Abstract
In Escherichia coli, SecA is a large, multifunctional protein that is a vital component of the general protein secretion pathway. In its membrane-bound form it functions as the motor component of the protein translocase, perhaps through successive rounds of membrane insertion and ATP hydrolysis. To understand both the energy conversion process and translocase assembly, we have used contrast-matched, small-angle neutron-scattering (SANS) experiments to examine SecA in small unilamellar vesicles of E. coli phospholipids. In the absence of nucleotide, we observe a dimeric form of SecA with a radius of gyration comparable to that previously observed for SecA in solution. In contrast, the presence of either ADP or a non-hydrolyzable ATP analog induces conversion to a monomeric form. The larger radius of gyration for the ATP-bound relative to the ADP-bound form suggests the former has a more expanded global conformation. This is the first direct structural determination of SecA in a lipid bilayer. The SANS data indicate that nucleotide turnover can function as a switch of conformation of SecA in the membrane in a manner consistent with its proposed role in successive cycles of deep membrane penetration and release with concommitant preprotein insertion.
Keywords: SecA, oligomerization, nucleotide, protein transport, small-angle neutron scattering
Many proteins that are synthesized in the cytoplasm of cells use the Sec translocase for routing to non-cytoplasmic locations. In Escherichia coli, SecYEG and SecA are critical components of this translocase that function in concert with the preprotein to ensure its passage through the membrane. SecYEG can be isolated and purified as a stable heterotrimeric complex,1-5 and constitutes the central component of the membrane-embedded translocase. SecA is a large, multifunctional protein that is found in both cytoplasmic and membrane-associated forms,6 and interfaces with SecYEG and the preprotein to propel preprotein translocation at the expense of ATP. Biochemical studies suggest that upon binding ATP7 and lipid,8 SecA undergoes a substantial conformational change9 and is deeply inserted into the membrane bilayer. This membrane association results in stimulation of SecA ATPase activity and preprotein translocation.10-12
A fundamental issue to be resolved is how SecA translates the chemical energy of ATP hydrolysis into the mechanical energy required for preprotein movement. To understand the mechanistic basis for this activity, the structural changes of membrane-associated SecA during nucleotide turnover must be elucidated. This is a complex problem to study, however, because the requirement for the membrane-bound form renders many classical methods of structural analysis of proteins in aqueous solution unfeasible. The SecA-nucleotide cycling event is a dynamic process, making studies in model systems useful for evaluating the species at different stages of nucleotide turnover, yet any such system needs to be physiologically relevant in terms of the content and concentration of components.
Several studies of SecA in aqueous solution indicate that a dimeric form predominates in the presence and in the absence of nucleotide,13-16 though conditions of lower temperature and higher ionic strength favor a shift toward monomer.17 The recently reported X-ray structures of Bacillus subtilis SecA at 2.7 Å resolution18 and Micrococcus tuberculosis SecA at 2.8 Å resolution19 also suggests that in aqueous media it forms a dimer that does not undergo a substantial conformational change upon nucleotide binding, consistent with small angle X-ray scattering (SAXS) analysis of SecA, also in an aqueous environment.14 However, earlier studies involving protease sensitivity, differential scanning calorimetry and dynamic light-scattering suggested that SecA in solution adopts different conformations, depending on the nature of nucleotide bound.9,20 Measurement of the molar ratio of bound ADP to SecA in solution indicates that only one high-affinity nucleotide-binding site exists per SecA dimer.21 This and a study involving heterodimers that include ATPase inactive subunits15 suggest that SecA maintains its dimeric form during translocation, yet recent analysis suggests that the association of SecA with negatively charged phospholipids22 or detergents23 shifts the equilibrium toward the monomeric form. The complicated nature of the analyses is illustrated by evidence that the oligomeric state of the SecYE components of the translocase involves a monomer,24 dimer,25 and higher-order structure.26
Here, we report for the first time the direct structural measurement of the size and shape of SecA in vesicles composed of E. coli phospholipids to model the biological membrane. To distinguish SecA features from those of the vesicle, our strategy employed contrast-matched, small-angle, neutron-scattering (SANS) to evaluate SecA in the absence and in the presence of nucleotide. We find that SecA without nucleotide exists in a predominantly dimeric form in this lipid environment that is converted almost exclusively to monomer by the presence of nucleotide. Significant shape differences are observed in the presence of ADP-Mg2+ or non-hydrolyzable ATP-Mg2+ analog.
