Abstract
Real-time tracking of kinase activity in living systems has revealed new modes of encoding signaling information into the spatiotemporal activity patterns and opened new avenues for screening kinase modulators. However, the sensitivity of kinase activity detection, which is commonly coupled to a FRET-based readout, has often been a limiting factor. Here we show a kinase-inducible bimolecular switch, consisting of a substrate for the kinase of interest and a phosphoamino acid binding domain, can be designed to sense different kinase activities and coupled to various readouts, thereby allowing for examination of dynamic kinase activity with increased sensitivity and versatility. Specifically, we demonstrate that bimolecular switches designed to sense Protein Kinase A (PKA) or Protein Kinase C (PKC) activities can turn on FRET as well as bioluminescence signals. Notably, the FRET-based sensors gain larger dynamic ranges when compared to their unimolecular counterparts; the novel bioluminescence-based reporters for PKA and PKC show high sensitivity and a unique capability to detect basal kinase activities and should enable new applications in in vivo imaging of kinase activity and high-throughput compound screening. Thus, this generalizable design advances the molecular toolkit of kinase activity detection and provides a means for versatile and sensitive detection of kinase activity in various biological systems.
Protein kinases are critical regulators of signal transduction in all eukaryotic organisms and help to translate an environmental cue into a specific cellular response. To understand how these key signaling molecules achieve their specific functions, powerful tools such as FRET-based kinase activity reporters have emerged for investigating the spatiotemporal regulation of protein kinases in the native context of the cell, tissue, or organism1–13. The kinase activity reporters (KARs) utilize a kinase-sensitive molecular switch flanked by a FRET pair as the reporting unit to indicate a phosphorylation event with a change in FRET14. While these probes have proven to be valuable tools to study dynamic kinase activity in living systems, they often suffer from limited dynamic range15,16, which is a critical parameter for optimal kinase activity detection2,17. For instance, using a biosensor with limited dynamic range makes it difficult to detect subtle yet physiologically relevant changes in kinase activity7, 10. Furthermore, kinase activity reporters that utilize readout other than FRET could offer significant advantages. For example, bioluminescence-based reporters, without the need of exogenous illumination, can eliminate background fluorescence signals and achieve superior sensitivity, thereby enabling new applications such as in vivo imaging of kinase activity18–22 and high-throughput compound screening18,23. Therefore, to achieve sensitive and versatile detection of endogenous kinase activity in various biological systems, we sought to develop a kinase-inducible bimolecular switch (KIBS) that could be coupled to both FRET and bioluminescence reporting units. We show that the FRET-based biosensors display enhanced dynamic range compared to their unimolecular counterparts and that the bioluminescence-based biosensors are sensitive at detecting basal kinase activities. Moreover, these KIBS-based designs are applicable to different kinases, thus providing a new general strategy for developing sensitive kinase activity reporters.
This kinase-inducible bimolecular switch consists of a kinase-specific substrate and a phosphoamino acid binding domain (PAABD) designed to bind to the phosphorylated form of the substrate, each fused to a separate reporting unit. Upon phosphorylation of the substrate, intermolecular binding of PAABD to the phosphorylated substrate brings Reporting Unit A and B into close proximity, generating a FRET or bioluminescence signal (Scheme 1). To test the KIBS design in the context of FRET-based biosensors, we generated an A-Kinase-inducible bimolecular switch (A-KIBS) from a previously developed unimolecular A-Kinase Activity Reporter, AKAR24, and utilized it to construct a FRET-based Bimolecular A-Kinase Activity Reporter (BimAKAR) (Figure 1a). This reporter consists of a PKA substrate fused to YPet, a YFP variant25, and a phosphothreonine-binding Forkhead-associated (FHA) domain fused to Cerulean, a CFP variant26, each targeted to the cytosol with a nuclear export sequence (NES). In HEK293T cells treated with the adenylyl cyclase agonist forskolin (Fsk), BimAKAR generated a marked increase in yellow/cyan emission ratio of 53.5% ± 3.0% (n = 9) (mean ± SEM, n = number of cells) (Figure 1b,c). In contrast, when BimAKAR expressing HEK293T cells were pretreated for 10 minutes with the relatively selective PKA inhibitor, H89, the Fsk-induced response was abolished (Figure 1b, red), confirming that the bimolecular switch is specific for PKA. The specificity of A-KIBS was further confirmed in HeLa cells where stimulation with the diacylglycerol mimic phorbol 12-myristate 13-acetate (PMA), a PKC activator, failed to generate a response from BimAKAR, yet Fsk stimulation produced a robust response (Supporting Figure 1). Furthermore, co-stimulation of BimAKAR expressing HeLa cells with Fsk and PMA induced a response comparable to that observed with Fsk stimulation alone (Supporting Figure 1c), suggesting presence of additional endogenously phosphorylated substrates does not restrict the response of the bimolecular biosensor. Finally, when the phospho-acceptor threonine of the PKA substrate was mutated to an alanine, application of Fsk did not elicit a detectable response (Figure 1b, black), suggesting that the FRET increase of BimAKAR results from PKA-dependent phosphorylation at this designed threonine, as in the case of unimolecular AKAR27.
