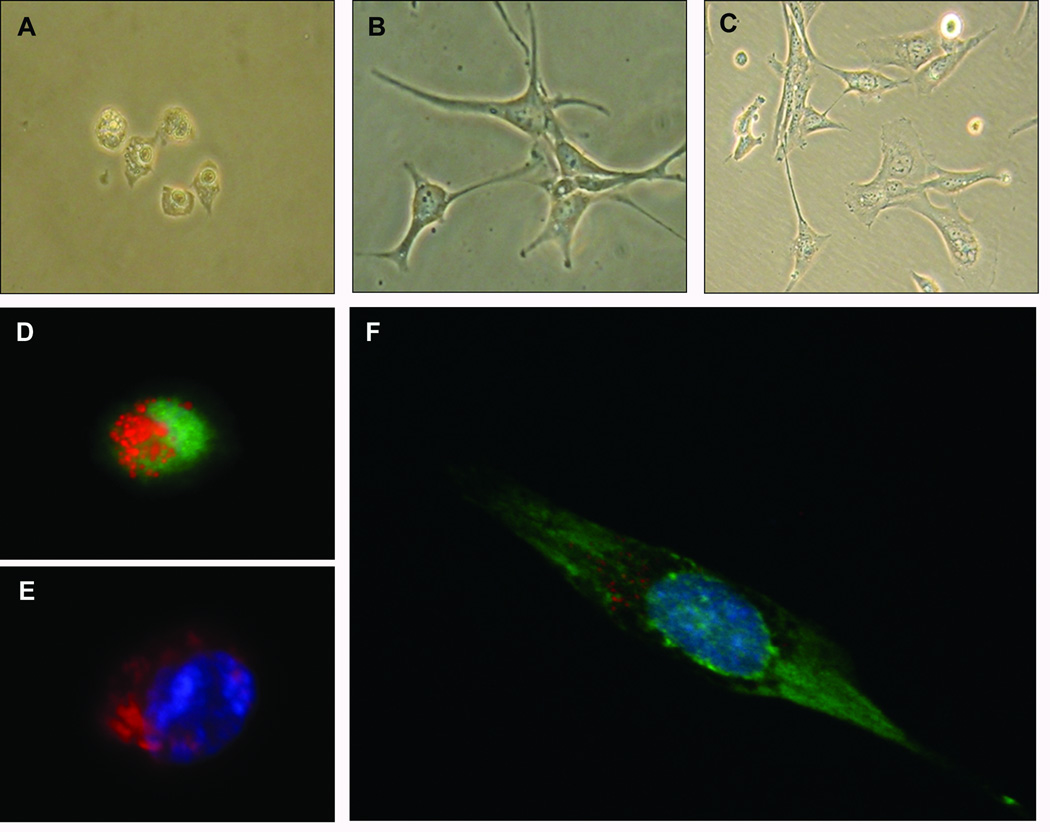
RLT-PSC is a human PSC line immortalized via transfection with SV40 large T antigen.12 When cultured on Matrigel plus N-acetyl cysteine treatment, RLT-PSCs exhibit a quiescent phenotype.12 We have found that RLT-PSCs cultured on collagen I coated cover slips or plates also maintain a quiescent phenotype denoted by morphology (Figure 1a), expression of GFAP (Figure 1d), presence of lipid droplets and absence of α-SMA (Figure 1e). The quiescent PSCs can then be activated by application of conditioned media harvested from MiaPaCa-2 pancreatic cancer cell lines. This activation is illustrated by morphology (Figure 1b), loss of oil red O staining, and expression of α-SMA (Figure 1f). PSCs cultured on uncoated tissue culture plates yielded an activated phenotype as well (Figure 1c). These results show that RLT-PSCs recapitulate features typical of primary human PSCs.
Figure 1.
Bright field microscopy demonstrating morphology of RLT-PSCs in (A) the quiescent state, and activated by (B) exposure to PCC conditioned media, or (C) growth on a tissue-culture treated plate. Fluorescent microscopy demonstrating (D) quiescent PSC lipid droplets (red), GFAP Expression (green), and nucleus (blue); (E) quiescent PSC lipid droplets (red), α-SMA (green), and nucleus (blue); and (F) activated PSC loss of lipid droplets (red), α-SMA Expression (green), and nucleus (blue). Cells in images A, B, D, E, and F were plated on collagen I coated coverslips, while those in image C were plated on tissue-culture treated plates. All cells were plated under serum-free conditions.
