Abstract
The conserved and essential eukaryotic protein Spt6 functions in transcription elongation, chromatin maintenance, and RNA processing. Spt6 has three characterized functions. It is a histone chaperone capable of reassembling nucleosomes, a central component of transcription elongation complexes, and is required for recruitment of RNA processing factors to elongating RNA polymerase II (RNAPII). Here, we report crystal structures of the 168 kDa Spt6 protein from Saccharomyces cerevisiae that together represent essentially all of the ordered sequence. Our two structures of the ~900 residue core region reveal a series of putative nucleic acid and protein-protein interaction domains that fold into an elongated form that resembles the bacterial protein Tex. The similarity to a bacterial transcription factor suggests that the core domain performs nucleosome-independent activities, and as with Tex we find that Spt6 binds DNA. Unlike Tex, however, the Spt6 S1 domain does not contribute to this activity. Crystal structures of the Spt6 C-terminal region reveal a tandem SH2 domain structure comprised of two closely associated SH2 folds. One of these SH2 folds is cryptic, while the other shares striking structural similarity with metazoan SH2 domains and possesses structural features associated with the ability to bind phosphorylated substrates including phosphotyrosine. Binding studies with phosphopeptides that mimic the RNAPII CTD revealed affinities typical of other RNAPII CTD-binding proteins but did not indicate a specific interaction. Overall, these findings provide a structural foundation for understanding how Spt6 encodes several distinct functions within a single polypeptide chain.
Introduction
Gene expression in eukaryotes relies on a synergistic relationship between transcription, RNA processing, and chromatin structure.1; 2 The specific positioning, composition, and posttranslational modification of nucleosomes defines a code for chromatin-templated transcriptional regulation. Moreover, transcription is intimately tied to mRNA processing, surveillance, and export from the nucleus. This coordination relies on precise cooperation among many proteins, with Spt6 being remarkable for playing multifaceted roles in several distinct processes.
SPT6 (Suppressor of Ty 6) was originally discovered in Saccharomyces cerevisiae as a gene that influences general transcription through manipulation of chromatin structure at upstream promoter elements.3 Subsequently, Spt6 has been implicated in a variety of biological processes in organisms ranging from yeasts to human, including embryogenesis in Zebrafish4, multiple stages of development in Drosophila5, gut morphogenesis in C. elegans6, signal transduction in mammals7; 8, and pathogenesis of HIV.9; 10 The broad utility of Spt6 stems from its ability to perform multiple functions as a histone chaperone, a transcription elongation factor, and a modulator of RNA transcript processing.
Spt6 is required for reassembly of nucleosomes in the wake of an elongating RNA Polymerase II (RNAPII), a function that has profound regulatory effects at both intergenic and intragenic start sites.11; 12 Spt6 binds directly to histones and nucleosomes in vitro,13; 14 and these activities may contribute to the nucleosome reassembly function. In addition, Spt6 recruits the H3K36 methyltransferase Set2 to the transcription complex15, providing a link between the processes of transcription and histone modification. While its roles in modifying and reassembling nucleosomes indirectly influence the elongation rate, Spt6 also directly affects RNAPII as it stimulates elongation on nucleosome-free DNA templates in vitro.9; 16 This role as an elongation factor independent of its effects on chromatin may also be significant in vivo as knocking down Spt6 caused a decrease in the RNAPII elongation rate even in regions where the chromatin was considered to be permissive to transcription.16 Yet another role as a modulator of transcript processing is indicated by the association of Spt6 with the Rrp6 subunit of the Drosophila exosome RNA processing complex17, and by the requirement for Spt6 to prevent premature 3’ processing at cryptic polyadenylation signals upstream of the appropriate sites.18 It has also been demonstrated that mammalian Spt6 can bind RNAPII CTD phosphorylated at Ser2 by the P-TEFb kinase and that this interaction can subsequently promote recruitment of RNA processing/export factors such as REF1/Aly.9; 15 Binding to the phosphorylated RNAPII CTD is mediated by a Src homology 2 (SH2) domain that is located near the C-terminus of Spt6 and is conserved from yeast to human.9 SH2 domains typically recognize phosphorylated tyrosine residues, are ubiquitous in metazoans, and are the primary recognition motif in phosphorylation-mediated signal transduction cascades.19 Strikingly, the Spt6 SH2 domain is the only SH2 domain predicted to occur in the yeast proteome.20 Spt6 therefore participates in a wide range of functions affecting transcription, with each activity requiring different subsets of its multiple distinct functional domains.
We have determined crystal structures of Spt6 from Saccharomyces cerevisiae and find that, consistent with the range of functional domains inferred from previous studies, it comprises a series of structural domains whose homologs are known to function in nucleic acid binding and/or protein-protein interactions. The core of the structure comprises several recognizable structural motifs and in composite resembles the bacterial transcription factor Tex21. A C-terminal Region that is tethered to the core by a flexible linker adopts a novel tandem SH2 domain comprising two closely associated SH2 folds, one of which corresponds to the previously predicted SH2 domain of Spt6 and contains many of the standard binding determinants characteristic of this family, while the other lacks these features but contributes to a putative specificity pocket of the more canonical SH2 domain. Our structure of the Spt6 tandem SH2 domain resembles two recently reported homologous Spt6 structures.22; 23 We also show that the Spt6 core domain has DNA-binding activity, and we examine the interaction between the Spt6 tandem SH2 domain and RNAPII-derived peptides for evidence of a phosphorylation-dependent interaction with the CTD.
Results and Discussion
Crystal structures of Spt6(236–1259), Spt6(239–1451), and Spt6(1247–1451)
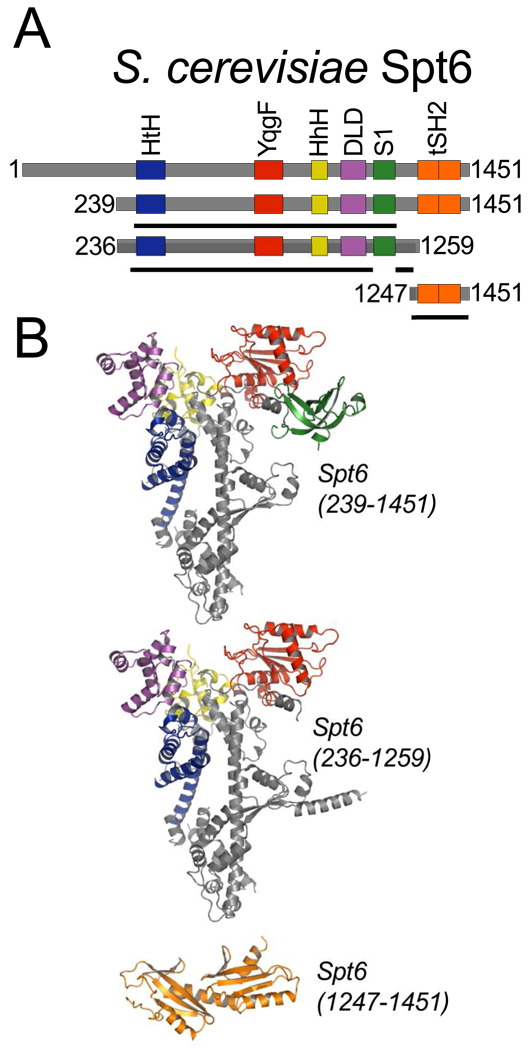
We have determined three crystal structures that together comprise the entire ordered region of the 1451-residue S.cerevisiae Spt6 protein (Figures 1, 2, and 3). Based on these structures and sequence analysis, Spt6 residues 1–297, 456–464, 485–500, 562–566, 1003–1008, 1211–1217, and 1441–1451 are likely to be disordered in the full-length protein, at least in the absence of binding partners. Spt6 displays multiple recognizable structural domains whose homologs have been implicated in binding of nucleic acids or proteins, although conservation at the sequence level is low and only three of these domains ((HhH)2, YqgF, S1) in the Spt6 core were predicted from the sequence24.
Figure 1. Spt6 structures.
(A) Schematic representation of full-length (top) Spt6 and constructs used for crystallization. The three crystallized constructs (239–1451, 236–1259, and 1247–1451) are shown below, with solid black lines indicating regions of the protein constructs visible in each of the crystal structures.
(B) Three independently determined Spt6 crystal structures colored by domain as in (A).
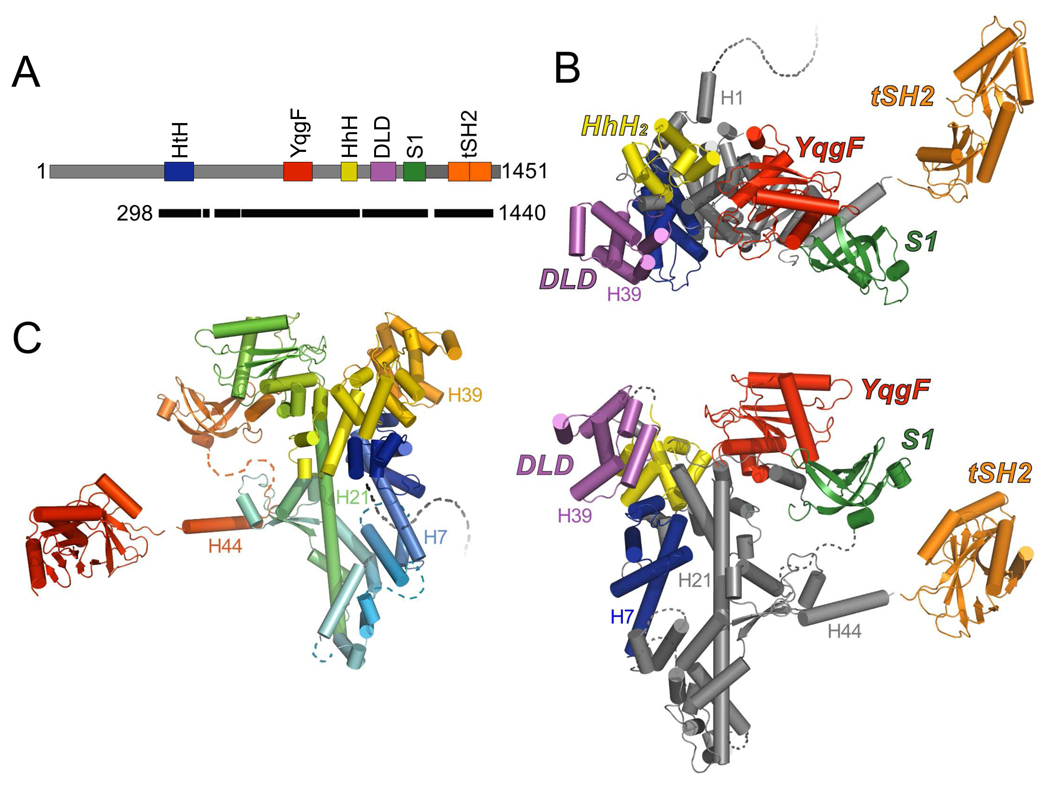
Figure 2. Composite model of Spt6.
