Abstract
Methods for chemical modifications of proteins have been crucial for the advancement of proteomics. In particular, site-specific covalent labeling of proteins with fluorophores and other moieties has permitted the development of a multitude of assays for proteome analysis. A common approach for such a modification is solvent-accessible cysteine labeling using thiol-reactive dyes. Cysteine is very attractive for site-specific conjugation due to its relative rarity throughout the proteome and the ease of its introduction into a specific site along the protein's amino acid chain. This is achieved by site-directed mutagenesis, most often without perturbing the protein's function. Bottlenecks in this reaction, however, include the maintenance of reactive thiol groups without oxidation before the reaction, and the effective removal of unreacted molecules prior to fluorescence studies. Here, we describe an efficient, specific, and rapid procedure for cysteine labeling starting from well-reduced proteins in the solid state. The efficacy and specificity of the improved procedure are estimated using a variety of single-cysteine proteins and thiol-reactive dyes. Based on UV/vis absorbance spectra, coupling efficiencies are typically in the range 70–90%, and specificities are better than ~95%. The labeled proteins are evaluated using fluorescence assays, proving that the covalent modification does not alter their function. In addition to maleimide-based conjugation, this improved procedure may be used for other thiol-reactive conjugations such as haloacetyl, alkyl halide, and disulfide interchange derivatives. This facile and rapid procedure is well suited for high throughput proteome analysis.
I. INTRODUCTION
Protein molecules are often targets for chemical modification by covalent conjugation (1, 2). These derivatized proteins can specifically recognize and bind target molecules, providing a handle for target detection, quantification, and analysis. Specific and efficient protein labeling techniques are therefore essential for biological research, diagnostics, and therapeutics. Site-specific labeling of proteins is also needed in single molecule fluorescence experiments (3–5). Many labeling methods have been developed and utilized, including covalent modification of amino acid side chains (6–8), covalent labeling by enzymes (9–12), conjugation to unnatural amino acids (13, 14), and affinity labeling (15, 16).
One of the most common specific covalent coupling methods is the conjugation of a moiety bearing a thiol-reactive group to a solvent-accessible cysteine. This method affords the site-specific labeling of a protein at a unique engineered (or native) surface cysteine. This cysteine could be specifically labeled with dye derivatives of haloacetyl compounds, disulfide, or maleimides. The maleimide is by far the most prevalent functional group to be coupled to a cysteine because the coupling reaction is highly specific and efficient.
The protein must be maintained in a reduced form prior to the maleimide-cysteine coupling reaction to prevent the formation of disulfide bridges and inactivation of the cysteines. Reducing agents such as dithiothreitol (DTT) and tris[2-carboxyethyl]phosphine (TCEP) can be used for this purpose. These reducing agents must be removed before conjugation so that their thiol groups will not compete with target thiols in proteins.
The labeling reaction must be performed shortly after the elimination of reducing agents to prevent reoxidation of the target thiols, resulting in decreased labeling efficiency. In order to circumvent this problem, we developed an alternative, rapid, efficient, and specific cysteine labeling technique; this procedure does not require time-consuming and cumbersome reduction/desalting steps because the labeling reaction is directly performed on highly reduced proteins in the solid state.
We demonstrate our new labeling method, which we dub “solid state-based labeling”, or SSL, on two small libraries of model proteins, all containing a single cysteine. The first library consists of mutants of the transcription initiation factor, σ70 subunit, of Escherichia coli RNA polymerase. The σ70 is required for promoter-specific transcription; σ70 binds to RNAP coenzyme, interacts with promoter DNA, plays roles in promoter unwinding, promoter escape, early elongation, and is a target for transcription regulators. The E. coli sigma factor has been utilized as a target protein for labeling in structural studies of bacterial transcription on the ensemble level (17, 18) and more recently on the single molecule level (19–23). Using SSL, several maleimide-linked dyes have been incorporated specifically and efficiently into single cysteine mutants of σ70, and fluorescence studies have showed that the labeled proteins are fully functional in transcription. The second library consists of a variety of proteins from the organism Shewanella oneidensis MR-1 (see (24–26) for reviews), either containing a single native surface-exposed cysteine or a single engineered cysteine added at the N-terminal region.