Nucleotide binding disrupts SecA dimer in lipid vesicles
We have used SANS to make direct measurements of the structure of SecA in vesicles composed of E. coli phospholipids. In addition to providing a model of the physiological membrane, the importance of this lipid composition in promoting ATPase activity27 and translocation28 in vitro has been demonstrated. Furthermore, the dependence of the signal peptide-stimulated SecA/lipid ATPase activity on a high lipid to protein ratio has been documented27 and is maintained in the studies here.
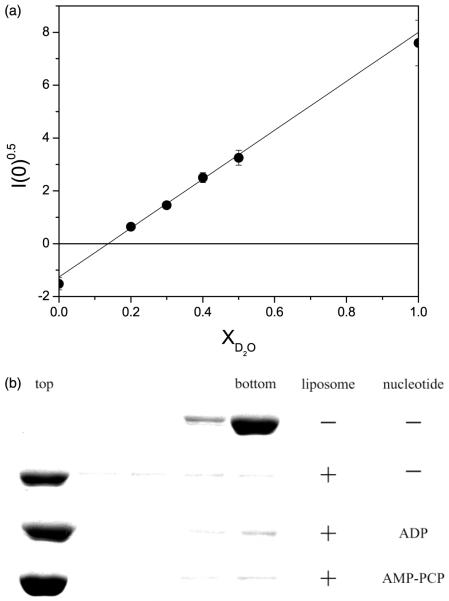
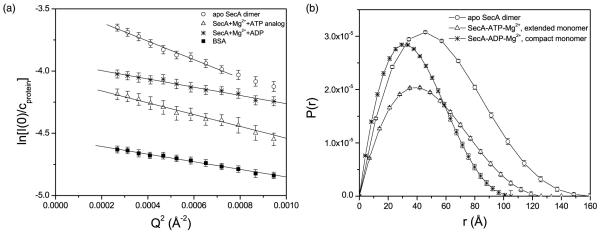
The contrast variation experiments require different volume fractions of 2H2O to determine the lipid contrast match point at which the scattering intensity from the small unilamellar vesicles (SUV) is zero. As shown in Figure 1(a), this occurs at 14.3% (v/v) 2H2O volume fraction. Flotation analysis shows that SecA is lipid-associated (Figure 1(b)). Figure 2(a) shows the Guinier plot of 2.9 mg/ml of SecA and 23 mg/ml of E. coli SUV in a 14.3% (v/v) 2H2O buffer solution. For the SecA dimer, the Guinier analysis was conducted in the region of Q ≤ 0.027 Å−1 corresponding to QRg ≤ 1.2. For the ATP-Mg2+ analog and ADP-Mg2+-bound forms of SecA, the Guinier analysis was conducted in the region of Q ≤ 0.030 Å−1 and Q ≤ 0.034 Å−1, corresponding to QRg ≤ 1.18 and QRg ≤ 1.08, respectively.
Figure 1.