PSC secretion is altered upon activation
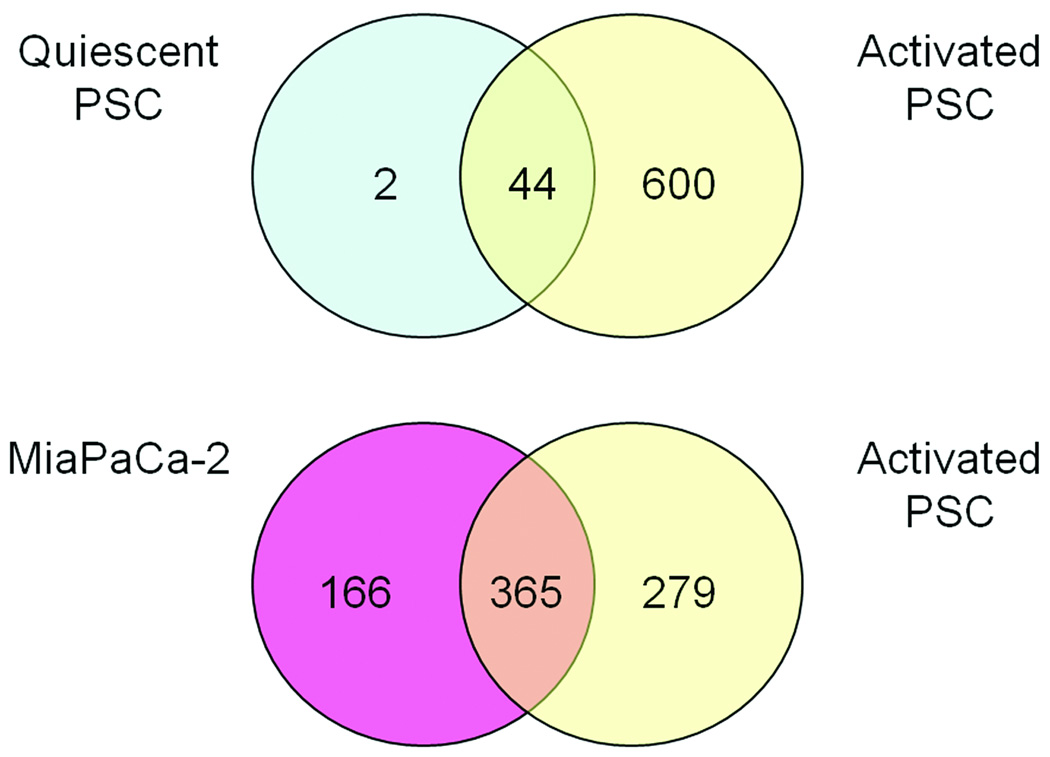
Proteomic profiling of the media harvested from cells in culture was undertaken to assess changes in the secretome of PSCs upon activation. Proteins identified by fewer than two different unique peptide sequences within a replicate were excluded. Reverse database analysis revealed a false discovery rate of less than 4 %. A list of all quiescent and activated PSC proteins and pancreatic cancer cell proteins is shown in Supplemental Table 1 (Supplemental Digital Content 1, http://links.lww.com/MPA/A36). Duplicate analyses were used in an effort to include as many potential biomarkers and therapeutic targets as possible, a reasonable approach given that any leads would be subject to further validation. A total of 641 unique proteins were identified in the secretome from activated PSCs. This is significantly higher than the 46 unique proteins indentified in the secretome from quiescent PSCs. The secretomes from CAPAN-2 and MiaPaCa-2 cancer cell lines contained 432 and 529 unique proteins, respectively. Proteins identified in the quiescent PSCs, activated PSCs, and cancer cell lines were compared to identify unique and shared proteins (Figure 2).
Figure 2.
Venn diagram of secreted proteins identified from pancreatic stellate cells in the quiescent and activated states. All identifications were made in the conditioned media derived from cells in culture.
The activated PSC data set was sorted by total number of spectra per protein to estimate relative abundance. A literature search was conducted on the most abundant proteins to identify those previously linked to cancer progression, including roles in angiogenesis, invasion, and apoptosis. These proteins (Table 1) were then chosen for further validation studies.
Table 1.
List of activated PSC proteins selected for further validation. The activated PSC secretome was sorted by total number of sequenced peptides per protein to estimate relative abundance. The most abundant proteins were screened via literature search to identify those most likely to play a role in cancer progression. These proteins were then chosen for further validation by western blot and IHC analysis.
| GI accession # | Abbreviation | Protein Name |
|---|---|---|
| gi|5453886 | SERPINI2 | serpin peptidase inhibitor, clade I (pancpin), member 2 |
| gi|4505863 | PLAU | urokinase plasminogen activator preprotein |
| gi|10835159 | SERPINE1 | plasminogen activator inhibitor - 1 |
| gi|11342666 | MMP2 | matrix metallopeptidase 2 |
| gi|4507467 | TGFBI | transforming growth factor, beta-induced, 68kDa |
| gi|5031863 | LGALS3BP | galectin-3-binding protein |
| gi|33286418 | PKM2 | pyruvate kinase, muscle isoform 1 |
| gi|66392203 | NME1 | nucleotide diphosphate kinase 1 |
| gi|161702986 | EZR | ezrin |
| gi|50845386 | ANXA2 | annexin A2 |
| gi|21361091 | UCHL1 | ubiquitin carboxyl-terminal esterase L1 |
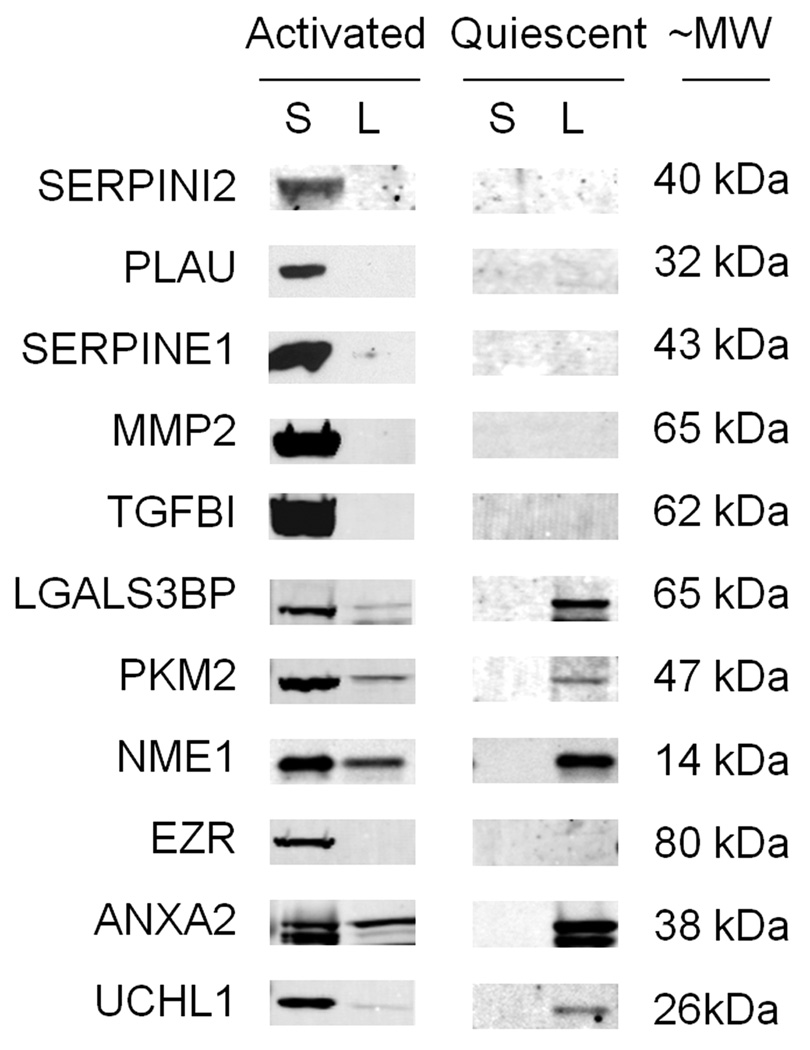
Western blot analysis confirms MS results
Expression of proteins secreted by activated RLT-PSCs (Table 1) was verified by Western blot analysis (Figure 3). Protein expression was also assessed in RLT-PSC whole cell lysate. Relative protein levels varied greatly. SERPINI2, PLAU, SERPINE1, MMP2, TGFBI, and EZR were undetected and LGALS3BP, PKM2, and UCHL1 showed faint bands in the activated RLT-PSC whole cell lysate. ANXA2 showed similar levels in the whole cell lysate and secretome. NME1 was expressed at higher abundance in the whole cell lysate than the secretome. All proteins were undetected in the quiescent RLT-PSC secretome. All proteins observed in the activated RLT-PSC whole cell lysate were also seen in the quiescent whole cell lysate.