Scheme 1.
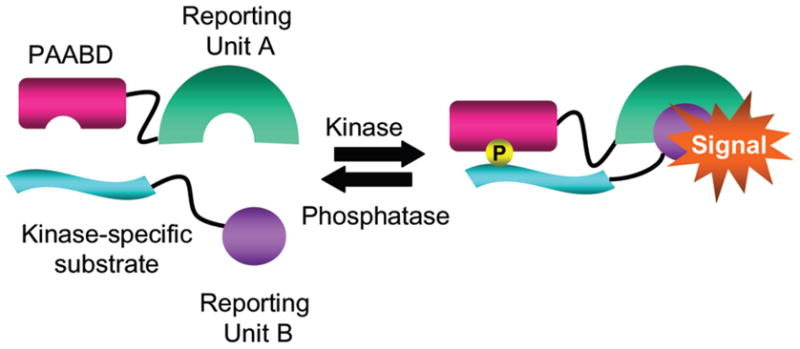
A Kinase-inducible Bimolecular Switch (KIBS) coupled to reporting units.
Figure 1.
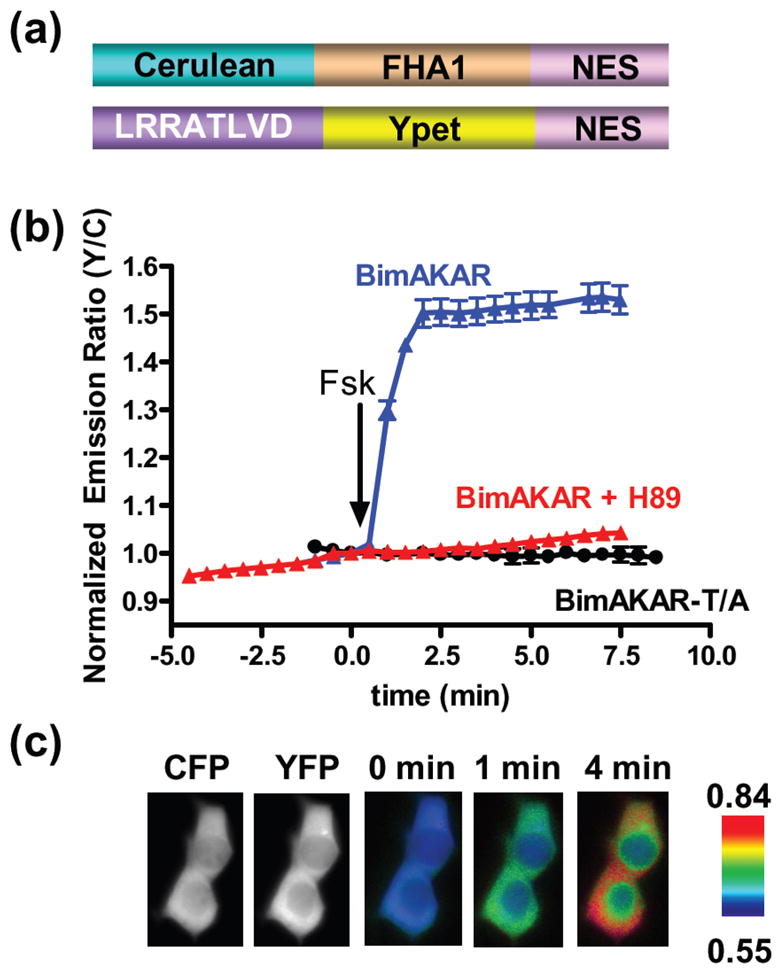
Design and characterization of BimAKAR. (a) Schematic representation of BimAKAR. (b) Time courses of Fsk (50 μM)-treated HEK293T cells expressing BimAKAR (n = 9), BimAKAR in the presence of 10 μM H89 (n = 12), and BimAKAR-T/A (n = 4); mean ± SEM. (c) CFP and YFP images show the localization of each portion of BimAKAR, and pseudocolor images show the response of BimAKAR to Fsk.