(a) Contrast variation experiments to determine the match point of E. coli SUV. To make SUV, 0.4 ml of 25 mg/ml of chloroform-dissolved E. coli polar lipid extract (Avanti Polar Lipids, Inc., AL) was transferred to a 12 ml Pyrex tube. A thin film of lipid was formed under a stream of filtered nitrogen and residual chloroform was removed by high vacuum overnight. The lipid film was then re-hydrated in 1 mM DTT, 50 mM potassium phosphate (pH 7.5), 50 mM KCl, 1 mM EDTA. SUV were formed by sonicating until the solution was optically clear. The contrast variation experiments required different volume fractions of 2H2O to determine the lipid contrast match point at which the scattering intensity from the SUV is zero. For contrast variation experiments, the lipid was resuspended separately in H2O and 2H2O buffer to make the two SUV solutions. The H2O and 2H2O SUV solutions were mixed in different proportions to produce six SUV solutions of different 2H2O volume fractions. The lipid concentration used for contrast variation experiments was 10 mg/ml. The square-root of scattering intensity I(0)0.5, from the SUV as a function of 2H2O volume fraction is shown. The contrast match point can be determined from this plot to be X2H2O = 0.143. At a 2H2O volume fraction of 0.143, the scattering intensity from the lipid vesicles is zero, indicating the scattering density contrast between the SUV and the buffer solution is zero. Further experiments on SecA/SUV solutions were therefore conducted at this 2H2O concentration to eliminate the scattering from the SUV. (b) Association of SecA with SUV analyzed by flotation gradient centrifugation. SecA (1.5 mg/ml) was mixed with E. coli SUV (12 mg/ml) in the aforementioned phosphate buffer with density of 1.29 g/ml adjusted with metrizamide. Magnesium acetate (60 μM) was added when ADP (60 μM) or AMP-PNP (2 mM) was included in buffer. Samples (200 μl) were layered below 400 μl of corresponding buffers with density of 1.27 g/ml. Samples were centrifuged (for 33 hours, at 4°C, at 35,000 rpm) in a Beckman SW50.1 rotor with adapters for 5 × 41 mm Beckman Ultraclear centrifuge tubes, six 100 μl fractions were harvested and the top three and bottom two fractions analyzed by SDS-PAGE and staining with Coomassie brilliant blue.
Figure 2.
| (1) |
| (2) |
| (3) |
| (4) |
The molecular mass standard bovine serum albumin (BSA 68 kDa) was dissolved with 23 mg/ml of lipid vesicles in 14.3% 2H2O (v/v) buffers. The protein concentration-normalized forward scattering intensity (I(0)/cprotein) was used to calibrate the molecular mass of the different forms of SecA. A calibration of I(0)cprotein of SecA against that of BSA gives a molecular mass of 204 kDa for SecA, indicating that it is a dimer at the concentration measured by SANS (see Table 1).
Table 1.
A summary of the size and oligomeric states of SecA measured by SANS
| I(0)/cprotein |
Mw (kDa) |
Rg (Å) |
|
|---|---|---|---|
| Apo SecA | 0.033 ± 0.002 | 204 | 45.1 ± 0.9 |
| SecA + Mg2+ + ATP analog | 0.017 ± 0.001 | 105 | 38.3 ± 0.8 |
| SecA + Mg2+ + ADP | 0.019 ± 0.001 | 117 | 31.1 ± 0.5 |
| BSA | 0.011 ± 0.001 | 68 | 31.6 ± 0.2 |
The error in protein concentration determination is about 10%. The error in molecular mass (calibrated against BSA) is also about 10%
The radius of gyration of SecA in the lipid vesicles determined by SANS is 45.1(±0.9) Å (Table 1), consistent with the previously reported value of 45.2 Å for SecA determined by SAXS in aqueous solution.14 This indicates that the radius of gyration of the SecA dimer observed by these methods does not change in the presence of E. coli lipid vesicles. Flotation analysis confirms that under these conditions SecA is nonetheless associated stably with the vesicles (and Figure 1(b)).10
The distance distribution function (P(r)) of apo SecA shows that SecA in the presence of lipid vesicles is a dimer, whereas the dimer dissociates upon binding ATP or ADP (Figure 2(b)). The asymmetric shape of P(r) suggests the SecA dimer is asymmetric. Our SANS results for the SecA dimer without nucleotide in lipid vesicles are thus in approximate agreement with the SAXS study of SecA in solution,14 although the shape of the SecA dimer seems to be more expanded in lipid relative to that in aqueous buffer (160 Å versus 150 Å).