Figure 3.
Western blotting of conditioned media supernatant (S) and lysate (L) from activated or quiescent PSCs demonstrating the presence of proteins of interest. Equal amounts of protein were added to each well.
IHC analysis confirms in vitro results
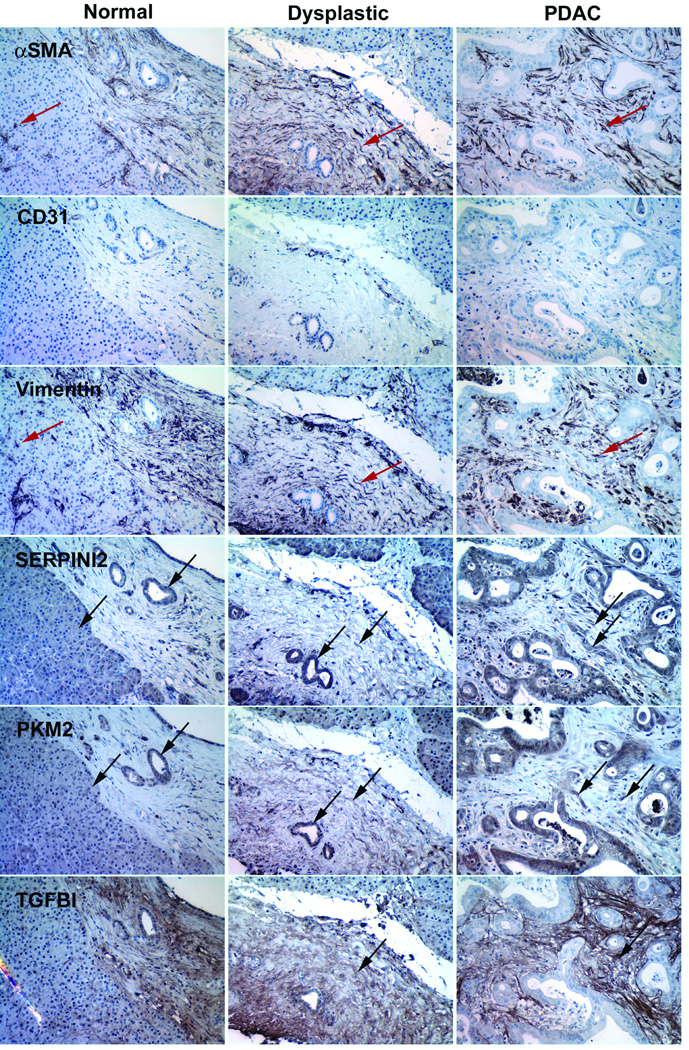
IHC analysis conducted on human tissue samples from patients with PDAC or normal controls was used to confirm and localize expression of selected proteins in relation to PSCs. PSCs were visualized by staining serial sections of tissue using PSC markers α-SMA and vimentin (Figure 4). To differentiate PSCs from endothelial cells, which also express α-SMA and vimentin, the endothelial cell marker, CD-31, was used.17 Therefore PSCs could be readily identified by the presence of α-SMA and vimentin and the absence of CD-31. Staining of serial sections was organized such that IHC staining of proteins of interest were performed in a tissue section immediately adjacent to a set of PSC markers (Table 2).
Figure 4.
Immunohistochemical staining of serial sections of normal human pancreas tissue (first column), dysplastic human pancreas tissue (middle column), and human pancreatic ductal adenocarcinoma (last column). Positive stain is in brown and indicated by black arrows. PSCs were visualized by the expression of markers α-SMA and vimentin and the absence of CD31. PSCs are indicated by red arrows.
Table 2.
Serial section staining scheme. IHC analysis conducted on human tissue samples from patients with pancreatitis, PDAC, or disease-free subjects was used to confirm and localize expression of selected proteins in relation to PSCs. PSCs were visualized by staining serial sections of tissue using PSC markers α-SMA and vimentin. To differentiate PSCs from endothelial cells, which also express α-SMA and vimentin, the endothelial cell marker, CD-31, was used. Therefore PSCs could be readily identified by the presence of α-SMA and vimentin and the absence of CD-31. Staining of serial sections was organized such that IHC staining of proteins of interest, listed in black, were performed in a tissue section immediately adjacent to a set of PSC markers, italicized.