One important advantage of genetically-encoded KARs is that they can be targeted, using subcellular localization signals, to distinct cellular compartments where they can detect local kinase activity24. By targeting the substrate-YPet unit of BimAKAR to the plasma membrane via a CAAX targeting sequence derived from K-Ras28 and leaving the Cerulean-FHAI portion cytoplasmic, we showed that this PKA-specific KIBS is still functional when separated into different cellular compartments and can be harnessed for detecting local PKA activity. Specifically, treatment of these cells with Fsk caused a 44.5% ± 4.5% (n = 9) increase in yellow/cyan emission (Supporting Figure 2). Markedly, the dynamic range of this sensor is over 5-fold larger than the equivalent unimolecular biosensor (AKAR4-Kras) which displays a maximal Fsk-stimulated response of 8.2 ± 0.9% in HEK293T cells28. The improvement in dynamic range over the unimolecular equivalent may be a result of lowering initial FRET due to physical separation of the donor and acceptor FPs16, and this data suggests that the bimolecular design may be a particularly effective method to study local kinase activity with enhanced sensitivity.
To test the generalizability of the KIBS design, we generated a C-Kinase inducible bimolecular switch (C-KIBS) by replacing the PKA-specific substrate in the A-KIBS with a PKC-specific substrate sequence (Supporting Figure 3a). In HeLa cells the resultant biosensor, Bimolecular C-Kinase Activity Reporter (BimCKAR), detects PKC activity by generating an emission ratio increase of 26.2 ± 2.1% (n = 7) in response to treatment with PMA, and this response is further enhanced to 34.0 ± 2.6% following treatment with the calcium ionophore Ionomycin (Iono) (Supporting Figure 3b, blue, 3c). In contrast, treatment with Fsk did not generate a FRET increase in HeLa cells expressing BimCKAR (Supporting Figure 3d) and pretreatment of HeLa cells expressing BimCKAR with the PKC inhibitor Gö6983 abolished the PMA-induced FRET increase (Supporting Figure 3b, red). Together these data demonstrate the specificity of BimCKAR for PKC. Furthermore, no PMA-induced response was observed in cells expressing BimCKAR-T/A (Supporting Figure 3b, black). Notably, the observed PMA-induced FRET change in BimCKAR is substantially larger than that of the unimolecular CKAR which shows an increase in cyan over yellow emission of less than 10% to a diacylglycerol mimic5. This improvement should alleviate the difficulty in using the current CKAR and facilitate the study of PKC activity dynamics in physiologically relevant systems. Together these data not only show that the KIBS design is generalizable, but also suggest that coupling a KIBS to a FRET pair might be a general method to improve the dynamic range of current unimolecular KARs.
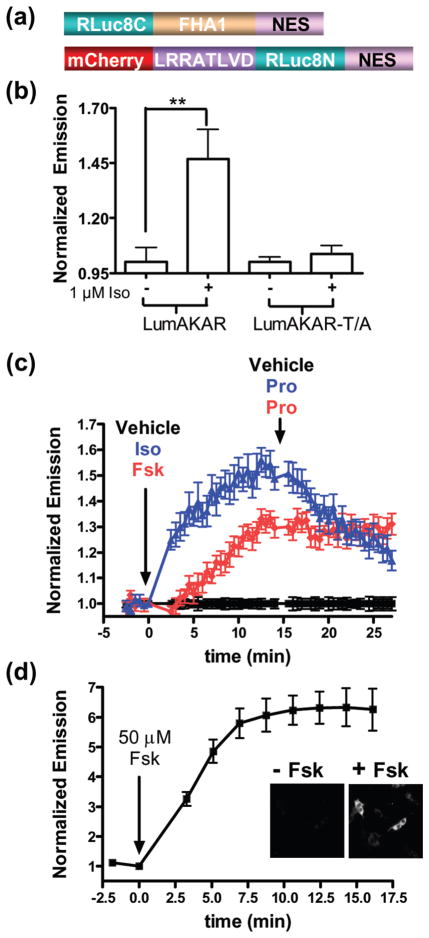
We next investigated whether this bimolecular switch can turn on bioluminescence as readout of kinase activity. We chose to couple the bimolecular switch to the fragment complementation of the bioluminescent protein Renilla luciferase (RLuc)29 for two reasons. First, the fragment complementation of RLuc displays high signal to noise and is sensitive enough to monitor signaling events in single cells29. Second, this complementation, unlike the complementation of many fluorescent proteins30, is reversible, and can thus track dynamic signaling events29. To generate a bioluminescence-based PKA activity biosensor19,29, FHA1 was tagged with the C-terminal fragment (a.a. 111–311) of the enhanced RLuc variant, RLuc831, 32, and the PKA-specific substrate with the N-terminal fragment of RLuc8 (a.a.1–110) (Figure 2a). mCherry33 was fused to the N-terminus of the PKA substrate as a marker for expression, and is not likely to be involved in resonance energy transfer with RLuc8 (data not shown). In the presence of benzyl-coelenterazine HEK293T cells expressing this bioluminescence-based AKAR (LumAKAR) treated with the β-adrenergic receptor (β-AR) agonist isoproterenol (Iso) for 10 minutes showed a 46.8% ± 13% (n = 4) increase in bioluminescence when compared to vehicle treated cells, whereas the threonine to alanine mutation abolished the sensitivity of LumAKAR to PKA (n = 4) (Figure 2b), thus validating the functionality of A-KIBS in the context of the bioluminescent biosensor.