II. EXPERIMENTAL PROCEDURES
II.1. Materials
Alexa-maleimides and tetramethylrhodamine-maleimide were purchased from Invitrogen Corp. (Carlsbad, CA), and Cy3b-maleimide was purchased from GE Healthcare (Piscataway, NJ). E. coli RNA polymerase core was purchased from Epicenter Biotechnologies (Madison, WI). Size-exclusion resin HW-50F was purchased from TOSOH Bioscience LLC (Montgomeryville, PA). Disposable polypropylene columns and Nickel-nitrilotriacetic acid (Ni-NTA) Agarose were purchased from Qiagen Inc. (Germany).
II.2. Production and Purification of Proteins
Plasmid pGEMD(-Cys), encoding a σ70 derivative with no Cys residues, and plasmids encoding σ70 derivatives with single Cys residues were described in Mukhopadhyay et al. (17). The expression construct of the single cysteine mutant (A59C) of σ70 was created by site-directed mutagenesis. The σ70 mutants were expressed and inclusion bodies were prepared according to a published protocol (17). The resulting proteins were refolded from the inclusion bodies and further purified by anion exchange chromatography (MonoQ) (GE Healthcare, USA), dialyzed against TGED buffer (20 mM Tris-HCl [pH 7.9]/0.1 mM Ethylenediamine tetraacetic acid [EDTA]/1 mM DTT/5% Glycerol) plus 200 mM NaClBacterial expression constructs for MR-1 σ24, σ32, and the integration host factor (IHF) were provided by Dr. Jeanne Perry of the Protein Expression Laboratory at UCLA and expressed and purified in our laboratory. Single cysteine Shewanella oneidensis MR-1 proteins were prepared by amplification of the open reading frame from genomic DNA with KOD DNA polymerase by using conditions and reagents provided by the vendor (Novagen, Madison, WI). The genes were cloned using a modified ligation-independent cloning protocol (27). This process generated an expression clone producing a fusion protein with an N-terminal His-tag and a V 5 epitope tag containing a single cysteine. Proteins were purified using immobilized metal affinity chromatography (28).
II.3. Ammonium Sulfate Precipitation
The purified proteins in TGED buffer were reduced with 10 mM DTT at 4 °C for 2 h. The 10 mM DTT is sufficient enough to reduce up to several hundred micromolar concentration of the protein sample. Ammonium sulfate powder was added to the reduced protein samples. Final concentration of the ammonium sulfate was 70% so that most proteins were precipitated. After addition of the salt, samples were gently agitated for less than 10 min, and stored at 4 °C. This solid form of proteins in the slurry could be stably stored without losing the structural and functional integrity at 4 °C for years. To estimate the quantity of proteins in the solid state, 50 uL of ammonium sulfate slurry was centrifuged, and the pellet was dissolved in TE buffer (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA). The dissolved sample was quantified by the Bradford assay.
II.4. Dye Conjugation
Four nanomoles of the sample in the slurry were taken and further reduced with 5–10 mM DTT for 2 h at 4 °C. We found that samples stored in the solid state for periods longer than 1 month at 4 °C were not in completely reduced state. Those samples were therefore treated with additional 5–10 mM DTT (at least 2 h incubation at 4 °C) before conjugation. Four nmoles of protein in the slurry were taken and centrifuged at 13 000 g for 2–5 min at 4 °C. After removal of the supernatant, the pellet was briefly washed with ice-cold Buffer A (100 mM Na2PO4[pH 7.3]/200 mM NaCl/1 mM EDTA) plus 70% (NH4)2SO4. Washing was achieved by gently inverting the samples several times and subsequent centrifugation for 2 min. The washed pellet was dissolved in 100 μL of Buffer A containing maleimide-linked fluorophores. Several dye probes were tested: Tetramethylrhodamine (TMR)-6-maleimide (Invitrogen, Eugene); Alexa Fluor488-C5-maleimide (Invitrogen, Eugene); Alexa Fluor647-C2-maleimide (Invitrogen, Eugene); and Cy3B-maleimide (GE Healthcare, Piscataway). Each fluorophore was dissolved in dimethyl sulfoxide (DMSO) and mixed with Buffer A. Four nanomoles of the dissolved protein were reacted with 20 nmol of dye (1:5 ratio) for 30 min at room temperature. The conjugation reaction was quenched by adding final 0.5% (72 mM) β-mercaptoethanol to the reaction mixture and incubating for 10 min at room temperature.