| Slide # | Protein |
|---|---|
| 1 | SERPINI2 |
| 2 | α-SMA |
| 3 | CD-31 |
| 4 | vimentin |
| 5 | PKM2 |
| 6 | TGFBI |
| 7 | α-SMA |
| 8 | CD-31 |
| 9 | vimentin |
| 10 | EZR |
| 11 | LGALS3BP |
| 12 | α-SMA |
| 13 | CD-31 |
| 14 | Vimentin |
| 15 | UCHL1 |
| 16 | NME1 |
| 17 | α-SMA |
| 18 | CD-31 |
| 19 | vimentin |
| 20 | ANXA2 |
In PDAC tissue sections, SERPINI2, PKM2 (Figure 4), LGALS3BP, and NME1 (Figure 5), were detected in the cytosol of cancer cells and PSCs (Figure 4). In the dysplastic human pancreas, SERPINI2, PKM2 (Figure 4), LGALS3BP, and NME1 (Figure 5) were detected in ductal cells and PSCs. These proteins were also detected within the cytosolic region of acinar and ductal cells in normal human pancreas. UCHL1 was detected distinctly in the nuclei of PSCs in PDAC tissue sections and at low levels in the nuclei of some ductal cells and PSCs in the dysplastic pancreas and some acinar and ductal cells in normal pancreas (Figure 5). TGFBI was the only protein detected distinctly in stromal regions, to the exclusion of cancer cells in PDAC tissue (Figure 4). In the dysplastic pancreas, TGFBI was found in the fibrotic region containing PSCs (Figure 4). Interestingly, TGFBI was also observed in vesicles in normal pancreatic tissue. In PDAC tissue, EZR was detected in the cytosol of cancer cells, but also was distinctly present at the luminal membrane of ductal cells and the nuclei of PSCs (Figure 5). EZR was also detected in the cytosol of ductal cells with strong localization to the luminal surface of ductal cells in dysplastic pancreas. In normal pancreas, EZR was detected in the centroacinar space and within a few acinar cells in the cytosolic region bordering the centroacinar space. In PDAC tissue, ANXA2 was detected in the cytosol of cancer cells and PSCs. Similar to EZR, ANXA2 was detected along the luminal membrane of ductal cells. ANXA2 was also detected in the cytosol of ductal cells and PSCs in the dysplastic pancreas. ANXA2 localization in normal pancreas was similar to that of EZR with strong staining observed in the centroacinar space and the bordering cytosol of acinar cells.
Figure 5.
Immunohistochemical staining of serial sections of normal human pancreas tissue (first column), dysplastic human pancreas tissue (middle column), and human pancreatic ductal adenocarcinoma (last column). Positive stain is in brown and indicated by black arrows.
Bioinformatics analysis
Of the 641 proteins identified in the RLT-PSC secretome 174 (27%) were implicated in at least one biological pathway according to analysis against the KEGG database (Table 3). Most classifiable proteins were involved in metabolism. As expected for secreted proteins, none were classified as involved with transcription pathways. The most highly populated pathway was cell communication followed closely by the immune system. Many proteins were also identified within the cell motility pathway. 17 proteins have been implicated in various cancers, of which 14 were implicated in pancreatic cancer.
Table 3.
Activated PSC proteins identified with KEGG pathways. The activated PSC secretome was subjected to pathway analysis using the KEGG database as a reference. The number of proteins identified within each pathway is presented.
| Pathways | Number of Gis |
|---|---|
| Metabolism | 48 |
| Genetic Information Processing | |
| Transcription | 0 |
| Translation | 7 |
| Folding, Sorting and Degradation | 18 |
| Proteasome | 11 |
| Replication and Repair | 10 |
| Environmental Information Processing | |
| Membrane Transport | 0 |
| Signal Transduction | 15 |
| MAPK Signaling pathway | 7 |
| Signaling Molecules and Interaction | 14 |
| Cellular Processes | |
| Cell Motility | 22 |
| Cell Growth and Death | 12 |
| Cell Communication | 31 |
| Focal adhesion | 21 |
| Insulin signaling pathway | 10 |
| Immune System | 30 |
| Leukocyte transendothelial migration | 5 |
| Nervous System | 6 |
| Sensory System | 2 |
| Development | 5 |
| Behavior | 0 |
| Human Diseases | |
| Cancers | 17 |
| Immune Disorders | 8 |
| Neurodegenerative Diseases | 9 |
| Metabolic Disorders | 2 |
| Infectious Diseases | 14 |
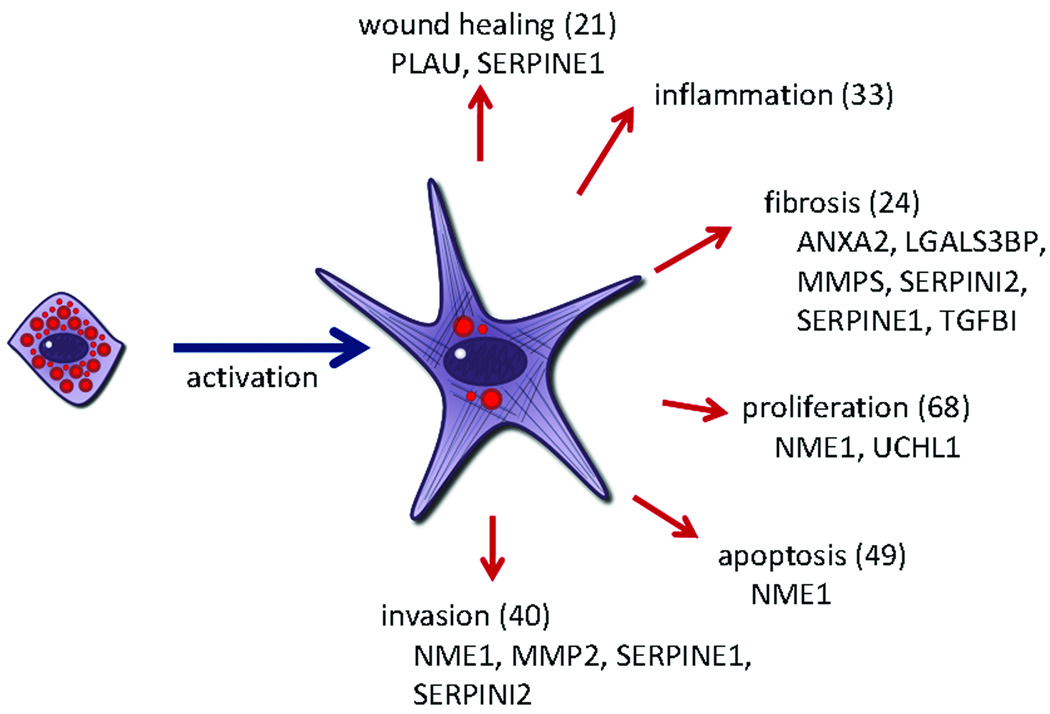
GO analysis was conducted to elucidate biological function (data not shown). As was seen with the KEGG analysis, the majority of proteins were related to cellular metabolic processes, however, a significant number of proteins were involved in fibrosis, invasion, inflammation, cell division, and apoptosis. PSCs are believed to play important roles in wound healing, inflammation, fibrosis, tumor progression and invasion through matrix remodeling (Figure 6). To identify activated PSC proteins that effect these changes, GO terms that relate to these functions were probed and the resulting list of proteins is presented in Supplemental Table 2 (Supplemental Digital Content 2, http://links.lww.com/MPA/A37). Some functions, such as wound healing, inflammation, and apoptosis relate readily to the GO terms available. Other important pathways, such as fibrosis, cannot be studied using GO terms currently available. Fibrotic tissue consists largely of ECM proteins and proteases control the remodeling of these proteins. For this reason, proteins relating to the ECM or protease inhibitors are listed under the category of fibrosis in Supplemental Table 2 (Supplemental Digital Content 2, http://links.lww.com/MPA/A37). To identify proteins associated with proliferation, all proteins classified as relating to the cell cycle, mitogens, or cell proliferation were grouped together (see Supplemental Table 2, Supplemental Digital Content 2, http://links.lww.com/MPA/A37). Finally, to identify proteins that play a role in invasion, proteins relating to cell motility and vasculature development were compiled and are presented in Supplemental Table 2 (Supplemental Digital Content 2, http://links.lww.com/MPA/A37).