(A) Schematic model of the Spt6 protein as in Figure 1A. Black bar indicates segments of the protein represented by the composite model.
(B) Two views of the composite model of Spt6 colored by domain. The C-terminal ~200 residues (tSH2 domain) are expected to be highly mobile with respect to the core. Secondary structure elements mentioned in the text are labeled. Dashed lines represent regions of the Spt6 protein that are not visible in our structures and are likely to be disordered, including the first 297 residues.
(C) Composite model of Spt6 colored from N to C terminus (blue to red). View orientation is rotated 180° from that of the lower image in panel B.
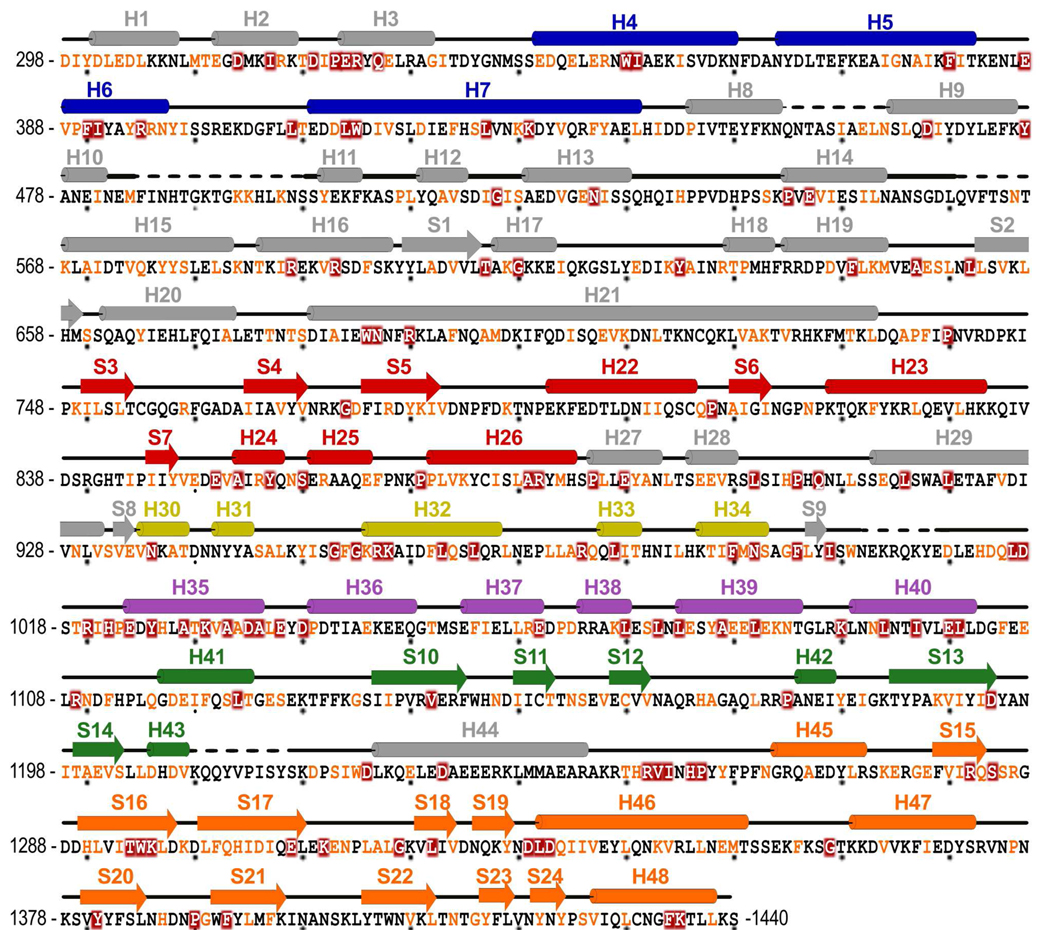
Figure 3. Spt6 Sequence.
Spt6 sequence present in the composite model with corresponding secondary structure elements colored by domain as for Figures 1 and 2. Coloring of sequence represents degree of conservation, dark red background (invariant), orange font (conserved), in an alignment (see Supplemental Figure S1) of proteins from Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, and Homo sapiens. Alignment performed using T-Coffee.65 Dashed lines indicate regions of disorder that are not included in the model(s).
Full length Spt6 expressed poorly in E. coli and did not yield crystals. In contrast, two different Spt6 constructs that lacked the first ~235 residues and either lacked the C-terminal 192 residues, Spt6(236–1259), or extended to the C-terminus, Spt6(239–1451), expressed well and the resulting proteins crystallized in different space groups (Table 1). The Spt6(236–1259) structure was determined by two-wavelength anomalous diffraction using data collected to 2.7 Å from crystals of selenomethionine-substituted (SeMet) protein, and was refined against 2.6 Å native data to R/Rfree values of 22.4/26.5%. The Spt6(239–1451) structure was determined by molecular replacement using the Spt6(236–1259) structure as a search model and refined against 3.3 Å data to R/Rfree values of 26.5/30.8%.
Table 1.
Crystallization Conditions
| Crystal | Spt6(236–1259) SeMet |
Spt6(236–1259) Native |
Spt6(239–1451) Native |
Spt6(1247–1451) SeMet |
Spt6(1247–1451) Native |
|---|---|---|---|---|---|
| Protein solutiona | 15mM Tris 5% Glycerol | 15mM Tris 5% Glycerol | 50mM Tris 10% Glycerol 5 mM BME | 15mM Tris 5% Glycerol | 15mM Tris 5% Glycerol |
| Protein conc. | 9 mg/ml | 8 mg/ml | 10 mg/ml | 26 mg/ml | 26 mg/ml |
| Reservoir solution | 50mM MES pH6.0 13% MPD 0.1 M KCl 5 mM MgCl2 | 50mM Mg(HCO2)2 10% Glycerol | 0.1M Na-MES pH6.5 13% PEG 4000 0.2 M MgCl2 12% Glycerol | 22% PEG 4000 0.3M (NH4)2SO4 | 0.1M Tris pH5.5 25% PEG 3350 0.2M (NH4)2SO4 |
| Drop ratio (µl: µl) Protein:reservoir | 2:2 | 3:1 | 3:1.5 | 2: 2 | 1:2 |
| Additive | - | 0.6 µl 1.0 M NDSB-256 | - | - | - |
| Temperature | 13°C | 13°C | 13°C | 21°C | 21°C |
| Cryo solutionb | 30% MPD | 35% Glycerol | 30% Glycerol | 30% Glycerol | 20% Glycerol |
All protein solutions were in 100 mM NaCl and Tris pH 7.5 (defined at room temperature).
Cryorprotectant solutions were made the same as for the reservoir solution, but including glycerol.
Together, Spt6(236–1259) and Spt6(239–1451) display ordered structure for the Spt6 core (residues 298–1248), which is primarily helical (54.6% Helical, 8.3% Strand, 37.1% Coil), has overall dimensions of 110 Å × 77 Å × 36 Å, and is quite similar between the two structures (RMSD = 1.2 Å over 747 out of 763 pairs of Cα atoms that are ordered in both structures). The core is built around an ~80 Å long central helix (H21, 680–733), with the rest of the protein chain wrapping around this helix at both ends to give an overall V shape. Additionally, Spt6(239–1451) displayed interpretable density for the S1 domain (residues 1128–1210) whereas the corresponding density in Spt6(236–1259) was poorly defined and the S1 domain was not included in this refined structure. As discussed below, we favor a model in which the S1 domain is highly mobile in solution. Interestingly, the C-terminal 240 residues of Spt6(239–1451), encompassing all of the residues absent from the shorter Spt6(236–1259) construct, are not visible in the electron density. The Spt6(239–1451) crystals have an estimated solvent content of 60% with cavities that could accommodate the C-terminal 240 residues, which suggests that this region is tethered by a flexible linker that is mobile both in solution and in the Spt6(239–1451) crystals.
Guided by secondary structure predictions and limited proteolysis experiments (data not shown), we expressed and crystallized the Spt6 C-terminal region, Spt6(1247–1451). This structure was determined by the selenomethionine SAD method using 2.7 Å resolution data and refined to R/Rfree values of 20.7/25.4%. A second native crystal form yielded 2.1 Å resolution data and this model was refined to R/Rfree values of 17.9/21.2%. The SeMet and native proteins crystallized in different space groups with one and four molecules in the asymmetric unit, respectively. Surprisingly, we found that the Spt6 C-terminal region comprises not one SH2 domain as anticipated20; 25, but two SH2 folds that are packed tightly against each other to form a tandem SH2 domain comprised of N-terminal (nSH2, residues 1250–1353) and C-terminal (cSH2, residues 1354–1440) folds. While this manuscript was in the final stages of preparation, equivalent tSH2 structures that overlap closely with our refined model were reported for Spt6 homologs from Candida glabrata22 (RMSD 1.0Å over 184 Cα atoms with 87% sequence identity) and Antonospora locustae23 (RMSD 1.6Å over 164 Cα atoms with 24% sequence identity).