To remove excess dye from labeled proteins, 2 mL of resin HW-50F were packed into a disposable column and equilibrated with Buffer B (40 mM Tris-HCl [pH 7.9]/200 mM NaCl/1 mM DTT/0.2 mM EDTA). The reaction mixture was loaded onto the column, eluted with Buffer B and collected in 0.1 mL fractions. Fractions of the labeled protein's peak were pooled together, glycerol was added to a final concentration of 50%, and the sample was stored at –20 °C. The column could be reused if it was thoroughly washed with Buffer C (20 mM Tris-HCl/6 M Guanidine HCl/pH 7.9) followed up by washing with deionized water (to get rid of the free dyes). Washed columns were equilibrated with 20% ethanol and stored at room temperature.
II.5. Determination of Labeling Efficiency and Functional Assays
Desalted proteins were quantified using Lambda 25 UV/vis spectrophotometer (PerkinElmer, Waltham). Dye coupling efficiency was calculated based on the extinction coefficient of each reagent: ∊280(σ70) = 41 370 M–1 cm–1; ∊495(Alexa Fluor 488) = 71 000 M–1 cm–1; ∊555(TMR) = 65 000 M–1 cm–1; ∊559(Cy3B) = 130 000 M–1 cm–1; ∊650(AlexaFluor 647) = 239 000 M–1 cm–1. Absorbance at 280 nm was corrected by a correction factor (CF) defined as CF280 = A280 (freedye)/Amax (freedye). The CF numerical values for each dye are CF280 (AlexaFluor 488) = 0.11; CF280(TetraMethylRhodamine) = 0.03; CF280(Cy3B) = 0.08; and CF280(AlexaFluor 647) = 0.03.
The coupling efficiency (CE) was calculated according to eq 1 (1):
| (1) |
Samples were run on 12% SDS-PAGE gels and visualized by Molecular ImagerFX (Bio-Rad, Hercules). A gel shift assay was carried out to confirm the complex-forming activity of derivatized σ70 according to a published protocol (29).
III. RESULTS AND DISCUSSION
III.1. Rationale of the Labeling Method
Although the maleimide reactivity toward a free thiol is robust, it is often difficult to achieve high conjugation efficiency due to a variety of issues. Labeling cysteine residues requires proteins be well-reduced by a strong reducing agent, such as dithiothreitol (DTT). However, DTT molecules need to be removed just before conjugation because their thiols are as reactive toward male-imides as cysteines’ thiols. Conventional desalting methods include dialysis and size-exclusion chromatography. Dialysis is lengthy and usually results in reoxidation of the free thiols. Size-exclusion chromatography permits faster desalting and is therefore more commonly used, but has its own drawbacks: (i) desalting is usually accompanied by a loss of proteins and undesired dilution of the sample, especially when a small quantity of proteins is labeled; (ii) desalted proteins need to be quantified before they are reacted with a stoichiometric amount of dye molecules; reoxidation cannot be avoided during this step, resulting in a lower labeling efficiency.
To increase the efficiency, methods for labeling after solid-phase reduction (17) and labeling in the presence of thiol-free reductant, tris(2-carboxyethyl)phosphine (TCEP) (30, 31) have been devised. TCEP, a thiol-free trialkylphosphine, was believed to not react with thiol-reactive compounds, thereby obviating the troublesome desalting step. However, it has been shown that TCEP interferes with labeling of proteins with a thiol-reactive dye (32). The authors of this report found that the reactivity of thiol-reactive dyes is inhibited by the presence of TCEP, and that dye-TCEP adducts are found in the sample (as evident from HPLC chromatograms). The dye concentration used for the labeling reaction therefore needs to be greater than the TCEP concentration and the concentration of the target cysteine. In the solid-phase reduction protocol, proteins are reduced by immobilized reducing agents (DTT or TCEP), are directly recovered from the reductant column, and are reacted with thiol-reactive dyes after quantification. Although this scheme eliminates the reductant removal step, intrinsic problems still remain: sample loss, dilution, and exposure to reoxidation during the quantitation step.