Figure 6.
Diagram illustrating proposed activated stellate cell functions. The number of proteins identified within a function is given in parentheses. Proteins selected for validation are listed below the function.
DISCUSSION
The crucial role of PSC activation in the pathobiology of PDAC has only recently begun to be appreciated. PSCs potentiate pancreatic tumors implanted in vivo,18,19 and conditioned media from activated PSCs enhances proliferation and invasion of PCCs in vitro.20,21 These observations support the hypothesis that PSCs secrete soluble factors that promote tumor progression. Therefore, this first characterization of the secretome from activated PSCs could provide insight into mechanisms of tumor progression.
While PSCs secrete few proteins in the quiescent state, activation results in a rich and complex mixture of secreted proteins. It is likely that the secretome of immortalized PSCs differ in some ways from freshly isolated PSCs. A comparison of gene expression profiles demonstrated few differences following immortalization, involving extracellular matrix proteins, integrins, intermediate filament proteins and cytokines.7 In our study the protein MMP2, which has previously been described as expressed at low levels in the quiescent PSC,22 was not identified in the quiescent RLT-PSC secretome. A number of factors are likely at play, such as differences resulting from immortalization and across species. Most importantly, previous studies have used cell lines which are only transiently quiescent, whereas the RLT-PSC cell line can be stably maintained in a quiescent state indefinitely. It is possible that the low level of MMP2 expression previously observed reflects low level PSC activation.
It is likely that the proteins secreted by activated PSCs are dependent upon the activating stimulus. Future studies will look at proteins secreted by PSCs activated in response to other stimuli, such as PDAC cells, to study these differences. Interestingly, many proteins secreted by activated PSCs are also secreted by the cancer cell lines studied. Approximately 60% of the proteins identified in the activated PSC secretome and one half to three-quarters of the cancer cell line secretomes were found common to both cell types (Figure 2).
Many proteins secreted by activated PSCs play important roles in proliferation, invasion, inflammation and fibrosis, pathways important for the proposed role of PSCs in supporting tumor growth and metastasis. The potential roles of individual proteins present within the identified PSC secretome are discussed below.
Wound healing
Wound healing is a primary function of PSCs. It has been hypothesized that cancer cells exploit this PSC function to support tumor growth. 21 proteins associated with wound healing were identified, including PLAU and SERPINE1. SERPINE1 inhibits tissue plasminogen activator (tPA) and urokinase (uPA), the activators of PLAU, a key enzyme in fibrinolysis. High levels of SERPINE1 have been correlated with a poor outcome in PDAC.23 Expression of PLAU and SERPINE1 have been previously reported in desmoplastic regions of PDAC tissue.24–27
Inflammation
33 proteins associated with inflammation were identified. Inflammation is one of the primary activators of PSCs in the pancreas. Chronic pancreatitis, a condition of prolonged inflammation, is one of the risk factors for PDAC. Inflammatory processes are thought to potentiate PDAC through classical mechanisms, such as oxidative-stress induced DNA damage. Another recently proposed mechanism implicates recruitment of immunosuppressive cells as a key pathway for evasion of immune surveillance early in pancreatic carcinogenesis.28 Our findings support the hypothesis that PSCs play a role in perpetuating a state of inflammation in the tumor microenvironment.
Proliferation
Accumulating evidence supports the presence of factors secreted by activated PSCs that enhance cancer cell proliferation. Among the 68 secreted PSC proteins involved in proliferation, NME1 and UCHL1 expression was verified in the current study. NME1, a nucleoside diphosphate kinase, was observed at higher levels in cervical, thyroid29,30 and metastatic ovarian tumors,31 suggesting a role in enhancing cancer cell proliferation.
UCHL1 is an enzyme whose function varies with dimerization and post-translational modifications.32 In some cancers, the gene for UCHL1 is methylated, and restoration of UCHL1 expression inhibited proliferation in hepatocellular carcinoma.33 Other studies have shown that UCHL1 is upregulated in lung cancer, invasive colorectal cancer, and PDAC.34–36
Apoptosis
Inhibition of apoptosis is a key mechanism in tumor growth. 49 proteins associated with apoptosis were identified, including NME1. NME1 has shown the ability to limit the expression of the apoptotic protein Bcl-2.37 Thus, in addition to promoting PCC proliferation, NME1 may support PDAC progression through inhibition of apoptosis.
Fibrosis and invasion
Activated PSCs are known to secrete ECM proteins such as collagens I, III, and IV, and fibronectin.1,2 This matrix then serves to promote cell proliferation and migration to heal wounded areas. Chronic activation, as seen in chronic pancreatitis or PDAC, results in the accumulation of fibrotic tissue. PSCs are also known to produce factors that break down ECM, which may play a role in promoting invasion. Indeed, PDAC lymph node metastases were observed to contain stroma and PSCs (see Supplemental Figure 1, Supplemental Digital Content 3, http://links.lww.com/MPA/A35). 24 ECM proteins and proteases and 40 proteins associated with cell motility and vasculature development were identified.