Figure 2(a) shows the Guinier plot of SecA at 2.9 mg/ml of SUV in the presence of non-hydrolyzable ATP analog and Mg2+ . The protein concentration-normalized forward scattering intensity is about half of that of the apo SecA, shown in Figure 2(a), indicating that the molecular mass of the ATP analog-bound form of SecA is half of the apo SecA in lipid. The molecular mass calculated from I(0)/cprotein against a molecular mass standard, BSA, shows that the ATP analog-bound form of SecA is a monomer at the concentration measured by SANS (Table 1). Similarly, the I(0)/cprotein of SecA in the presence of ADP and Mg2+ is about half of that of SecA, indicating that the ADP-bound form of SecA is a monomer. In contrast, SAXS indicated that SecA in aqueous solution remained dimeric even in the presence of nucleotide.14
The ATP-bound SecA monomer is more expanded than the ADP-bound SecA monomer
Based on a Kd of 0.8 μM ADP for SecA in solution,21 and an estimate of the Kd for the non-hydolyzable ATP analog,29 we have studied SecA in the presence of nucleotide sufficient to yield >97% occupancy of the high-affinity nucleotide-binding site on SecA. From the Guinier plots (Figure 2(a)), the radius of gyration of SecA–ATP-Mg2+ is Rg = 38.3(±0.8) Å, larger than the radius of gyration of SecA–ADP-Mg2+ of Rg = 31.1(±0.5) Å. This suggests that the ATP-bound form of the monomeric SecA has a more expanded global conformation than the ADP-bound, monomeric form. The P(r) of the ATP-bound and the ADP-bound forms of SecA show that the SecA dimer dissociates upon binding ATP or ADP (Figure 2(b)), and the maximum dimension of the SecA–ATP-Mg2+ form is larger than that of the SecA–ADP-Mg2+ form (about 125 Å and 105 Å, respectively).
The Kd for the monomer-dimer equilibrium of SecA in aqueous solution in the absence of nucleotide is estimated to be in the submicromolar to low micromolar range.13,17 The corresponding values in the presence of lipid vesicles and/or nucleotides remain undefined. A recent study of SecA using fluorescence resonance energy transfer suggests that in the presence of membranes containing acidic phospholipids, SecA dissociates into monomers.22 The concentration of SecA used in the fluorescence resonance energy transfer experiment was about 1.5 μM, while the concentration used in the SANS experiment was about 28 μM, suggesting that these experiments were run just below and above, respectively, the Kd for dimer dissociation in the absence of nucleotide. Collectively, the data suggest that the presence of the lipid environment shifts the Kd upward into the micromolar range and the presence of nucleotide increases the Kd value even further, resulting in a species that is predominantly monomeric. The physiological concentration of SecA is estimated at about 10 μM for the bulk volume of the bacterium16,22 yet is effectively higher at the translocon due to the concentrating effect of binding to specific sites comprised of SecYEG. Therefore, our SANS experiments likely provide a good physiological representation of the local concentration in this environment. Furthermore, our observation that SecA forms a dimer in the absence of nucleotide is consistent with the finding that the dimeric species can associate with the biological membrane in vitro.15 However, since those experiments involved ongoing protein translocation and continual nucleotide turnover, individual SecA states formed during that process could not be distinguished. An advantage of the strategy we have employed here is the ability to distinguish distinct structural states under defined conditions. Moreover, we have been able to characterize the dimensions of SecA in the presence of a bilayer composed of E. coli phospholipids. In this environment, nucleotide modulates substantial SecA conformation changes, consistent with previous biochemical analyses in membranes.30-32 This underscores the role of the ATPase activity in the membrane interactive species and the possibility that the accompanying changes in conformation serve to propel the preprotein through the translocon.