The improved labeling protocol presented here is based on performing the labeling reaction from a precipitated form of target protein. We reasoned that thiols of protein in the solid state may not be easily oxidized and converted to disulfide forms because of their restricted movement in the state. To test this hypothesis, we chose the well-established ammonium sulfate precipitation through the salting-out effect (33). Ammonium sulfate is highly soluble, inexpensive, and apparently unharmful to proteins’ function. Target proteins were precipitated by a final concentration of 70–75% ammonium sulfate in a highly reducing environment and stored as a slurry at 4 °C until needed. Precipitation and storage were done in the presence of 5 to 10 mM DTT in order to break any existing disulfide bonds between surface cysteines and to maintain reducing environment. The efficiency of the maleimide-dye conjugation to the protein precipitate was evaluated according to the protocol described in section II.5.
III.2. Labeling of E. coli σ70 Mutants and Shewanella oneidensis MR-1 Proteins
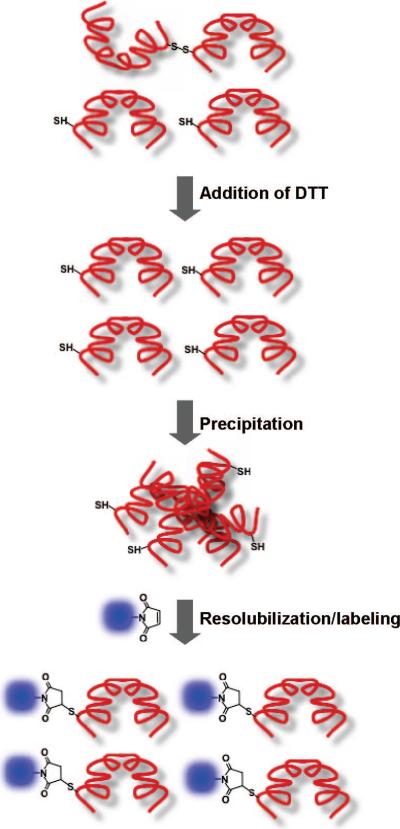
Figure 1 illustrates the labeling strategy. Prior to conjugation, the sample slurry was pelleted by centrifugation and the precipitate was briefly washed with a reductant-free wash buffer (buffer A). The washing step eliminates most of the residual DTT. The wash buffer contains 70% ammonium sulfate to prevent resolubilization of the protein. After washing, the precipitate is immediately reacted with thiol-reactive dyes. Another commonly encountered problem of protein labeling is the separation of labeled proteins from unreacted excess dyes. This step is crucial for fluorescence-based assays since free dyes contribute to a significant background signal that obscures the measurement. However, it is usually not easy to achieve a complete excess dye removal by a single desalting step. We found that the Toyopearl size-exclusion resin (consists of a modified hydrophilic methacrylate polymer) is superior to other commercially available agaroseor polyacrylamide-based resins in separating excess dyes from labeled proteins. The size exclusion separation step was performed using a 2 mL resin volume, under isocratic buffer conditions at neutral pH (pH 7–8) with a moderate ionic strength (100–500 mM NaCl). This desalting step was sufficient in removing >98% of the free-dye in a single step (data not shown).
Figure 1.
Strategy of solid state-based cysteine labeling. The purified protein is reduced by DTT and stored as an ammonium sulfate precipitate under reducing conditions. A brief centrifugation removes DTT in the supernatant and retrieves the protein as a pellet. Upon redissolving the proteins in a buffer containing thiol-reactive dyes, they are immediately and efficiently labeled.