ANXA2, a phospholipid binding protein, is believed to play a role in angiogenesis, invasion, and metastasis, and its expression has been best described in colon cancer.38,39 ANXA2 has also been reported to serve as a receptor for plasminogen40 and as such may play a role in the plasminogen activator pathway The urokinase plasminogen activator pathway is believed to play a role in cancer cell invasion in the pancreas24,26 and includes the proteins MMP2 and SERPINE1, along with PLAU. SERPINI2 is a protein specific to the pancreas and to breast cancer stroma.41,42 The function of SERPINI2 is poorly understood, however its sequence identifies it as a serine protease inhibitor in the same family as SERPINE1, suggesting a role in the plasminogen activator pathway, as well. LGALS3BP, a protein expressed in advanced gastric cancer,43 binds the receptor galectin-3, a protein highly expressed in PDAC metastases.44 LGALS3BP modulates cell-integrin interactions to either promote or inhibit cell attachment.45 NME1 has been implicated as a suppressor of tumor metastasis in melanoma and breast cancer.46,47 In vitro studies have shown that NME1 suppresses tumor metastatic potential and negatively regulates cell migration, possibly via regulation of integrin expression or trafficking.48–50 TGFBI is a secreted RGD-containing protein and has anti-adhesive properties.51,52 TGFBI is upregulated in renal cell carcinoma and esophageal adenocarcinoma53,54 and has been reported to inhibit cell attachment in vitro.51
Finally, UCHL1 and EZR are believed to enhance invasion. UCHL1 is thought to modify cell morphology through modulation of the Akt-signaling pathway.55 EZR is a mediator of cell-integrin interactions and high expression of EZR has been associated with prostate and pancreatic cancer progression.56,57 By organizing the interaction between the cytoskeleton and the cell membrane EZR regulates cell adhesion, migration and invasion.58 Furthermore, EZR has been found to be essential to the metastatic potential of some cell lines.59,60 Taken together, our results demonstrate PSCs are an important source of proteins that regulate ECM remodeling, cell motility and invasion.
The proteins discussed here are a sampling of the many secreted proteins identified from the activated PSC secretome. It is clear that the PSC secretome is an important source of proteins that regulate proliferation, inflammation, and ECM remodeling as well as cell motility and invasion. Characterization of the activated PSC secretome is an important first step in elucidating the complex role of PSCs in PDAC progression. This work lays the foundation for future studies, for example, characterizing the proteins involved in paracrine signaling with PDAC cells. Further studies will determine whether secreted PSC proteins identified here can serve as biomarkers of PDAC progression or potential new therapeutic targets.
Supplementary Material
Acknowledgements
We would like to thank Ralf S. Jesenofsky of the Division of Molecular Gastroenterology, Faculty of Medicine of the University of Heidelberg for the generous donation of the RLT-PSC line. Special thanks to Gary Swain and staff of the University of Pennsylvania Morphology Core Center for Molecular Studies in Digestive and Liver Diseases in the preparation of human tissue slides for IHC analysis and microscopy.
Funding: Supported by NIH grants R25CA101871, P30ES013508, P30DK050306, an Institute for Translational Medicine and Therapeutics Research Fellowship and the Foundation for Digestive Health and Nutrition Bernard L. Schwartz Designated Research Award in Pancreatic Cancer.
Abbreviations
- α-SMA
alpha smooth muscle actin
- ANXA2
annexin A2
- DAPI
4',6-diamidino-2-phenylindole
- ECM
extra-cellular matrix
- EZR
ezrin
- GFAP
glial fibrillary acidic protein
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LGALS3BP
galectin-3-binding protein
- MMP2
matrix metallopeptidase 2
- NME1
non-metastatic cells 1, protein (NM23A)
- PCC
pancreatic cancer cell
- PDAC
pancreatic ductal adenocarcinoma
- PIR
Protein Information Resource
- PKM2
pyruvate dehydrogenase kinase, muscle isoform
- PLAU
urokinase plasminogen activator
- PSC
pancreatic stellate cell
- SERPINE1
plasminogen activator inhibitor-1
- SERPINI2
serpin peptidase inhibitor, clade I (pancpin), member 2
- SILAC
stable isotope labeling of amino acids in cell culture
- TBST
TBS with 1% Tween 20
- TGFBI
transforming growth factor, beta-induced, 68kDa
- UCHL1
ubiquitin carboxyl-terminal esterase L1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental digital contents are available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the Journal’s Web site (www.pancreasjournal.com).
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
Contributor Information
Angela Y. Wehr, Centers for Cancer Pharmacology and Excellence in Environmental Toxicology, University of Pennsylvania School of Medicine, Philadelphia, PA.
Emma E. Furth, Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine, Philadelphia, PA.
Vineet Sangar, Centers for Cancer Pharmacology and Excellence in Environmental Toxicology, University of Pennsylvania School of Medicine, Philadelphia, PA.
Ian A. Blair, Centers for Cancer Pharmacology and Excellence in Environmental Toxicology, University of Pennsylvania School of Medicine, Philadelphia, PA.
Kenneth H. Yu, Gastrointestinal Oncology Service, Memorial Sloan Kettering Cancer Center, New York, NY.