N-terminal region
The first ~300 amino acids of Spt6 are extremely acidic, with an overall charge of −62 and predicted pI of 4.3, and are also predicted to be disordered.26 This is consistent with our Spt6(236–1259) and Spt6(239–1451) structures, which lacked discernible density prior to residue 298. Despite the lack of inherent order, this region of Spt6 is functionally important. Residues 239–268 bind the essential Spn1/Iws1 protein and overlap with residues required for nucleosome binding14. Furthermore, the spt6-1002 allele, a deletion of residues 2–122, displays synthetic lethality with deletion of the gene encoding the transcription factor Paf1.18
Helix-turn-helix domain
Residues 336–442 resemble a DNA-binding Helix-turn-Helix (HtH) motif, as seen in transcription factors such as c-Myb and the bacterial sigma factors.27 Nevertheless, the structure is not consistent with binding DNA in a canonical manner; binding of Spt6 H7 in the major groove of DNA in the manner predicted for a canonical HtH domain would cause steric clashes between bound DNA and the rest of the structure. It is possible that Spt6 undergoes conformational changes upon binding DNA, or that the Spt6 HtH domain serves as a protein-protein interaction motif, as occurs with members of the PWI subgroup of HtH domains.28 The Spt6 HtH overlaps with the U4/U6 Ribonucleoprotein Prp3 PWI (RMSD 2.7 Å, 69 Cα, pdbcode 1×4q) and the Nab2 PWI domains (RMSD 2.9 Å, 73 Cα, pdbcodes 2v75 and 3lcn), conserves the eponymous PWI motif as NWI (Asn349-Trp350-Ile351), and could utilize the equivalent protein-binding surface without invoking a conformational change in Spt6.
YqgF homologous domain
The Spt6 YqgF domain (residues 735–887) resembles members of the YqgFc superfamily, such as the E. coli protein YqgF and the RuvC class of Holliday junction resolvases.24 The alignment is especially close with E. coli RuvC (RMSD 2.9 Å, 117 Cα, pdb code 1hjr). Despite this similarity, the putative Spt6 YqgF catalytic site lacks carboxylate side chains that are critical for coordinating magnesium ions that mediate phosphodiester bond hydrolytic cleavage29. Thus, it does not appear that the Spt6 YqgF fold is capable of nuclease activity using a catalytic mechanism similar to RuvC or related RNAseH-fold nucleases.
Helix-hairpin-helix domain
Residues 933–1002 form two consecutive Helix-hairpin-Helix (HhH) motifs that pack together through highly conserved hydrophobic residues at an ~90° angle to form a (HhH)2 domain that resembles known dsDNA binding domains.30 These include proteins such as E. coli RNA polymerase α CTD (RMSD 2.3 Å, 56 Cα, pdb code 1lb2) and the Holliday junction-binding protein RuvA (RMSD 2.2 Å, 60 Cα, pdb code 1bvs). The first Spt6 HhH represents a characteristic HhH motif in the relative angle of the antiparallel helices and the presence of the Gly-hydrophobic-Gly motif within the hairpin loop.30 The second HhH motif is more variant, as also observed in other (HhH)2 domains, including DNA Polymerase β and 5’ to 3’ exonucleases. Though (HhH)2 folds are primarily found in proteins that interact with DNA, they also occur in proteins that mediate protein-protein interactions, such as the sterile α motif (SAM) proteins.31 Notably, the first 28 ordered residues (~298–325) of Spt6 wrap around the (HhH)2 domain in a fashion that would occlude binding of a canonical (HhH)2 domain to a dsDNA ligand, although this interaction could be transient. The absence of corresponding density for H1 and part of H2 in the initial SeMet Spt6(236–1259) and Spt6(239–1451) maps suggests that these N-terminal residues might adopt a different conformation to allow binding of a physiological partner in vivo.
Death-like-domain
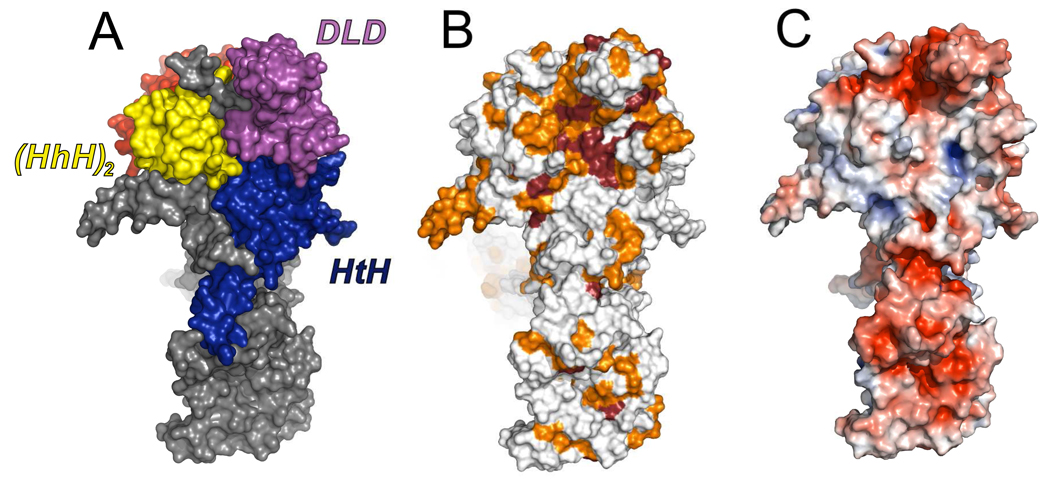
Residues 1019–1104 form a prominent lobe of the structure that resembles members of the Death-domain superfamily. Death domains typically serve as recognition modules in proteins that assemble and activate inflammatory and apoptotic complexes.32 The Spt6 Death-like-Domain (DLD) maintains the characteristic overall topology of death domains, consisting of a six helix bundle with three stacked antiparallel helices, but with an additional helix inserted between the final two helices of the bundle (H39 in Figures 2 and 3). Spt6 aligns reasonably well with several known death domain superfamily proteins, including the caspase-2-activating PIDDosome PIDD protein subunit component (RMSD 3.0 Å, 60 cα, pdb code 2of5). Although it is unlikely that the Spt6 DLD functions in an apoptotic process in yeast, its prominent location and the observation that it displays the most highly conserved region of the Spt6 surface, suggests that it mediates important intermolecular interactions (Figure 4).
Figure 4. The most conserved surface of the Spt6 core.
(A) View of the Spt6 core showing the interface between HtH, DLD, and (HhH)2 domains.
(B) Same orientation as panel (A) but colored by conservation to illustrate the high level of surface conservation at the intersection of these domains, especially on the DLD. Coloring represents degree of conservation as described in Figures 3 and S1.
(C) Same as panel B, but colored by electrostatic potential (−5 to +5 kT/e).
S1 domain
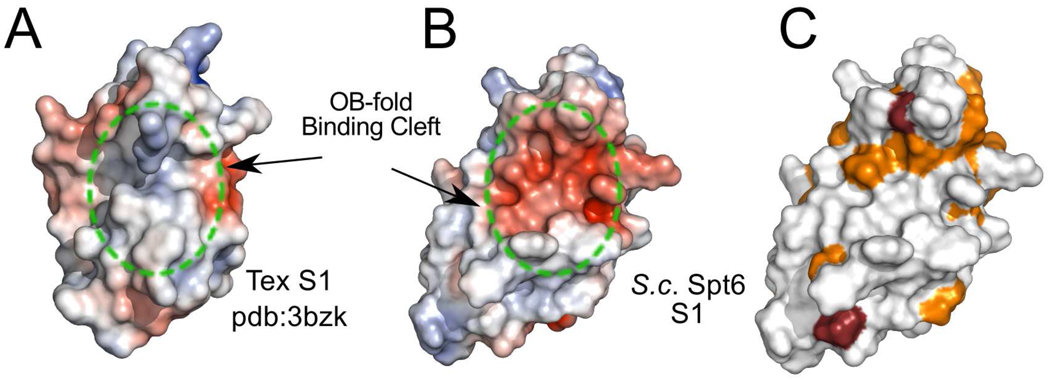
A mostly unstructured linker of 15 residues leads to the S1 domain (residues 1129–1219), which adopts the canonical S1/OB-fold of a β-barrel composed of two three-stranded β-sheets where strand 1 (S10) is shared by both sheets.33 Despite the structural similarity, Spt6 lacks the typical S1 binding cleft residues that are important for binding nucleic acids33. In addition, the predicted electrostatic potential surface does not appear conducive to nucleic acid binding, shows a low level of conservation and, as discussed below, is not required for dsDNA binding (Figure 5). This is in contrast to the distantly related bacterial Tex protein, which loses its capacity to bind DNA or RNA in the absence of the S1 domain21. OB folds are used to bind partners other than nucleic acids, including oligosaccharides and proteins33; 34, so it remains possible that the Spt6 S1 domain is used for an important interaction that does not involve nucleic acids.
Figure 5. Surface representations of the Spt6 and Tex S1 domains.
(A) Electrostatic surface representation (−5 to +5 kT/e) of the S1 domain from P. aeruginosa Tex (pdbcode 3bzk) with the position of the nucleic acid binding OB-fold cleft approximated by the circle with the dotted green line.
(B) Electrostatic surface representation (−5 to +5 kT/e) of the S. cerevisiae Spt6 S1 domain in the same orientation as in panel A showing a clustering of negative charge in the putative OB-fold binding cleft.
(C) S. cerevisiae Spt6 S1 domain in the same orientation as B but colored by conservation in the same color scheme as Figure 3 to illustrate the low level of conservation within the region equivalent to the binding cleft of canonical S1 domains.