III.3. Labeling Efficiencies and Specificities
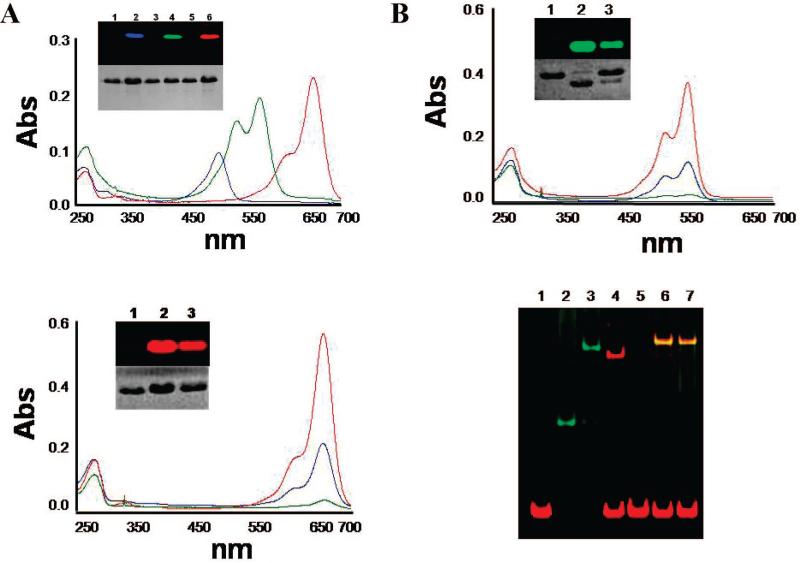
Labeling efficiencies were evaluated by measuring the UV/vis absorbance spectra of the samples (see section II.5.). In a first experiment, several dyes were conjugated to a σ70 mutant that has a unique cysteine at residue 596 (arginine to cysteine - R596C) (18). Figure 2A shows the absorbance spectra of SSL-labeled σ70 (R596C) with Alexa 488 (blue), Cy3b (green), and Alexa 647 (red). The corresponding calculated labeling efficiencies are 91% for Alexa 488 and 70% for Alexa 647. In the case of Cy3b, there was a significant spectral change of the dye after conjugation (appearance of an additional emission peak around 540 nm), precluding an accurate estimation of coupling efficiency to be made. We note that spectral properties of dyes could be significantly altered after conjugation due to their local environment. Nonetheless, the efficiency is likely to be high since the calculated value based on the major peak's absorption only (around 560 nm) is ~65%. SDS-PAGE of these samples analyzed by fluoroimaging and Coomassie-staining indicates the labeling is very specific (Inset of Figure 2A).
Figure 2.
Efficient and specific cysteine labeling: (A) UV/vis spectra of σ70 (R596C) labeled with Alexa 488 (blue), Cy3b (green), and Alexa 647 (red). Inset: SDS-PAGE gel of the labeled σ70: the upper panel is a gel image acquired from fluoroimager scan. Each signal, Alexa 488, Cy3b and Alexa 647, was separately imaged by Molecular Imager FX (Bio-Rad, Hercules) and the images were merged: lane 1, 3 and 5 are cysteine-free (Δcys) σ70 controls that were subjected to SSL procedure with Alexa 488, Cy3b and Alexa 647, respectively. The fluorescent signals of σ70 (R596C) labeled with Alexa 488 (lane 2), Cy3b (lane 4) and Alexa 647 (lane 6) are shown in blue, green and red, respectively. After fluoroimaging, the gel was Coomassie-stained (lower panel). (B) Comparison of SSL to conventional cysteine labeling: UV/vis spectra of Cy3b-σ70 (A59C) labeled by SSL (red), conventionally (blue) and a Δcys σ70 control (green). Inset: SDS-PAGE gel of the labeled σ70: the upper panel is a gel image from fluoroimager scan: Cy3b-labeled protein bands are shown in green. After fluoroimaging, the gel was Coomassie-stained (lower panel). Lane 1 is cysteine-free (Δcys) σ70 control that was subjected to SSL procedure with Cy3b. Lanes 2 and 3 are Cy3b-σ70 labeled by SSL and conventional method, respectively. (C) Comparison of SSL to conventional cysteine labeling of Alexa 647-σ70 (A59C) (line colors and lane numbers are the same as in B). (D) Electrophoretic mobility shift assay of SSL-labeled Cy3b-σ70 (S59C). Holoenzymes were added to Alexa 647-labeled 65 base pairs long lacCONS-11 promoter DNAs (17) (a derivative of the lacUV5–11 promoter) to a final concentration of 77 nM core, 308 nM Cy3b-σ70, and 77 nM DNA fragment. Following a 30 min incubation at 37 °C, the complexes were challenged with final 0.1 mg/mL heparin, the samples were then incubated for another 5 min at 37 °C before loading onto a 5% polyacrylamide gel (29:1 acrylamide/bisacrylamide; 13.5 × 16 × 0.75 cm) and electrophoresed in 0.5× TBE buffer at 150 V for 2.5 h: lane 1: Alexa 647-DNA only; lane 2: Cy3b-σ70 only; lane 3: RNA polymerase core (RNAPc) and Cy3b-σ70; lane 4: RNAPc + Alexa 647-DNA; lane 5: RNAPc + Alexa 647-DNA + heparin; lane 6: RNAPc + Cy3b-σ70 + Alexa 647-DNA; lane 7: RNAPc + Cy3b-σ70 + Alexa 647-DNA + heparin. Each signal, Cy3b and Alexa 647, was separately imaged, and the images were merged. The yellow bands in lane 6 and 7 are comigrating signals of DNA (red) and Cy3b-σ70 (green), indicating that the labeled protein is fully functional for promoter binding.