REFERENCES
- 1.Apte MV, Haber PS, Applegate TL, et al. Periacinar stellate shaped cells in rat pancreas: identification, isolation, and culture. Gut. 1998;43(1):128–133. doi: 10.1136/gut.43.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachem MG, Schneider E, Gross H, et al. Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology. 1998;115(2):421–432. doi: 10.1016/s0016-5085(98)70209-4. [DOI] [PubMed] [Google Scholar]
- 3.Crnogorac-Jurcevic T, Efthimiou E, Nielsen T, et al. Expression profiling of microdissected pancreatic adenocarcinomas. Oncogene. 2002;21(29):4587–4594. doi: 10.1038/sj.onc.1205570. [DOI] [PubMed] [Google Scholar]
- 4.Omary MB, Lugea A, Lowe AW, et al. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117(1):50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apte MV, Park S, Phillips PA, et al. Desmoplastic reaction in pancreatic cancer: role of pancreatic stellate cells. Pancreas. 2004;29(3):179–187. doi: 10.1097/00006676-200410000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Aoki H, Ohnishi H, Hama K, et al. Cyclooxygenase-2 is required for activated pancreatic stellate cells to respond to proinflammatory cytokines. Am J Physiol Cell Physiol. 2007;292(1):C259–C268. doi: 10.1152/ajpcell.00030.2006. [DOI] [PubMed] [Google Scholar]
- 7.Bachem MG, Zhou Z, Zhou S, et al. Role of stellate cells in pancreatic fibrogenesis associated with acute and chronic pancreatitis. J Gastroenterol Hepatol. 2006;21 Suppl 3:S92–S96. doi: 10.1111/j.1440-1746.2006.04592.x. [DOI] [PubMed] [Google Scholar]
- 8.Gao R, Brigstock DR. Connective tissue growth factor (CCN2) in rat pancreatic stellate cell function: integrin alpha5beta1 as a novel CCN2 receptor. Gastroenterology. 2005;129(3):1019–1030. doi: 10.1053/j.gastro.2005.06.067. [DOI] [PubMed] [Google Scholar]
- 9.Phillips PA, Wu MJ, Kumar RK, et al. Cell migration: a novel aspect of pancreatic stellate cell biology. Gut. 2003;52(5):677–682. doi: 10.1136/gut.52.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shek FW, Benyon RC, Walker FM, et al. Expression of transforming growth factor-beta 1 by pancreatic stellate cells and its implications for matrix secretion and turnover in chronic pancreatitis. Am J Pathol. 2002;160(5):1787–1798. doi: 10.1016/s0002-9440(10)61125-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu KH, Barry CG, Austin D, et al. Stable isotope dilution multidimensional liquid chromatography-tandem mass spectrometry for pancreatic cancer serum biomarker discovery. J Proteome Res. 2009;8(3):1565–1576. doi: 10.1021/pr800904z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jesnowski R, Furst D, Ringel J, et al. Immortalization of pancreatic stellate cells as an in vitro model of pancreatic fibrosis: deactivation is induced by matrigel and N-acetylcysteine. Lab Invest. 2005;85(10):1276–1291. doi: 10.1038/labinvest.3700329. [DOI] [PubMed] [Google Scholar]
- 13.Yocum AK, Yu K, Oe T, et al. Effect of immunoaffinity depletion of human serum during proteomic investigations. J Proteome Res. 2005;4(5):1722–1731. doi: 10.1021/pr0501721. [DOI] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 16.Huang dW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Michalski CW, Gorbachevski A, Erkan M, et al. Mononuclear cells modulate the activity of pancreatic stellate cells which in turn promote fibrosis and inflammation in chronic pancreatitis. J Transl Med. 2007;5:63. doi: 10.1186/1479-5876-5-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachem MG, Schunemann M, Ramadani M, et al. Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology. 2005;128(4):907–921. doi: 10.1053/j.gastro.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 19.Schneiderhan W, Diaz F, Fundel M, et al. Pancreatic stellate cells are an important source of MMP-2 in human pancreatic cancer and accelerate tumor progression in a murine xenograft model and CAM assay. J Cell Sci. 2007;120(Pt 3):512–519. doi: 10.1242/jcs.03347. [DOI] [PubMed] [Google Scholar]
- 20.Bachem MG, Zhou S, Buck K, et al. Pancreatic stellate cells-role in pancreas cancer. Langenbecks Arch Surg. 2008;393(6):891–900. doi: 10.1007/s00423-008-0279-5. [DOI] [PubMed] [Google Scholar]
- 21.Hwang RF, Moore T, Arumugam T, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68(3):918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Philips PA, McCarroll JA, Park S, et al. Rat pancreatic stellate cells secrete matrix metalloproteinases: implications for extracellular matrix turnover. Gut. 2003;52(2):275–282. doi: 10.1136/gut.52.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith R, Xue A, Gill A, et al. High expression of plasminogen activator inhibitor-2 (PAI-2) is a predictor of improved survival in patients with pancreatic adenocarcinoma. World J Surg. 2007;31(3):493–502. doi: 10.1007/s00268-006-0289-9. [DOI] [PubMed] [Google Scholar]
- 24.Friess H, Cantero D, Graber H, et al. Enhanced urokinase plasminogen activation in chronic pancreatitis suggests a role in its pathogenesis. Gastroenterology. 1997;113(3):904–913. doi: 10.1016/s0016-5085(97)70186-0. [DOI] [PubMed] [Google Scholar]
- 25.Wojtukiewicz MZ, Rucinska M, Zacharski LR, et al. Localization of blood coagulation factors in situ in pancreatic carcinoma. Thromb Haemost. 2001;86(6):1416–1420. [PubMed] [Google Scholar]
- 26.Cantero D, Friess H, Deflorin J, et al. Enhanced expression of urokinase plasminogen activator and its receptor in pancreatic carcinoma. Br J Cancer. 1997;75(3):388–395. doi: 10.1038/bjc.1997.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi Y, Nakao A, Harada A, et al. Expression of plasminogen activators and their inhibitors in human pancreatic carcinoma: immunohistochemical study. Am J Gastroenterol. 1993;88(11):1928–1933. [PubMed] [Google Scholar]
- 28.Clark CE, Hingorani SR, Mick R, et al. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67(19):9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 29.Wang PH, Ko JL, Chang H, et al. Clinical significance of high nm23-H1 expression in intraepithelial neoplasia and early-stage squamous cell carcinoma of the uterine cervix. Gynecol Obstet Invest. 2003;55(1):14–19. doi: 10.1159/000068951. [DOI] [PubMed] [Google Scholar]
- 30.Ferenc T, Lewinski A, Lange D, et al. Analysis of nm23-H1 protein immunoreactivity in follicular thyroid tumors. Pol J Pathol. 2004;55(4):149–153. [PubMed] [Google Scholar]