Tandem SH2 domain
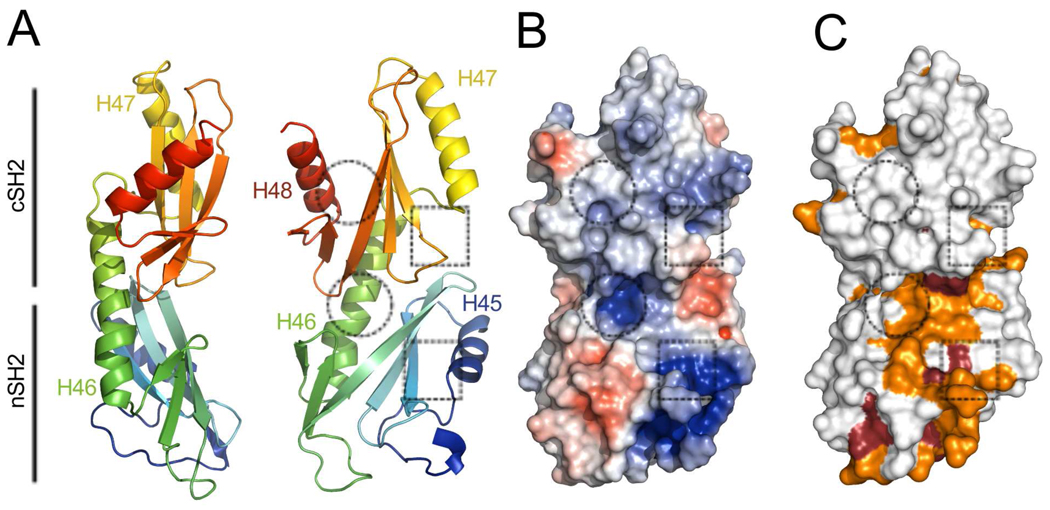
The S1 domain is followed by an unstructured ~10 residue segment and a ~30 Å helix (H44; 1227–1247) that buries ~440 Å2 of accessible surface area against the core in the Spt6(236–1259) structure (Figure 2). While some density is present for H44 in the Spt6(239–1451) maps, this region is too disordered for reliable model building, which may indicate that this interface is not always formed in solution. H44 links the core to a tandem SH2 domain (tSH2; residues 1250–1440)) that comprises N-terminal (nSH2; residues 1250–1353) and C-terminal (cSH2; 1353–1440) folds that associate through an ~800 Å2 interface to form a single structural unit (Figure 6). Both nSH2 and cSH2 conform to the standard SH2 domain fold (standard SH2 nomenclature in parenthesis) of an N-terminal helix (αA), a central three-stranded beta sheet (βB–βD), a small two-strand extension to the beta sheet (βE–βF), and a second helix (αB)35. Intervening loops are labeled based on their relative position between these elements (e.g. the BC loop connects the βB and βC strands). The Spt6 nSH2 and cSH2 superimpose well with each other (RMSD 2.1 Å, 69 Cα; Figure 7) and with the multitude of other characterized SH2 domains, such as those from v-Src kinase (nSH2: RMSD 2.0 Å, 82 Cα; cSH2: RMSD 2.1 Å, 80 Cα, pdb code 1sps; Figure 7) and Nck2 (nSH2: RMSD 2.1 Å, 83 Cα; cSH2: RMSD 1.9 Å, 73 Cα, pdb code 2cia).
Figure 6. The S. cerevisiae Spt6 tSH2 domain.
(A) Two views of a cartoon representation of the tSH2 domain colored from N to C terminus (blue to red; residues 1247–1440 shown). Dotted squares and circles show approximate positions of the canonical SH2 domain pTyr and specificity pockets, respectively.
(B) Surface representation colored by electrostatic potential surface (−5 to +5 kT/e).
(C) Same as panel B but colored by residue conservation in the same color scheme as Figures 3 and 4.
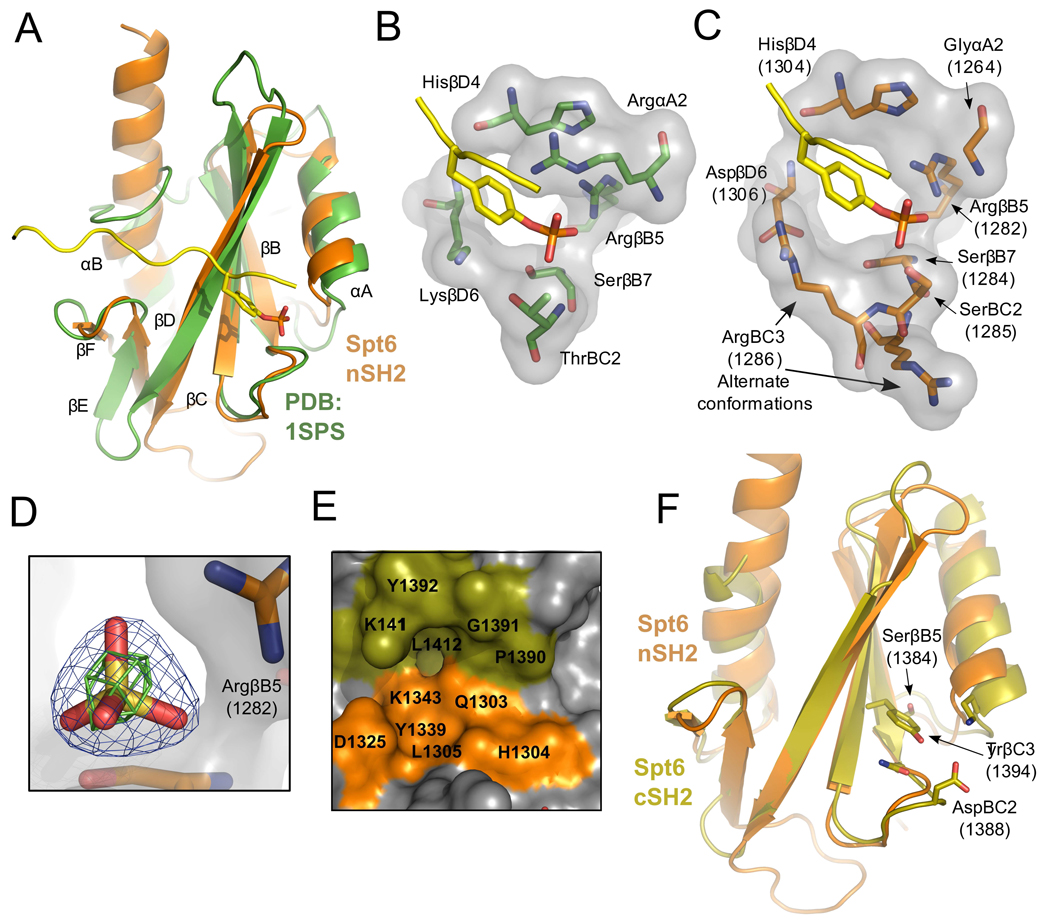
Figure 7. tSH2 binding pockets.
(A) Overlay of Spt6 nSH2 (orange) and the v-Src Kinase SH2 domain (green; pdb:1sps) bound to a pTyr ligand (yellow). Secondary structure elements labeled based using the standard SH2 nomenclature.35
(B) Detailed view of the pTyr binding pocket of 1sps. Residues contributing to coordination of the pTyr ligand are shown.
(C) Same as panel B but for the Spt6 nSH2, with the pTyr peptide from 1sps positioned after overlap on the SH2 protein domains.
(D) Electron density for a sulfate bound in the tSH2 crystal structures. Blue density represents the 2mFo-DFc map contoured at 2.0 sigma and green density an anomalous difference Fourier map contoured at 3.0 sigma.
(E) The putative nSH2 specificity pocket. Residues from both the nSH2 (orange) and cSH2 (olive) line the nSH2 specificity pocket.
(F) Alignment of the Spt6 nSH2 (orange) and cSH2 (olive) folds. Residues that protrude into the typical location of the pTyr binding pocket of the cSH2 fold are shown.
The relative orientation of the two SH2 folds that comprise the Spt6 tSH2 domain is unlike previously reported tandem SH2 domains from other proteins. The αB helix of nSH2 undergoes a ~20° kink where the αB helix would end in a canonical SH2 domain and extends along the backside of the cSH2 fold to form an extensive hydrophobic packing interface with the cSH2 central β-sheet. The relative orientation of nSH2 and cSH2 folds therefore appears to be constrained, consistent with the observation that superposition of the five crystallographically independent tSH2 domains observed in our various crystal forms (one in SeMet Spt6(1247–1451), four in Spt6(1247–1451)) indicates a maximum relative rotation of ~8° between nSH2 and cSH2 folds.
Implications for ligand binding by the tSH2 domain
The extent to which Spt6 nSH2 corresponds to a prototypic SH2 domain is seen in a comparison with v-Src (pdbcode 1sps) (Figure 7A–C). The primary determinants of phosphate binding are preserved in nSH2 whereas the positively charged Arg/Lys side chains of classical SH2 domains that flank the aromatic ring of phosphotyrosine ligands are absent from their usual positions. Conserved residues include the consensus FLVRES (FVIRQS, 1279–1284 in Spt6) sequence motif that contributes the phosphate coordinating Arg1282 and Ser1284 side chains (Spt6 numbering). Moreover, the following Ser1285 Spt6 side chain is also well positioned to hydrogen bond the phosphate and functionally substitute the ThrBC2 of the Src SH2 domain. Other positions within SH2 domains that are important for binding phosphate include HisβD4 (Spt6 H1304) and Ser/ThrβC4 (Spt6 Thr1294), whose side chains hydrogen bond the Arg1282 side chain in an optimum orientation for phosphate binding and, in cognate SH2-ligand complexes, also form a main chain to main chain hydrogen bond with the ligand residue following the pTyr.
Classical SH2 domains typically have a basic residue at the αA2 position that binds against one side of the tyrosine ligand aromatic ring where it forms an amino-aromatic interaction with the π ring and also hydrogen bonds with both the phosphate and the tyrosine main chain carbonyl. In contrast, the Spt6 nSH2 has a glycine at this position, G1264, which can make none of the same ligand interactions. This does not argue strongly against binding of phosphotyrosine by Spt6, however, because a number of other SH2 domains that bind phosphotyrosine ligands have a variety of substitutions at this position, including the PTPN11/SHPTP2/Syp phosphatase (pdb code 1ayc) that, like Spt6, has a Gly at αA2, and is known to bind a phosphotyrosine-containing peptide.36 On the other side of a canonical phosphotyrosine ligand side chain, classical SH2 domains typically have a basic residue in the βD6 position. This residue is a lysine in Src but is an aspartate (D1306) in Spt6. Interestingly, in three of the five crystallographically independent Spt6 tSH2 molecules in our structures, the space typically occupied by the basic βD6 side chain is filled by R1286 in the BC3 position (Figure 7C). Thus, Spt6 retains the ability to provide a positively charged basic group in this position, consistent with the potential to bind phosphotyrosine.
Ammonium sulfate was present in the crystallization solutions for both native and SeMet tSH2 domain structures, and all five crystallographically independent molecules displayed a sulfate ion at the putative nSH2 phosphate binding site, where it forms hydrogen bonds with R1282, S1284, and S1285 in the same manner as the phosphate of pTyr-SH2 complexes (Figure 7D). Assignment of the density as sulfate was confirmed in anomalous difference Fourier maps for the native data, which showed peaks that were similar in size to those of cysteine and methionine sulfur atoms for some of the sulfates. This further suggests that nSH2 binds a phosphorylated ligand and that it might accommodate a phosphotyrosine side chain in a suboptimal binding pocket.