In a second experiment, conventional cysteine labeling (1) was compared to the SSL method. Maleimide-Cy3b and maleimide-Alexa 647 were conjugated to a σ70 mutant that has a unique cysteine at residue 59 (alanine to cysteine - A59C). Normalized spectra of Cy3b-A59C (Figure 2B) and Alexa 647-A59C (Figure 2C) labeled by SSL (red) and conventionally (blue). A cysteine-free Δcys σ70 control was also subjected to the same SSL procedure (green). Labeling yields were calculated from these spectra. For SSL labeling, the calculated yields were 76% (Cy3b) and 65% (Alexa 647). For conventional labeling, the calculated yields were 33% (Cy3b) and 24% (Alexa 647).
The specificity of SSL labeling was estimated by a comparison of the UV/vis absorbance spectra of the labeled protein with a cysteine-free protein (Δcys σ70). The labeling of the cysteine-free mutant was less than 5% efficient. The labeling specificity was also confirmed by a cysteine-specific cleavage assay (29), which did not cleave the conjugated cysteine residue. Figure S1 shows an example, Cy3b-A59C, that indicates the conjugation is specific to the cysteine residue.
The labeled proteins were also analyzed by SDS-PAGE followed by fluorescence imaging. In the case of Cy3b-maleimide labeling at residue 59, the mobility of the labeled protein was faster than the unlabeled protein (inset of Figure 2B). The reason for the difference in mobilities is unknown. We note, though, that the region containing residue 59 is highly negatively charged, and the hydrophobic Cy3B dye might change some local conformation even in the presence of SDS. Although unexplained, this mobility shift provides another assessment of the labeling efficiency via the intensity ratio of the labeled/unlabeled bands in the Coomassie-stained gel (inset of Figure 2B).
SSL-labeled σ70 molecules with Cy3b were analyzed for functionality by a gel-shift assay (Figure 2D). The molecules were added to core enzyme RNA polymerase (RNAP) molecules to form holoenzyme molecules. Holoenzymes were added to promoter DNAs and challenged with heparin to compete off nonspecific protein-DNA interactions (34). Lane 3 of Figure 2D clearly shows that the labeled σ70 is competent for forming holoenzyme. Lanes 6 and 7 of the same figure clearly show that holoenzyme is fully functional for binding promoter DNA. Labeled σ70 proteins have been successfully applied to single molecule and ensemble biophysical assays and showed high activity (21). In addition, SSL method was also successfully applied to conjugation of E. colicatabolite activator protein (CAP) as well as σ70 with benzophenone-maleimide (Y. Kim, Y. Ebenstein and S. Weiss, unpublished result).
To demonstrate the general applicability of the approach, SSL was also used to label a small library of single-cysteine Shewanella oneidensis MR proteins with several maleimide dyes (Table 1). σ24, which contained an inaccessible cysteine in the wild-type sequence, was modified such that the inaccessible cysteine was replaced with serine, and an accessible cysteine was added to the N-terminus. σ32 contained a unique surfaced exposed cysteine in the wild type sequence that was used for labeling. The remainder of the proteins (originally cysteine-free in the wild type) were labeled via introduction of a unique single cysteine in the N-terminal fusion tag. As shown in Table 1, the proteins were efficiently labeled, and more importantly, multiple samples at a time were labeled simultaneously with high efficiencies because the crucial steps were easily synchronized by the brief centrifugation. These results demonstrate that SSL could be implemented in a titer-plate format, allowing the site-specific and stoichiometric labeling of a large library of proteins in a high throughput fashion.