- 31.Leary JA, Kerr J, Chenevix-Trench G, et al. Increased expression of the NME1 gene is associated with metastasis in epithelial ovarian cancer. Int J Cancer. 1995;64(3):189–195. doi: 10.1002/ijc.2910640308. [DOI] [PubMed] [Google Scholar]
- 32.Setsuie R, Wada K. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int. 2007;51(2–4):105–111. doi: 10.1016/j.neuint.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Yu J, Tao Q, Cheung KF, et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48(2):508–518. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]
- 34.Hibi K, Westra WH, Borges M, et al. PGP9.5 as a candidate tumor marker for non-small-cell lung cancer. Am J Pathol. 1999;155(3):711–715. doi: 10.1016/S0002-9440(10)65169-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamazaki T, Hibi K, Takase T, et al. PGP9.5 as a marker for invasive colorectal cancer. Clin Cancer Res. 2002;8(1):192–195. [PubMed] [Google Scholar]
- 36.Tezel E, Hibi K, Nagasaka T, et al. PGP9.5 as a prognostic factor in pancreatic cancer. Clin Cancer Res. 2000;6(12):4764–4767. [PubMed] [Google Scholar]
- 37.Curtis CD, Likhite VS, McLeod IX, et al. Interaction of the tumor metastasis suppressor nonmetastatic protein 23 homologue H1 and estrogen receptor alpha alters estrogen-responsive gene expression. Cancer Res. 2007;67(21):10600–10607. doi: 10.1158/0008-5472.CAN-07-0055. [DOI] [PubMed] [Google Scholar]
- 38.Sharma MC, Sharma M. The role of annexin II in angiogenesis and tumor progression: a potential therapeutic target. Curr Pharm Des. 2007;13(35):3568–3575. doi: 10.2174/138161207782794167. [DOI] [PubMed] [Google Scholar]
- 39.Duncan R, Carpenter B, Main LC, et al. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer. 2008;98(2):426–433. doi: 10.1038/sj.bjc.6604128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hajjar KA, Jacovina AT, Chacko J. An endothelial cell receptor for plasminogen/tissue plasminogen activator. I. Identity with annexin II. J Biol Chem. 1994;269(33):21191–21197. [PubMed] [Google Scholar]
- 41.Ozaki K, Nagata M, Suzuki M, et al. Isolation and characterization of a novel human pancreas-specific gene, pancpin, that is down-regulated in pancreatic cancer cells. Genes Chromosomes Cancer. 1998;22(3):179–185. [PubMed] [Google Scholar]
- 42.Xiao G, Liu YE, Gentz R, et al. Suppression of breast cancer growth and metastasis by a serpin myoepithelium-derived serine proteinase inhibitor expressed in the mammary myoepithelial cells. Proc Natl Acad Sci U S A. 1999;96(7):3700–3705. doi: 10.1073/pnas.96.7.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park YP, Choi SC, Kim JH, et al. Up-regulation of Mac-2 binding protein by hTERT in gastric cancer. Int J Cancer. 2007;120(4):813–820. doi: 10.1002/ijc.22369. [DOI] [PubMed] [Google Scholar]
- 44.Berberat PO, Friess H, Wang L, et al. Comparative analysis of galectins in primary tumors and tumor metastasis in human pancreatic cancer. J Histochem Cytochem. 2001;49(4):539–549. doi: 10.1177/002215540104900414. [DOI] [PubMed] [Google Scholar]
- 45.Hughes RC. Galectins as modulators of cell adhesion. Biochimie. 2001;83(7):667–676. doi: 10.1016/s0300-9084(01)01289-5. [DOI] [PubMed] [Google Scholar]
- 46.Florenes VA, Aamdal S, Myklebost O, et al. Levels of nm23 messenger RNA in metastatic malignant melanomas: inverse correlation to disease progression. Cancer Res. 1992;52(21):6088–6091. [PubMed] [Google Scholar]
- 47.Youn B, Kim HD, Kim J. Nm23-H1/nucleoside diphosphate kinase as a key molecule in breast tumor angiogenesis. Expert Opin Ther Targets. 2008;12(11):1419–1430. doi: 10.1517/14728222.12.11.1419. [DOI] [PubMed] [Google Scholar]
- 48.MacDonald NJ, de la RA, Benedict MA, et al. A serine phosphorylation of Nm23, and not its nucleoside diphosphate kinase activity, correlates with suppression of tumor metastatic potential. J Biol Chem. 1993;268(34):25780–25789. [PubMed] [Google Scholar]
- 49.Murakami M, Meneses PI, Lan K, et al. The suppressor of metastasis Nm23-H1 interacts with the Cdc42 Rho family member and the pleckstrin homology domain of oncoprotein Dbl-1 to suppress cell migration. Cancer Biol Ther. 2008;7(5):677–688. doi: 10.4161/cbt.7.5.5665. [DOI] [PubMed] [Google Scholar]
- 50.Waikel RL, Kawachi Y, Waikel PA, et al. Deregulated expression of c-Myc depletes epidermal stem cells. Nat Genet. 2001;28(2):165–168. doi: 10.1038/88889. [DOI] [PubMed] [Google Scholar]
- 51.Skonier J, Bennett K, Rothwell V, et al. beta ig-h3: a transforming growth factor-beta-responsive gene encoding a secreted protein that inhibits cell attachment in vitro and suppresses the growth of CHO cells in nude mice. DNA Cell Biol. 1994;13(6):571–584. doi: 10.1089/dna.1994.13.571. [DOI] [PubMed] [Google Scholar]
- 52.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 53.Hourihan RN, O'Sullivan GC, Morgan JG. Transcriptional gene expression profiles of oesophageal adenocarcinoma and normal oesophageal tissues. Anticancer Res. 2003;23(1A):161–165. [PubMed] [Google Scholar]
- 54.Ivanov SV, Ivanova AV, Salnikow K, et al. Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem Biophys Res Commun. 2008;370(4):536–540. doi: 10.1016/j.bbrc.2008.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim HJ, Kim YM, Lim S, et al. Ubiquitin C-terminal hydrolase-L1 is a key regulator of tumor cell invasion and metastasis. Oncogene. 2009;28(1):117–127. doi: 10.1038/onc.2008.364. [DOI] [PubMed] [Google Scholar]
- 56.Valdman A, Fang X, Pang ST, et al. Ezrin expression in prostate cancer and benign prostatic tissue. Eur Urol. 2005;48(5):852–857. doi: 10.1016/j.eururo.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 57.Akisawa N, Nishimori I, Iwamura T, et al. High levels of ezrin expressed by human pancreatic adenocarcinoma cell lines with high metastatic potential. Biochem Biophys Res Commun. 1999;258(2):395–400. doi: 10.1006/bbrc.1999.0653. [DOI] [PubMed] [Google Scholar]
- 58.Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3(8):586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- 59.Khanna C, Wan X, Bose S, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10(2):182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 60.Yu Y, Khan J, Khanna C, et al. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10(2):175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.