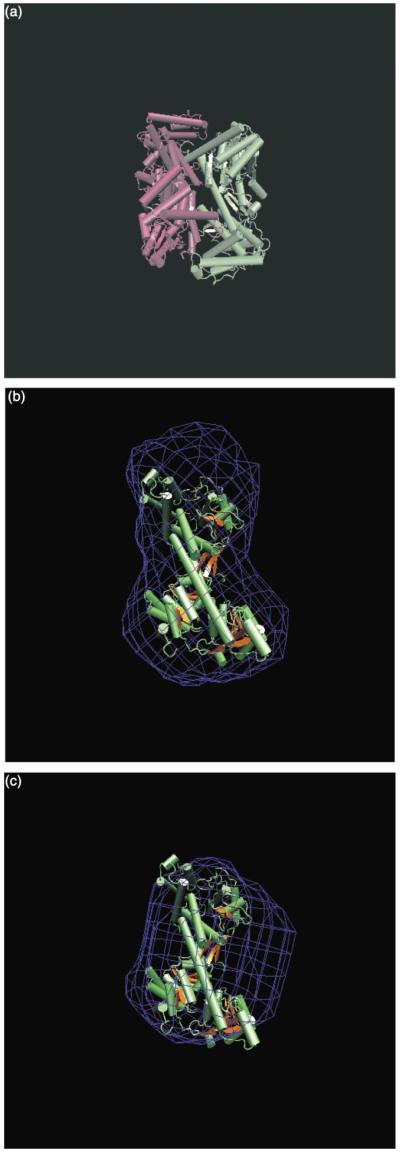
The maximum dimension (Dmax) of the SecA–ATP-Mg2+ form is about 125 Å and the Dmax of SecA–ADP-Mg2+ form is about 105 Å (Figure 2(b)). We have used the program DAMMIN33 to calculate the envelopes of the different forms of SecA. Figure 3(a) is the calculated envelope based on the crystal structure of B. subtilis SecA in an aqueous environment18 published while we were preparing this manuscript, and Figure 3(b) and (c) are envelopes of the ATP analog-bound and ADP-bound forms, respectively, of SecA calculated from SANS data. The envelopes shown in Figure 3 provide a direct view of the structural changes of SecA upon binding to nucleotides.
Figure 3.
Models of different forms of SecA. (a) The crystal structure of SecA dimer.18 (b) The SANS-determined SecA–ATP-Mg2+ monomer structure represented by a blue wire model. This blue wire model was calculated from the SANS P(r) function using the programs DAMMIN33 and Situs.44 The crystal structure of the apo SecA monomer was docked to the SANS model using the program Situs44 to show that the ATP-bound monomer is more expanded than the apo SecA structure. (c) The model of SecA–ADP-Mg2+ calculated from SANS data (blue wire) shows that the ADP-bound monomer is more compact than the apo SecA. These images were made with VMD and are owned by the Theoretical and Computational Biophysics Group, an NIH Resource for Macromolecular Modeling and Bioinformatics, at the Beckman Institute, University of Illinois at Urbana-Champaign.
The crystal structure18,19 of SecA can be considered to contain three domains: the preprotein-binding domain, the first nucleotide-binding fold (NBFI), and the second nucleotide-binding fold (NBFII). The actual nucleotide-binding site is in the cleft between NBFI and NBFII. In this spirit, the structure of SecA resembles the structure of a single subunit of GroEL which is composed of three domains: the apical domain that binds to unfolded protein, the intermediate domain and the equatorial domain. The nucleotide-binding site on GroEL is in the cleft formed between the intermediate and the equatorial domains.34,35 When bound to ATP, the intermediate domain of GroEL orients in such a way as to push the apical domain further from the equatorial domain through rigid body movement.34-39 In the ADP-bound form, the intermediate domain reorients itself to bring the apical domain and the equatorial domain closer into a more compact form. Based on our analysis of the structural analogy of SecA with a single subunit of GroEL, we hypothesize that nucleotide-binding induces similar conformation changes in both proteins and these two proteins may share common features in the reaction cycles of nucleotide binding and hydrolysis, and interacting with the unfolded proteins. In the case of SecA, these conformational changes are consistent with models for processive chain movement;40,41 the more expanded ATP-bound form is well suited for traversing the membrane carrying a portion of the preprotein, while subsequent hydrolysis triggers the release of the preprotein, generation of the more compact form of SecA and its retraction to the cytoplasmic face of the membrane. ADP-ATP exchange could then serve to initiate another cycle of structural changes with insertion and de-insertion to processively transport successive segments of the preprotein. Further, that nucleotide does not induce these conformational changes in the absence of membranes serves to dedicate its function in preprotein processivity to the locale of the translocon and inhibit the likelihood that SecA in the cytoplasm participates non-productively in this activity.