Typical SH2 domain ligands bind through a two-prong mechanism that, in addition to binding pTyr, also involves binding of the side chain three residues C-terminal to the pTyr into the “specificity pocket”.35 Binding partner preference is typically defined in the specificity pocket by the BG and EF loops and the βD3 and βD5 residues, which usually favors binding of hydrophobic residues. In contrast, the Spt6 nSH2 fold predominantly displays charged and polar residues in this site, and there is no BG loop due to the extension of nSH2 αB to the cSH2 fold. Instead, cSH2 residues such as the DE loop and a βD side-chain (K1411) protrude into the pocket forming the top portion of the nSH2 specificity pocket (Figure 7E). This indicates that if the nSH2 fold binds substrate in the typical “two-pronged” manner common to SH2 domains, the cSH2 fold would make significant contributions to binding.
In contrast to nSH2, the cSH2 fold appears to be cryptic and unlikely to bind a phosphorylated ligand because residues critical for phosphate binding are substituted to display a very different chemical environment, and the Y1394 side chain fills the space where a phosphate would typically bind (Figure 7F). Moreover, the region of the specificity pocket lacks even a shallow depression as it is filled by bulky, aromatic side-chains (F1397, Y1406, W1408, and F1434). Thus, in contrast to nSH2, and consistent with the lack of sequence conservation at cSH2 (Figure 6C), it seems unlikely that cSH2 binds ligands in a manner reminiscent of SH2 domains.
Binding of Spt6 tSH2 domain with phosphorylated peptides
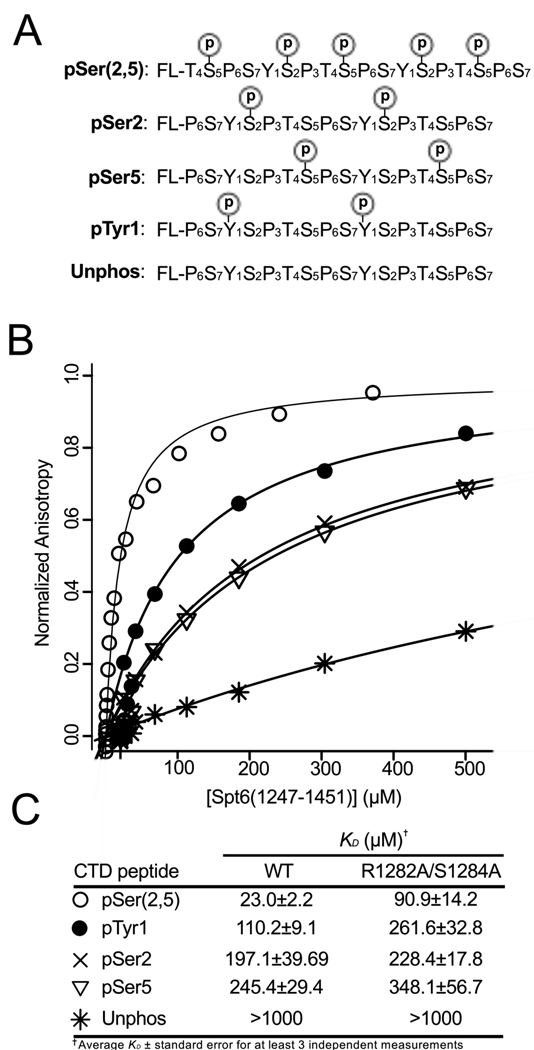
The human Spt6 C-terminal domain has been reported to bind the heptad repeat sequences of the mammalian RNAPII large subunit when the RNAPII CTD is treated with P-TEFb, a kinase that phosphorylates Ser2 during transcriptional elongation9. In order to investigate this interaction more quantitatively, we used Fluorescence Anisotropy (FA) to measure binding of S. cerevisiae Spt6(1247–1451) to di-heptad repeat peptides representing various phosphoisoforms of the RNAPII CTD (Figure 8). Peptides tested include sequences representing pSer2, pSer5, pTyr1, and pSer(2,5). pSer5 and pSer(2,5) peptides were included to test specificity and because these modifications also occur on the RNAPII heptad repeats. A pTyr1 peptide was included because SH2 domains typically bind phosphotyrosine peptides and this modification occurs in mammals37, although it has not been reported to occur in yeast.
Figure 8. Spt6 tSH2 binds RNAPII-CTD phosphopeptides.
(A) Peptides used in binding studies with positions of phosphoserine or phosphotyrosine residues indicated.
(B) Representative fluorescence anisotropy binding isotherms for Spt6(1247–1451) binding to various peptides with symbols defined in (C).
(C) Binding affinities for WT and R1282A/S1284A Spt6(1247–1451) proteins based on fluorescence anisotropy experiments.
All of the peptides assayed bound with affinities in the range of ~20–250 µM, which is similar to the affinity of other RNAPII CTD interactions with isolated binding domains38, but is ~10–100-fold weaker than is typically found for the interaction of SH2 domains with phosphotyrosine ligands39. Of the ligands we assayed, the pSer(2,5) peptide bound Spt6(1247–1451) with the highest affinity (23 µM). This may indicate that RNAPII CTD sequences phosphorylated on both Ser2 and Ser5 are the authentic in vivo ligands for the yeast Spt6 tSH2 domain, which would be consistent with the reports that Ser2 phosphorylated sequences are preferred9 and that localization of Ser2 and Ser2/5 phosphorylation overlaps substantially in average transcription units40; 41. On the other hand, interpretation is complicated by the fact that our pSer2/5 peptide has considerably more negative charge than the other peptides assayed and so may be more prone to non-specific effects. Interestingly, the pTyr1 peptide binds with a KD of 110 µM, which is tighter than that of the pSer2 (197 µM) and pSer5 (245 µM) peptides bearing an equivalent number of phosphate groups. A higher affinity for pTyr1 peptide with the same overall charge as pSer2 or pSer5 peptides is consistent with the similarity between nSH2 and well-characterized pTyr binding SH2 domains, but the physiological relevance of this result is not clear given the lack of observed pTyr modification of the RNAPII CTD in yeast.
The conclusion that the Spt6 tSH2 domain interacts specifically with phospho-CTD peptides is reinforced by our observations that an unphosphorylated form of the RNAPII-CTD showed negligible (Kd >1,000 µM) binding and that similar results were obtained when the assays were performed in a phosphate-buffered saline condition (data not shown). As a further test of specific interactions, we measured binding to a mutant form of Spt6(1247–1451) in which residues R1282 and S1284, which are important for phosphate binding in typical SH2 domains, were both substituted with alanine. Binding to the pSer(2,5) peptide was decreased by ~4 fold while binding to the pTyr1, pSer2, and pSer5 peptides was decreased by 2.4, 1.2, and 1.4 fold, respectively. These modest effects are consistent with the putative nSH2 phosphate binding site contributing to the binding interaction, but not performing a dominant role for interaction with the peptides assayed.
Deletion of the entire tSH2 region by truncation of the SPT6 gene leads to defects in growth attributed to suboptimal transcription elongation.22; 23 To examine the importance of tSH2 residues implicated in phosphotyrosine binding in vivo more carefully, we mutated the single genomic copy of SPT6 to produce proteins with R1282H, S1284D, R1286A, Q1303E, EN1313/1314AA, or K1343E mutations (nSH2 domain), or P1390A or K1411E mutations (cSH2 domain). The effects of these mutations were quite mild, failing to recapitulate the severe defects caused by truncation of the gene (data not shown; see Materials an Methods for a list of phenotypes screened). The C-terminal region of Spt6 therefore appears to have some activity that is not interrupted when residues within the tSH2 domain expected to be important for binding phosphorylated substrates are mutated.
Our data are consistent with a recent report that concluded that pSer2 RNAPII CTD peptides bound the tSH2 domain of the Candida glabrata Spt6 homolog with 10 µM affinity22. The tighter affinity observed in that study could reflect differences between the proteins, but is more likely to be due to the very low (10 mM) concentration of NaCl used in the binding assays. Our data also extend the earlier work by showing that the S. cerevisiae Spt6 tSH2 domain displays little discrimination for binding CTD peptides with different modifications, but does show a small preference for a peptide with a single phosphorylated tyrosine (pTyr1). Our findings suggest that Spt6 activities in vivo may be modulated by phosphorylation of binding partners, and that the ligands of the tSH2 domain may include phosphorylated tyrosine residues.
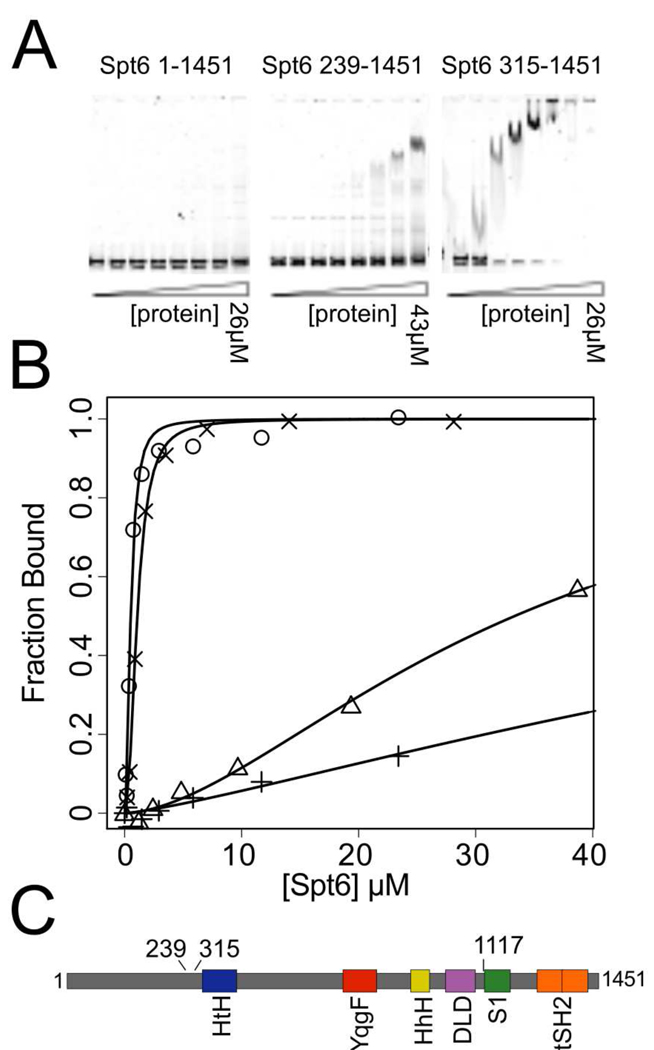
Binding to dsDNA
To test whether Spt6 is capable of binding dsDNA, we performed electrophoretic mobility gel shift assays (EMSA) using a 177 bp dsDNA fragment. We tested several different Spt6 constructs and found that binding was tighter (KD ± standard deviation) for Spt6(315–1451) (KD 0.53 ± 0.07 µM) than for Spt6(239–1451) (KD 33.7 ± 3.8 µM) or Spt6(1–1451) (KD 106.7 ± 38.9 µM) (Figure 9), demonstrating that the disordered and negatively charged N-terminal residues diminish dsDNA binding. Unlike Tex, which requires the S1 domain for nucleic acid binding21, an Spt6 construct lacking the S1 domain, Spt6(315–1117), retains the ability to bind dsDNA with a KD of 1.08 ± 0.06 µM (Figure 9B).