Figure 2.
Design and characterization of LumAKAR in HEK293T cells. (a) Schematic representation of LumAKAR. mCherry serves as a transfection marker. (b) Average response of LumAKAR and LumAKAR-T/A to Iso (1 μM) or vehicle DMSO (n = 4 for all treatments) ** p < 0.01. Each sample was normalized to the basal signal before drug addition. (c) Representative response curves of LumAKAR to vehicle control (black), Iso (1 μM) then Pro (10 μM) (blue) or Fsk (50 μM) then Pro (red) (n = 7–8 for all treatments). Each sample was normalized to the basal signal before drug addition and then normalized to the average vehicle control reading at each time point. (d) Average response curve of cells expressing LumAKAR in response to Fsk (50 μM) (n = 5) from live-cell imaging experiments. The inset shows the bioluminescence intensity images of single cells expressing LumAKAR showing the cytoplasmic localization of LumAKAR before (left) and after (right) drug addition. All assays are in the presence of benzyl-coelenterazine (5 μM) and all values are mean ± SEM.
To further test the capacity of LumAKAR, transiently transfected HEK293T cells were treated with different drugs and tracked over time to monitor dynamic PKA activity. While addition of the vehicle DMSO caused little change in detectable PKA activity (Figure 2c, black), treatment of cells with Iso or Fsk caused a bioluminescence increase of 48.9 ± 4.4% (n = 7) or 29.6 ± 3.9% (n = 7), respectively. Importantly, the kinetics of the increase in signal induced by the different compounds corresponds to the expected increases in cAMP, and thus onset of PKA activity, triggered by the different treatments28,34. Specifically, Iso induces more rapid cAMP accumulation than the AC activator Fsk34, and this discrepancy can be detected with LumAKAR (Fig. 2c) suggesting that this method, like its FRET-based counterpart, has the desired temporal resolution for distinguishing different signals that activate the signaling pathway. Next, to investigate the reversibility of LumAKAR, we added the β-AR antagonist propranolol (Pro) to the cells that were first treated with Iso or Fsk. Since Fsk functions downstream of the β-AR, there was no reduction in signal after Pro addition (Figure 2c, red). In contrast, in cells that were first treated with Iso, Pro treatment dramatically decreased the bioluminescence signal, reversing it to basal levels within 20 minutes (Figure 2c, blue, Supporting Figure 4), thus demonstrating that LumAKAR is a reversible biosensor. Together, this data shows that LumAKAR can accurately detect dynamic PKA activity in a population of living cells in real time.