Acknowledgements
We thank Sharyn Rusch for helpful discussions and critically reading this manuscript. This research was supported, in part, by National Institutes of Health grant GM37639 (to D.A.K.). This work utilized facilities supported, in part, by the National Science Foundation under agreement no. DMR-9986442.
Abbreviations used
- AMP-PNP
5′-adenylylimidodiphosphate
- BSA
bovine serum albumin
- SUV
small unilamellar vesicles
- SANS
small-angle neutron-scattering
- SAXS
small angle X-ray scattering
- SUV
small unilamellar vesicles
Footnotes
Present address: Z. Bu, Fox Chase Cancer Center, Philadelphia, PA 19111, USA.
References
- 1.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 2.Brundage L, Fimmel CJ, Mizushima S, Wickner W. SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J. Biol. Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- 3.Douville K, Price A, Eichler J, Economou A, Wickner W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 4.Hanada M, Nishiyama KI, Mizushima S, Tokuda H. Reconstitution of an efficient protein translocation machinery comprising SecA and the three membrane proteins, SecY, SecE, and SecG (p12) J. Biol. Chem. 1994;269:23625–23631. [PubMed] [Google Scholar]
- 5.Akimaru J, Matsuyama S, Tokuda H, Mizushima S. Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc. Natl Acad. Sci. USA. 1991;88:6545–6549. doi: 10.1073/pnas.88.15.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabelli RJ, Dolan KM, Qian LP, Oliver DB. Characterization of membrane-associated and soluble states of SecA protein from wild-type and SecA51(TS) mutant strains of Escherichia coli. J. Biol. Chem. 1991;266:24420–24427. [PubMed] [Google Scholar]
- 7.Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol. Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- 8.Ulbrandt ND, London E, Oliver DB. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J. Biol. Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 9.Shinkai A, Mei LH, Tokuda H, Mizushima S. The conformation of SecA, as revealed by its protease sensitivity, is altered upon interaction with ATP, presecretory proteins, everted membrane vesicles, and phospholipids. J. Biol. Chem. 1991;266:5827–5833. [PubMed] [Google Scholar]
- 10.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 11.Lill R, Cunningham K, Brundage LA, Ito K, Oliver D, Wickner W. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham KW, Wickner WT. Specific recognition of the leader region of precursor proteins is required for the activation of translocation ATPase of Escherichia coli. Proc. Natl Acad. Sci. USA. 1989;86:8630–8634. doi: 10.1073/pnas.86.22.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle SM, Braswell EH, Teschke CM. SecA folds via a dimeric intermediate. Biochemistry. 2000;39:11667–11676. doi: 10.1021/bi000299y. [DOI] [PubMed] [Google Scholar]
- 14.Shilton B, Svergun DI, Volkov VV, Koch MH, Cusack S, Economou A. Escherichia coli SecA shape and dimensions. FEBS Letters. 1998;436:277–282. doi: 10.1016/s0014-5793(98)01141-7. [DOI] [PubMed] [Google Scholar]
- 15.Dreissen AJM. SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry. 1993;32:13190–13197. doi: 10.1021/bi00211a030. [DOI] [PubMed] [Google Scholar]
- 16.Akita M, Shinkai A, Matsuyama S, Mizushima S. SecA, an essential component of the secretory machinery of Escherichia coli, exists as homodimer. Biochem. Biophys. Res. Commun. 1991;174:211–216. doi: 10.1016/0006-291x(91)90507-4. [DOI] [PubMed] [Google Scholar]
- 17.Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 19.Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, et al. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc. Natl Acad. Sci. USA. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.den Blaauwen T, Fekkes P, de Wit JG, Kuiper W, Driessen AJM. Domain interactions of the peripheral preprotein translocase subunit SecA. Biochemistry. 1996;35:11994–12004. doi: 10.1021/bi9605088. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt M, Ding H, Ramamurthy V, Mukerji I, Oliver D. Nucleotide binding activity of SecA homodimer is conformationally regulated by temperature and altered by prlD and azi mutations. J. Biol. Chem. 2000;275:15440–15448. doi: 10.1074/jbc.M000605200. [DOI] [PubMed] [Google Scholar]