Figure 9. dsDNA binding studies.
(A) Representative gel shift assays for three different constructs of Spt6.
(B) Representative binding isotherms used to calculate dissociation constants for various Spt6 constructs binding to the 177bp Widom 601 dsDNA62. Symbols used to indicate isotherms for different constructs: (+)=Spt6(1–1451) KD= 106.7 ± 38.9 µM, (△)=Spt6(239–1451) KD= 33.7 ± 3.8 µM, (×)=Spt6(315–1117) KD= 1.08 ± 0.06 µM, (○)=Spt6(315–1451) KD= 0.53 ± 0.07 µM.
(C) Schematic diagram indicating endpoints for constructs used in DNA binding studies.
Overall Structure and Functional Implications
Our composite model of Spt6 (Figure 10) features a core region (residues ~298–1117) that has multiple recognizable domains whose packing in the crystal likely reflects, to a large extent, their organization in solution, at least in the absence of binding partners. The N-terminal residues 1–297 display considerable overall negative charge and are expected to be highly mobile, while residues 239–263 also comprise the Spn1/Iws1-binding determinant and overlap with the nucleosome binding site14. This high degree of mobility may provide a flexible tether for bridging binding partners, such as Spn1/IwsI, RNAPII, and nucleosomes9, and the negative charge may modulate histone and DNA interactions.
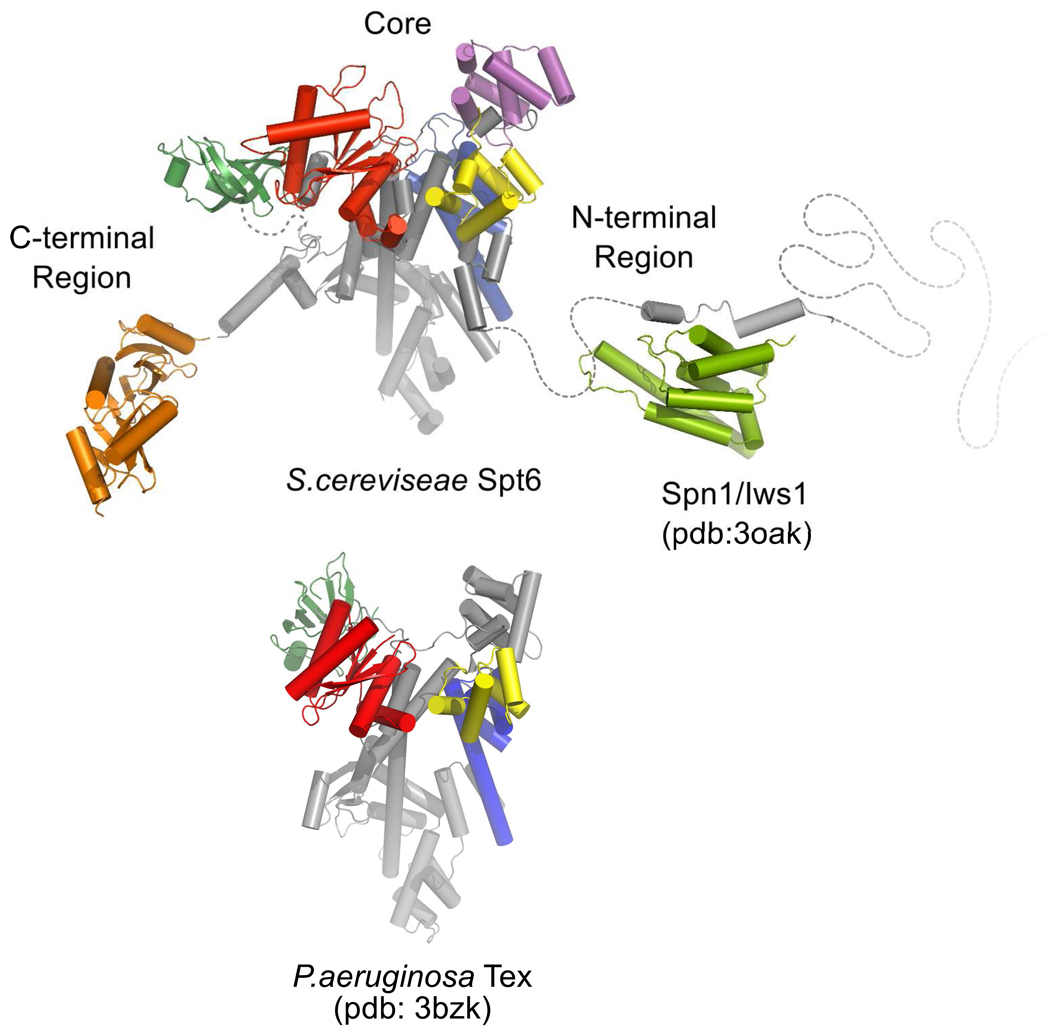
Figure 10. Comparison of Spt6 and Tex structures.
The overall structure of the Spt6 core resembles that of the prokaryotic Tex protein. This similarity implicates the Spt6 core in nucleosome-independent functions, such as transcriptional elongation on naked template DNA. The N-terminal region includes determinants essential for binding Spn1/Iws1 or nucleosomes, which appear to bind competitively with each other14. The negatively charged N-terminal region may also be important for modulating binding to nucleic acids. The C-terminal region has been implicated in binding RNAPII.9
Inherent flexibility is also a feature of the C-terminal S1 domain, H44, and tSH2 domain. The S1 domain is loosely associated with the core, lacks density in the Spt6(236–1259) structure, and may be visible in the Spt6(239–1451) structure only because of ordering by a crystal lattice contact. Whereas the core of the distantly related Tex protein clearly resembles that of Spt6, the Tex and Spt6 S1 domains are displaced by a rotation of ~80° and ~25–30 Å translation with respect to each other, and the S1 domain appears to be shifted by 14 Å in different crystal forms of Tex21. Therefore, it is likely that a highly mobile S1 domain is an important feature of both proteins, although one notable difference is that the Tex S1 binds nucleic acids whereas our DNA binding data and consideration of surface amino acid residues indicate that the Spt6 S1 domain does not. Although the low conservation of residues in the putative binding cleft of the S1 domain suggests that this region is unlikely to have a highly conserved binding partner, the electrostatic characteristics of this surface (Figure 5) are consistent with potential binding partners that are positively charged, such as histones.
H44 is only visible in the Spt6(236–1259) structure, where it also appears to be ordered by a lattice contact, and the C-terminal tSH2 domain is expected to be highly dynamic with respect to the rest of the protein. The leading model is that this domain binds RNAPII that is phosphorylated on Ser2 of its C-terminal domain9. Our binding data are consistent with this view, provided other determinants contribute to binding/specificity, but also indicate the possibility that other binding partners might be functionally important. For example, Spt5 (discussed below) has been shown to co-localize with Spt6 and contains a phosphorylated C-terminal repeat domain similar to the RNAPII CTD,42 which could be a physiological ligand for the Spt6 tSH2 domain.
Spt6 has been shown to be functionally associated with a large number of proteins involved in transcription elongation, chromatin maintenance, and RNA processing. The structure of Spt6 presented here will serve as a foundation for more precise mapping of protein binding partners. Along with protein binding partners, Spt6 is also expected to interact with nucleic acids in the transcription complex. DNA binding is probably important for nucleosome reassembly and potentially for transcription elongation. Association with nascent RNA transcripts could be important for enhancing the elongation rate or for organizing interactions with RNA modification/export factors such as REF/Aly. Future studies will be needed to see if Spt6 binds RNA and to further map nucleic acid binding to different domains of Spt6. For example, the YqgF or the (HhH)2 domain may mediate binding to specific DNA structures such as 4-way (Holliday) junction DNA, structures similar to those found at the DNA entry/exit points of nucleosomes.43 The tSH2 domain may also contribute to nucleic acid interactions as it is likely to bind negatively charged substrates containing phosphate groups. A simple electrostatic surface analysis indicates that each of these domains retains properties found in homologous domains with known functions. Further, examination of the histone binding activity of the various Spt6 domains will be of significant interest in furthering our understanding of nucleosome assembly/reassembly.
The structural similarity between Spt6 and the prokaryotic Tex protein is limited to the core and S1 domains. Consistent with the extent of structural similarity, N- and C-terminal regions that are unique to Spt6 are required for eukaryotic-specific interactions with nucleosomes11; 13; 14, hyperphosphorylated forms of the RNAPII CTD9, Spn1/IwsI14, and mRNA processing/export factors9. An attractive possibility is that the core region provides activities that are conserved among prokaryotes and eukaryotes. In this regard, it is striking that most of the Spt6 core domains belong to structural families whose members function in nucleic acid binding, an activity that is likely to be a key component of transcription factors such as Spt6 and Tex that are capable of stimulating elongation on nucleosome-free DNA templates9; 16. Consistent with this possibility, our data indicate that the Spt6 core can bind dsDNA. Curiously, some of the putative nucleic acid binding surfaces of Spt6 domains are occluded in the structure, although conformational changes might displace residues 298–320 (H1 and H2) to expose a DNA binding activity on the (HhH)2 domain. Consistent with this model, we find that truncation of the N-terminal ~314 residues leads to tighter dsDNA binding. One attractive model is that conformational changes of this nature are induced by binding partners such as histones or Spn1/Iws1. The relationship between Spt6 and Tex proposed here is reminiscent of that between another eukaryotic transcription elongation factor, Spt5, and its bacterial counterpart NusG. These proteins also display similar core domains while Spt5 has an acidic N-terminal extension and a C-terminal extension44 that confers eukaryote-specific functions such as binding to RNAPII44, interaction with mRNA capping enzymes45, and extensive phosphorylation by RNAPII CTD kinases such as P-TEFb46. Therefore, like Spt5, Spt6 is likely to have built on the fundamental transcription activities of its core to accommodate the additional complexities of eukaryotic gene regulation.