Table 1.
SSL labeling Efficiencies of Several Single-Cysteine Shewanella oneidensis MR-1 Proteins
| protein | labeling efficiency | dye |
|---|---|---|
| MutT/nudix family proteinaa | 81% | TMR |
| Nitrogen regulatory protein P-II 1 (glnB-1)a | 75% | Cy3B |
| Peptidyl-prolyl cis–trans isomerase FklBa | 75% | TMR |
| dnaK, chaperone proteina | 80% | TMR |
| rpoE, sigma-24 factora | 80%, 94% | Cy3B, Alexa488 |
| rpoH, sigma-32 factorb | 75% | Cy3B |
| Integration host factor alphaa | 90% | Cy3B |
| Integration host factor betaa | 85% | Cy3B |
A unique single cysteine was introduced near the N-terminus (wild type is cystene-free).
Contains a unique surface exposed single cysteine in the wild type.
In conclusion, we developed an improved general procedure for maleimide-based labeling of cysteine containing proteins. This procedure features high labeling efficiency, specificity and simplicity. It is cost-effective and potentially could be used to label a large number of proteins in a high throughput format, offering a platform for proteome-wide analysis. Our approach could be further expanded to other types of thiol labeling such as haloacetyl, alkyl halide, and disulfide interchange derivatives. Current efforts in our laboratory include the utilization of this technique to the studies of protein–protein and protein-DNA interactions of E. coli and Shewanella oneidensis MR-1 transcription machineries by single molecule fluorescence spectroscopy (19, 22).
ACKNOWLEDGMENT
We thank Dr. Ekaterina Korthonjia, Mr. Yun Cheol Kim, Dr. Uljana Mayer-Cumblidge and Dr. Margie Romine for valuable discussions, and Janet Chiang and Dr. Jeanne Perry for the preparation of plasmids for this project. This work was supported by the U.S. Department of Energy, Grant number DE-FG02-04ER63938 and the NIH, Grant number GM069709-01.
Footnotes
Supporting Information Available: Cysteine-specific cleavage results showing the specificity of labeling. This material is available free of charge via the Internet at http://pubs.acs.org/ BC.
LITERATURE CITED
- 1.Hermanson GT. Bioconjugate techniques. Academic Press; San Diego: 1996. [Google Scholar]
- 2.Aslam M, Dent A. Bioconjugation: Protein coupling techniques for biomedical sciences. MacMillan Reference Ltd.; London: 1998. [Google Scholar]
- 3.Kapanidis AN, Weiss S. Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules. J. Chem. Phys. 2002;117:10953–10964. [Google Scholar]
- 4.Schuler B. Single-molecule fluorescence spectroscopy of protein folding. ChemPhysChem. 2005;6:1206–1220. doi: 10.1002/cphc.200400609. [DOI] [PubMed] [Google Scholar]
- 5.Michalet X, Weiss S, Jager M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem. ReV. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holmes KL, Lantz LM. Protein labeling with fluorescent probes. Methods Cell Biol. 2001;63:185–204. doi: 10.1016/s0091-679x(01)63013-9. [DOI] [PubMed] [Google Scholar]
- 7.Gentle IE, De Souza DP, Baca M. Direct production of proteins with N-terminal cysteine for site-specific conjugation. Bioconjugate Chem. 2004;15:658–63. doi: 10.1021/bc049965o. [DOI] [PubMed] [Google Scholar]
- 8.Scheck RA, Francis MB. Regioselective labeling of antibodies through N-terminal transamination. ACS Chem. Biol. 2007;2:247–51. doi: 10.1021/cb6003959. [DOI] [PubMed] [Google Scholar]
- 9.Lin CW, Ting AY. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 2006;128:4542–3. doi: 10.1021/ja0604111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jager M, Nir E, Weiss S. Site-specific labeling of proteins for single-molecule FRET by combining chemical and enzymatic modification. Protein Sci. 2006;15:640–646. doi: 10.1110/ps.051851506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–9. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 12.Duckworth BP, Zhang Z, Hosokawa A, Distefano MD. Selective labeling of proteins by using protein farnesyltransferase. ChemBioChem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 13.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–7. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 14.Lin FL, Hoyt HM, van Halbeek H, Bergman RG, Bertozzi CR. Mechanistic investigation of the staudinger ligation. J. Am. Chem. Soc. 2005;127:2686–95. doi: 10.1021/ja044461m. [DOI] [PubMed] [Google Scholar]