- 22.Or E, Navon A, Rapoport T. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benach J, Chou YT, Fak JJ, Itkin A, Nicolae DD, Smith PC, et al. Phospholipid-induced monomerization and signal-peptide-induced oligomerization of SecA. J. Biol. Chem. 2003;278:3628–3638. doi: 10.1074/jbc.M205992200. [DOI] [PubMed] [Google Scholar]
- 24.Yahr TL, Wickner WT. Evaluating the oligomeric state of SecYEG in preprotein translocase. EMBO J. 2000;19:4393–4401. doi: 10.1093/emboj/19.16.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessonneau P, Besson V, Collinson I, Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21:995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manting EH, van der Does C, Remigy H, Engel A, Driessen AJ. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 2000;19:852–861. doi: 10.1093/emboj/19.5.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller A, Wang L, Kendall DA. Synthetic signal peptides specifically recognize SecA and stimulate ATPase activity in the absence of preprotein. J. Biol. Chem. 1998;273:11409–11412. doi: 10.1074/jbc.273.19.11409. [DOI] [PubMed] [Google Scholar]
- 28.Henrick JP, Wickner W. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J. Biol. Chem. 1991;266:24596–24600. [PubMed] [Google Scholar]
- 29.Matsuyama S, Kimura E, Mizushima S. Complementation of two overlapping fragments of SecA, a protein translocation ATPase of Escherichia coli, allows ATP binding to its amino-terminal region. J. Biol. Chem. 1990;265:8760–8765. [PubMed] [Google Scholar]
- 30.Ramamurthy V, Oliver D. Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J. Biol. Chem. 1997;272:23239–23246. doi: 10.1074/jbc.272.37.23239. [DOI] [PubMed] [Google Scholar]
- 31.Eichler J, Wickner W. Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc. Natl Acad. Sci. USA. 1997;94:5574–5581. doi: 10.1073/pnas.94.11.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn T, Kim H. SecA of Escherichia coli traverses lipid bilayer of phospholipid vesicles. Biochem. Biophys. Res. Commun. 1994;203:326–330. doi: 10.1006/bbrc.1994.2185. [DOI] [PubMed] [Google Scholar]
- 33.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Braig K, Otwinowski Z, Hegde R, Boisvert DC, Joachimiak A, Horwich AL, Sigler PB. The crystal structure of the bacterial chaperonin GroEL at 2.8 Å. Nature. 1994;371:578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- 35.Boisvert DC, Wang J, Otwinowski Z, Horwich AL, Sigler PB. A crystal structure of the bacterial chaperonin GroEL complexed with ATP gamma S. Nature Struct. Biol. 1996;3:170–177. doi: 10.1038/nsb0296-170. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, Roseman AM, Hunter AS, Wood SP, Burston SG, Ranson NA, et al. Location of a folding protein and shape changes in GroEL–GroES complexes imaged by cryo-electron microscopy. Nature. 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 37.Roseman AM, Chen S, White H, Braig K, Saibil HR. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 1996;87:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- 38.Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 39.Ma J, Sigler PB, Xu Z, Karplus M. A dynamic model for the allosteric mechanism of GroEL. J. Mol. Biol. 2000;302:303–313. doi: 10.1006/jmbi.2000.4014. [DOI] [PubMed] [Google Scholar]
- 40.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 41.Economou A, Pogliano JA, Beckwith J, Oliver D, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 42.Glinka C, Barker J, Hammouda B, Krueger S, Moyer J, Orts W. The 30m SANS instruments at NIST. J. Appl. Crystallog. 1998;31:430–445. [Google Scholar]
- 43.Bu Z, Engelman DM. A method for determining transmembrane helix association and orientation in detergent micelles using small angle X-ray scattering. Biophys. J. 1999;77:1064–1073. doi: 10.1016/S0006-3495(99)76956-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wriggers W, Chacón P. Using situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J. Appl. Crystallog. 2001;34:773–776. [Google Scholar]