Materials and Methods
Protein Expression and Purification
The protein constructs were expressed from pET151-D/TOPO vectors (Invitrogen) in BL21 codon plus (RIL) E. coli cells (Stratagene). Cultures were grown in autoinduction media47 in baffled 1.8L flasks at 37°C with continuous shaking. After 4–8 hours, the cultures were shifted to 23°C and grown for an additional 16–24 hours. Harvested cells were stored at −80°C. Cells were thawed and lysed in buffer containing lysozyme and protease inhibitors, followed by sonication and centrifugation (25,000–30,000xg). The soluble fraction was applied to Nickel agarose resin (Qiagen) and eluted in buffer containing 300mM Imidazole and 100mM NaCl, immediately followed by application to a heparin column (5ml HiTrap Heparin, GE Healthsciences) and elution over a NaCl gradient. Fractions containing Spt6 were pooled and processed overnight at room temperature in buffer containing TEV protease. A Nickel agarose column was used to remove the tagged TEV protease and unprocessed Spt6 protein, and the flow-through was concentrated and loaded onto a size-exclusion column (Superdex 200, or S75 (for 1247–1451 constructs), GE Healthsciences). Selenomethionine-substituted protein was expressed using an autoinduction protocol47 for seleomethionine incorporation and purified by the same protocol as native protein. All crystals were grown in sitting drops, transferred briefly to a cryoprotection solution, suspended in a nylon loop, and plunged into liquid nitrogen (Table 1).
Crystal structure determinations and refinements
Data were processed with HKL200048. Spt6(236–1259) was determined by the MAD method. SOLVE49 was used to locate Selenium atoms and RESOLVE50 was used for density modification and preliminary model building. AutoSol in Phenix51 was used to determine the Spt6(1247–1451) structure by the SAD method. The Spt6(239–1451) structure was determined by molecular replacement using AutoMR in Phenix51 to a resolution of 3.3Å. A homology model built by TASSER52 of the Spt6 S1 domain was used as a guide for model building. The native Spt6(1247–1451) structure was determined by molecular replacement using PHASER53. Phenix51 and TLSMD54 were used for refinement, Coot55 was used for model building, and Molprobity56 was used for structure validation. The following residues were ordered and included in the refined models: Spt6(236–1259) 297–455, 464–484, 501–561, 567–1002, 1009–1128, and 1219–1248; Spt6(239–1451) 312–455, 464–489,509–552, 567–649, 653–1001, and 1014–1210; Spt6(1247–1451) 1247–1440. Structural alignments were performed using DALI57 and SSM58. PyMOL59 was used to create figures. Electrostatic surface representations were calculated using PDB2PQR and APBS tools60; 61 using the AMBER force field and colored from red (−5 kT/e) to blue (+5 kT/e).
DNA binding experiments
177 bp Widom 601 DNA with a 5’ Cy3 fluorophore was generated as described62 followed by precipitation, gel purification, and electroelution. Electrophoretic mobility shift binding experiments were performed and KD values were determined as described previously21. In short, 2-fold serial dilutions of the respective purified protein construct was mixed with nucleic acid substrate at room temperature in binding buffer [15 mM Tris–HCl, pH 7.5, 100 mM NaCl, 10% glycerol, 0.5 mM EDTA] where the final concentration of dsDNA was 10–20 fold below the estimated KD for the interaction. After incubation for 30 minutes, samples were run on 4–20% TBE native gels (Bio-Rad Laboratories) and imaged and quantified using a TYPHOON imaging system with ImageQuant software (GE Health Sciences). The fraction bound was calculated by quantifying the DNAtotal (total fluorescence in entire lane) and DNAfree. DNA of slower mobility than the DNAfree were considered bound. The fraction bound = 1−([DNA]free/[DNA]total). Dissociation constants (KD values) were calculated by plotting data points and curve fitting in the program R63 using the Hill formalism where fraction bound = 1/(1+(KDn/[P]n)). In all cases standard deviations are calculated from at least three measurements, except for the Spt6(315–1117) construct, which was repeated twice.
Fluorescence Anisotropy binding experiments
Peptides were synthesized by the University of Utah Core Facility or purchased commercially through AnaSpec Inc. (San Jose, CA) purified to >98% purity by HPLC, and confirmed by MALDI-TOF mass spectrometry. Purified Spt6(1247–1451) was titrated in 1.5–2.0 fold serial dilutions against a constant concentration of fluorescein labeled peptide (10–20 fold below estimated KD) in 20mM Tris-Cl pH 7.5, 100mM NaCl, and 5% Glycerol. Samples were incubated at room temperature for at least 15 minutes prior to reading. Parallel and perpendicular fluorescence intensity was measured in a multi-well format using a Tecan Infinite 200 microplate reader using excitation/emission wavelengths of 485/535nm. Anisotropy values were calculated, normalized, and plotted as a function of protein concentration. KD values were determined by fitting the data using the equation64 A = ((AT*([pro]/KD))/(1+[pro]/KD)) where A = measured anisotropy, AT = total change in anisotropy, and [pro] = protein concentration.
Genetic analysis of tSH2 mutations
The following alleles of SPT6 were screened for phenotypes in strains isogenic with the A364a genetic background: WT, spt6-R1282H, spt6-S1284D, spt6-R1286A, spt6-Q1303E, spt6-E1313A, N1314A, spt6-K1343E, spt6-P1390A, and spt6-K1411E. Mutations were introduced into the genomic copy of SPT6 such that expression was from the native promoter at the normal locus except for the introduction of a URA3 or TRP1 marker downstream of the ORF. Strains were tested for growth on rich medium at 30°C and 38°C; on medium lacking lysine at 30°C and 37°C (all strains had the lys2-128∂ allele, so growth would reveal an Spt- phenotype); and on media containing 150 mM hydroxyurea, 75 µg/ml 6-azauracil, 0.6 µg/ml 4-nitroquinolone, 10 mM caffeine, 3% formamide, 1.2 M NaCl, 45 µg/ml mycophenolic acid, or 6% ethanol (all at 30°C). None of the mutants were sensitive to any of the stress conditions relative to the WT strain. spt6-S1284D and spt6-Q1303E strains were somewhat more resistant to 3% formamide than the WT, and spt6-Q1303E strains displayed a very weak Spt- phenotype (faint growth after 7 days). Mutants were not tested for a defect in cryptic initiation.
Protein Data Bank Accession Numbers
Coordinates and structure factors for Spt6(236–1259), Spt6(239–1451), and SeMet-Spt6(1247–1451), and native Spt6(1247–1451) have been deposited in the PDB with accession numbers 3psf, 3psi, 3psj, and 3psk, respectively.
Supplementary Material
Table 2.
Data Collection and Refinement Statistics
| Se-Spt6 (236–1259) peak |
Se-Spt6 (236–1259) inflection |
Spt6 (236–1259) |
Spt6 (239–1451) |
Se-Spt6 (1247–1451) peak |
Spt6 (1247–1451) |
|
|---|---|---|---|---|---|---|
| Data Collection | ||||||
| Space group | P212121 | P212121 | P3121 | I4122 | P212121 | |
| Unit cell dimensions (Å) |
a = 114.0 b = 116.4 c = 122.7 |
a = 115.1 b = 116.2 c = 117.4 |
a = 118.7 b = 118.7 c = 214.4 |
a = 97.5 b = 97.5 c = 132.4 |
a = 78.8 b = 105.2 c = 119.5 |
|
| Molecules/ASU | 1 | 1 | 1 | 1 | 4 | |
| Solvent Content (%) | 63.7 | 62.4 | 60.0 | 63.5 | 53.1 | |
| Beamlinea | SSRL 11-1 | SSRL 11-1 | SSRL 7-1 | NSLS X29 | SSRL 9-1 | Home |
| Wavelength (Å) | 0.97886 | 0.97922 | 0.97773 | 1.10000 | 0.97908 | 1.54178 |
| Resolution (Å) | 50-2.7 | 50-2.7 | 32-2.6 | 46-3.3 | 35-2.7 | 30-2.1 |
| High resolution shell (Å) | (2.8-2.7) | (2.8-2.7) | (2.7-2.6) | (3.42-3.3) | (2.8-2.7) | (2.18-2.1) |
| No. unique reflections | 83,751 | 82,946 | 49,595 | 27,025 | 8,481 | 58,489 |
| No. total reflections | 261,886 | 257,334 | 434,557 | 182,336 | 649,419 | 258,498 |
| Mean I/σI | 22.1 (2.4) | 22.1 (2.1) | 29.8 (4.3) | 39.8 (3.3) | 25.1 (3.7) | 16.8 (3.2) |
| Completeness (%) | 95.8 (75.8) | 94.7 (70.1) | 99.8 (99.9) | 99.5 (99.9) | 92.2 (74.5) | 99.5 (99.2) |
| Rsym (%)b | 4.8 (35.7) | 4.7 (38.3) | 6.5 (43.9) | 5.8 (56.9) | 7.0 (29.6) | 6.6 (47.8) |
| Refinement | ||||||
| Rcrystc/Rfreed (%) | 22.4/26.5 | 26.5/30.8 | 20.7/25.4 | 17.9/21.2 | ||
| No. non-H atoms: Protein | 6788 | 6876 | 1575 | 6988 | ||
| Solvent | 48 | 0 | 19 | 491 | ||
| < B > (Å2) | 103.7 | 167.6 | 74.7 | 44.2 | ||
| RMSD Bond lengths (Å) | 0.006 | 0.013 | 0.008 | 0.004 | ||
| RMSD Bond angles (°) | 0.895 | 1.59 | 1.12 | 0.757 | ||
| Ramachandran outliers (%) | 0.0 | 0.8 | 0.0 | 0.40 | ||
| Ramachandran favored (%) | 95.6 | 93.6 | 97.9 | 97.5 | ||
| Rotamer outliers (%) | 0.94 | 0.3 | 0.6 | 1.4 | ||
Values in parentheses correspond to the high resolution shell.