- 15.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–76. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 16.Kapanidis AN, Ebright YW, Ebright RH. Site-specific incorporation of fluorescent probes into protein: hexahistidine-tag-mediated fluorescent labeling with (Ni(2+): nitrilotriacetic Acid (n)-fluorochrome conjugates. J. Am. Chem. Soc. 2001;123:12123–5. doi: 10.1021/ja017074a. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH. Translocation of sigma(70) with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell. 2001;106:453–63. doi: 10.1016/s0092-8674(01)00464-0. [DOI] [PubMed] [Google Scholar]
- 18.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 19.Kapanidis AN, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, Kortkhonjia E, Mekler V, Ebright RH, Weiss S. Retention of transcription initiation factor s 70 in transcription elongation: single-molecule analysis. Mol. Cell. 2005;20:347–356. doi: 10.1016/j.molcel.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 20.Margeat E, Kapanidis AN, Tinnefeld P, Wang Y, Mukhopadhyay J, Ebright RH, Weiss S. Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys. J. 2006;90:1419–1431. doi: 10.1529/biophysj.105.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee NK, Kapanidis AN, Koh HR, Korlann Y, Ho SO, Kim Y, Gassman N, Kim SK, Weiss S. Three-color alternating-laser excitation of single molecules: monitoring multiple interactions and distances. Biophys. J. 2007;92:303–12. doi: 10.1529/biophysj.106.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapanidis AN, Margeat E, Ho SO, Kortkhonjia E, Weiss S, Ebright RH. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–7. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8936–41. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gralnick JA, Hau HH. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 2006 doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 25.Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, Ringelberg DB, White DC, Nishijima M, Sano H, Burghardt J, Stackebrandt E, Nealson KH. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 1999;49:705–24. doi: 10.1099/00207713-49-2-705. [DOI] [PubMed] [Google Scholar]
- 26.Tiedje JM. Shewanella--the environmentally versatile genome. Nat. Biotechnol. 2002;20:1118–23. doi: 10.1038/nbt1102-1093. [DOI] [PubMed] [Google Scholar]
- 27.Dieckman LJ, Hanly WC, Collart ER. Strategies for high-throughput gene cloning and expression. Genet. Eng. (NY) 2006;27:179–90. doi: 10.1007/0-387-25856-6_10. [DOI] [PubMed] [Google Scholar]
- 28.Lin CT, Moore PA, Auberry DL, Landorf EV, Peppler T, Victry KD, Collart FR, Kery V. Automated purification of recombinant proteins: combining high-throughput with high yield. Protein Expr. Purif. 2006;47:16–24. doi: 10.1016/j.pep.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH. Structural organization of the RNA polymerase-promoter open complex. Cell. 2000;101:601–11. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 30.Levison ME, Josephson AS, Kirschenbaum DM. Reduction of biological substances by water-soluble phosphines: gamma-globulin (IgG). Experientia. 1969;25:126–7. doi: 10.1007/BF01899076. [DOI] [PubMed] [Google Scholar]
- 31.Getz EB, Xiao M, Chakrabarty T, Cooke R, Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal. Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- 32.Tyagarajan K, Pretzer E, Wiktorowicz JE. Thiol-reactive dyes for fluorescence labeling of proteomic samples. Electrophoresis. 2003;24:2348–58. doi: 10.1002/elps.200305478. [DOI] [PubMed] [Google Scholar]
- 33.Englard S, Seifter S. Precipitation techniques. Methods Enzymol. 1990;182:285–300. doi: 10.1016/0076-6879(90)82024-v. [DOI] [PubMed] [Google Scholar]
- 34.Cech CL, McClure WR. Characterization of ribonucleic acid polymerase-T7 romoter binary complexes. Biochemistry. 1980;19:2440–2447. doi: 10.1021/bi00552a023. [DOI] [PubMed] [Google Scholar]