Data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL), the National Synchrotron Light Source (NSLS), or on a Rigaku MicroMax-007HF rotating anode X-ray generator with a copper anode and VariMax confocal optics and a Rigaku R-AXIS IV image plate detector (home).
Rsym = (Σ|(I−<I>)|)/(Σ I), where <I> is the average intensity of multiple measurements.
Rcryst = (Σ|Fobs−Fcalc|)/(Σ |Fobs|).
Rfree is the Rcryst based on ~1000 (at least 10%) of the reflections that were excluded from refinement.
Acknowledgements
We thank Hua Xin and Charisse Kettelkamp for technical assistance, and Heidi Schubert for advice with the crystallographic analysis. Portions of this work were performed in Core Facilities at the University of Utah, which were supported by P30CA042014 from the National Cancer Institute. Some of the X-ray diffraction data for this study were measured at the National Synchrotron Light Source (NSLS). Financial support for NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. Portions of this research were performed at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health (NIH), National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences. S.J.J. was supported by a postdoctoral fellowship (GM074368). This work was supported by the NIH RO1 GM076242.
Abbreviations used
- BME
β-mercaptoethanol
- CTD
C-terminal Domain
- DLD
Death-like-Domain
- dsDNA
double-stranded DNA
- EMSA
electrophoretic mobility gel shift assay
- FA
Fluorescence Anisotropy
- HhH
helix-hairpin-helix
- HtH
helix–turn–helix
- NSLS
National Synchrotron Light Source
- OB
Oligonucleotide/oligosaccharide binding
- PDB
Protein Data Bank
- pSer
phosphoserine
- pTyr
phosphotyrosine
- RMSD
Root Mean Squared Deviation
- RNAPII
RNA polymerase II
- SAD/MAD
single/multiple-wavelength anomalous diffraction
- SH2
Src homology 2
- Spt6
Suppressor of Ty 6
- SSRL
Stanford Synchrotron Radiation Laboratory
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 2.Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- 3.Clark-Adams CD, Winston F. The SPT6 gene is essential for growth and is required for delta-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:679–686. doi: 10.1128/mcb.7.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kok FO, Oster E, Mentzer L, Hsieh JC, Henry CA, Sirotkin HI. The role of the SPT6 chromatin remodeling factor in zebrafish embryogenesis. Dev Biol. 2007;307:214–226. doi: 10.1016/j.ydbio.2007.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardehali MB, Yao J, Adelman K, Fuda NJ, Petesch SJ, Webb WW, Lis JT. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009;28:1067–1077. doi: 10.1038/emboj.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishiwaki K, Sano T, Miwa J. emb-5, a gene required for the correct timing of gut precursor cell division during gastrulation in Caenorhabditis elegans, encodes a protein similar to the yeast nuclear protein SPT6. Mol Gen Genet. 1993;239:313–322. doi: 10.1007/BF00276929. [DOI] [PubMed] [Google Scholar]
- 7.Shen X, Xi G, Radhakrishnan Y, Clemmons DR. Identification of novel SHPS-1-associated proteins and their roles in regulation of insulin-like growth factor-dependent responses in vascular smooth muscle cells. Mol Cell Proteomics. 2009;8:1539–1551. doi: 10.1074/mcp.M800543-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baniahmad C, Nawaz Z, Baniahmad A, Gleeson MA, Tsai MJ, O'Malley BW. Enhancement of human estrogen receptor activity by SPT6: a potential coactivator. Mol Endocrinol. 1995;9:34–43. doi: 10.1210/mend.9.1.7760849. [DOI] [PubMed] [Google Scholar]
- 9.Yoh SM, Cho H, Pickle L, Evans RM, Jones KA. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007;21:160–174. doi: 10.1101/gad.1503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanti M, Gallastegui E, Respaldiza I, Rodriguez-Gil A, Gomez-Herreros F, Jimeno-Gonzalez S, Jordan A, Chavez S. Yeast genetic analysis reveals the involvement of chromatin reassembly factors in repressing HIV-1 basal transcription. PLoS Genet. 2009;5:e1000339. doi: 10.1371/journal.pgen.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell. 2006;21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites. Science. 2003;301:1096–1099. doi: 10.1126/science.1087374. [DOI] [PubMed] [Google Scholar]
- 13.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 14.McDonald SM, Close D, Xin H, Formosa T, Hill CP. Structure and Biological Importance of the Spn1–Spt6 Interaction, and Its Regulatory Role in Nucleosome Binding. Mol Cell. 2010 doi: 10.1016/j.molcel.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes & Development. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endoh M, Zhu W, Hasegawa J, Watanabe H, Kim DK, Aida M, Inukai N, Narita T, Yamada T, Furuya A, Sato H, Yamaguchi Y, Mandal SS, Reinberg D, Wada T, Handa H. Human Spt6 stimulates transcription elongation by RNA polymerase II in vitro. Mol Cell Biol. 2004;24:3324–3336. doi: 10.1128/MCB.24.8.3324-3336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan CD, Holland MJ, Winston F. Interaction between transcription elongation factors and mRNA 3'-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J Biol Chem. 2005;280:913–922. doi: 10.1074/jbc.M411108200. [DOI] [PubMed] [Google Scholar]
- 19.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 20.Maclennan AJ, Shaw G. A yeast SH2 domain. Trends Biochem Sci. 1993;18:464–465. doi: 10.1016/0968-0004(93)90006-9. [DOI] [PubMed] [Google Scholar]
- 21.Johnson SJ, Close D, Robinson H, Vallet-Gely I, Dove SL, Hill CP. Crystal structure and RNA binding of the Tex protein from Pseudomonas aeruginosa. J Mol Biol. 2008;377:1460–1473. doi: 10.1016/j.jmb.2008.01.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun M, Lariviere L, Dengl S, Mayer A, Cramer P. A tandem SH2 domain in transcription elongation factor Spt6 binds the phosphorylated RNA polymerase II CTD. J Biol Chem. 2010 doi: 10.1074/jbc.M110.144568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diebold ML, Loeliger E, Koch M, Winston F, Cavarelli J, Romier C. A non-canonical tandem SH2 enables interaction of elongation factor SPT6 with RNA polymerase II. J Biol Chem. 2010 doi: 10.1074/jbc.M110.146696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–3652. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dengl S, Mayer A, Sun M, Cramer P. Structure and in vivo requirement of the yeast Spt6 SH2 domain. J Mol Biol. 2009;389:211–225. doi: 10.1016/j.jmb.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 26.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Grant RP, Marshall NJ, Yang JC, Fasken MB, Kelly SM, Harreman MT, Neuhaus D, Corbett AH, Stewart M. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. J Mol Biol. 2008;376:1048–1059. doi: 10.1016/j.jmb.2007.11.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito A, Iwasaki H, Ariyoshi M, Morikawa K, Shinagawa H. Identification of four acidic amino acids that constitute the catalytic center of the RuvC Holliday junction resolvase. Proc Natl Acad Sci U S A. 1995;92:7470–7474. doi: 10.1073/pnas.92.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shao X, Grishin NV. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res. 2000;28:2643–2650. doi: 10.1093/nar/28.14.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 32.Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waksman G, Shoelson SE, Pant N, Cowburn D, Kuriyan J. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell. 1993;72:779–790. doi: 10.1016/0092-8674(93)90405-f. [DOI] [PubMed] [Google Scholar]
- 36.Lee CH, Kominos D, Jacques S, Margolis B, Schlessinger J, Shoelson SE, Kuriyan J. Crystal structures of peptide complexes of the amino-terminal SH2 domain of the Syp tyrosine phosphatase. Structure. 1994;2:423–438. doi: 10.1016/s0969-2126(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 37.Duyster J, Baskaran R, Wang JY. Src homology 2 domain as a specificity determinant in the c-Abl-mediated tyrosine phosphorylation of the RNA polymerase II carboxyl-terminal repeated domain. Proc Natl Acad Sci U S A. 1995;92:1555–1559. doi: 10.1073/pnas.92.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunde BM, Reichow SL, Kim M, Suh H, Leeper TC, Yang F, Mutschler H, Buratowski S, Meinhart A, Varani G. Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2010;17:1195–1201. doi: 10.1038/nsmb.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladbury JE, Lemmon MA, Zhou M, Green J, Botfield MC, Schlessinger J. Measurement of the binding of tyrosyl phosphopeptides to SH2 domains: a reappraisal. Proc Natl Acad Sci U S A. 1995;92:3199–3203. doi: 10.1073/pnas.92.8.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 41.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol. 2009;29:4852–4863. doi: 10.1128/MCB.00609-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlatanova J, van Holde K. Binding to four-way junction DNA: a common property of architectural proteins? FASEB J. 1998;12:421–431. [PubMed] [Google Scholar]
- 44.Guo M, Xu F, Yamada J, Egelhofer T, Gao Y, Hartzog GA, Teng M, Niu L. Core structure of the yeast spt4-spt5 complex: a conserved module for regulation of transcription elongation. Structure. 2008;16:1649–1658. doi: 10.1016/j.str.2008.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 46.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W, Carter CW, MS JR, editors. Methods in Enzymology. New York: Academic Press; 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Vol. Volume 276: Macromolecular Crystallography. [DOI] [PubMed] [Google Scholar]
- 49.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 52.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 59.DeLano WL. The Pymol Molecular Graphics System. Palo Alto, CA: Delano Scientific; 2002. [Google Scholar]
- 60.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 63.Team, R. D. C. R: A Language and Environment for Statistical Computing. 2010 [Google Scholar]
- 64.LiCata VJ, Wowor AJ. Applications of fluorescence anisotropy to the study of protein-DNA interactions. Methods Cell Biol. 2008;84:243–262. doi: 10.1016/S0091-679X(07)84009-X. [DOI] [PubMed] [Google Scholar]
- 65.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.