We next sought to test if LumAKAR could detect PKA activity with subcellular resolution. When HEK293T cells expressing nuclear excluded LumAKAR were imaged in the presence of benzyl-coelenterazine, PKA activity was detected specifically in the cytoplasm such that treatment of the cells with Fsk caused a six-fold increase in signal above baseline (n = 5) (Figure 2d). Moreover, no bioluminescence signal was detected with a 4 min exposure from HEK293T cells expressing the threonine to alanine mutant of LumAKAR (data not shown), suggesting that the increase in LumAKAR bioluminescence results from PKA-dependent phosphorylation at the designed threonine. This data further suggests that the baseline bioluminescence signal of LumAKAR prior to any stimulation mainly results from basal PKA activity. Consistent with this observation, the basal signal detected on a platereader from a population of cells expressing WT LumAKAR is on average five-fold higher than that of cells expressing LumAKAR-T/A. Compared with other kinase activity biosensors, LumAKAR appears to detect even the low amount of basal PKA activity that exists in un-stimulated cells, which was often undetectable with the previous FRET-based AKARs except at specific subcellular locations such as mitochondria and membrane rafts24, 28. Accordingly, this biosensor will be an important addition to the molecular toolkit of PKA activity detection as it can be useful in characterizing the basal states of the cAMP/PKA pathway in different cell types. In summary, the KIBS design has led to development of a novel bioluminescence-based kinase activity biosensor for detecting PKA activity with several valuable features including high sensitivity, large dynamic range, desirable kinetic properties, reversibility and subcellular resolution in living cells.
To demonstrate that LumAKAR, like BimAKAR, can be adopted to detect the activity of other kinases, we replaced the PKA-specific substrate in LumAKAR with a PKC-specific substrate. In HEK293T cells expressing bioluminescence-based CKAR (LumCKAR) (Supporting Figure 5a), treatment with PMA plus ionomycin (Iono) generated a 24.1± 5.6 % (n = 5) increase in bioluminescence compared to control (Supporting Figure 5b). Conversely, cells treated with PKC inhibitor Gö6983 showed a 10.1 ± 3.3 % (n = 5) decrease in bioluminescence signal relative to control. This Gö6983-induced decrease in bioluminescence signal not only illustrates the dependence of the LumCKAR response on PKC activity but also shows that LumCKAR, like LumAKAR, detects basal kinase activity in HEK293T cells. Importantly, HEK293T cells expressing LumCKAR in which the target threonine is mutated to alanine showed no change in bioluminescence relative to vehicle treated cells when treated with either PMA plus Iono or with Gö6983, further confirming that the LumCKAR signal results from PKC-dependent phosphorylation at this designed threonine (Supporting Figure 5b).
In conclusion we have enhanced the molecular toolkit of kinase activity detection via the design of a versatile kinase-inducible bimolecular switch that can be adopted to detect real-time kinase activity dynamics with either fluorescence or bioluminescence readout in living cells. This biosensor design has led to the development of FRET biosensors with significantly improved dynamic range as well as novel bioluminescence-based PKA and PKC activity biosensors with superior sensitivity and enhanced capabilities. Importantly, these KIBS-based bioluminescence kinase activity biosensors represent a new class of activity biosensors that open new avenues for studying dynamic kinase activities in living systems and for developing and characterizing kinase inhibitors and activators. For instance, in vivo imaging with kinase activity biosensors provides a powerful assay for in vivo validation of drug-target interactions10, 35, but it has been challenging to apply FRET-based kinase activity biosensors in vivo due to various factors including autofluorescence from cells and tissues. In the same vein, interference of fluorescent compounds limits the use of FRET-based kinase activity reporter in live-cell high-throughput compound screening. Therefore, bioluminesence-based AKAR and CKAR should find great use in both of these applications. Also in this report, we show that this KIBS design is generalizable both in terms of kinase specificity and readout. Consequently, in much the same way that BimCKAR and LumCKAR were generated from their AKAR counterparts, it is likely that novel luminescent KARs could easily be generated by using a KIBS specific for a kinase of interest. Thus this new technology provides a generalizable method to diversify the readout of kinase activity detection, and should have an impact in facilitating the study of specific kinase activities in their native environments.
Supplementary Material
Acknowledgments
This work was funded by NIH R01 DK073368, DP1 OD006419, and 3M (to J. Z.).
Footnotes
Supporting Information Available: Experimental methods and supplementary figures can be found online free of charge via the internet at http://pubs.acs.org.
References
- 1.Herbst KJ, Ni Q, Zhang J. IUBMB Life. 2009:61. doi: 10.1002/iub.232. spcone. [DOI] [PubMed] [Google Scholar]
- 2.VanEngelenburg SB, Palmer AE. Curr Opin Chem Biol. 2008;12:60–65. doi: 10.1016/j.cbpa.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Ma Y, Taylor SS, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuller BG, Lampson MA, Foley EA, Rosasco-Nitcher S, Le KV, Tobelmann P, Brautigan DL, Stukenberg PT, Kapoor TM. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Violin JD, Zhang J, Tsien RY, Newton AC. J Cell Biol. 2003;161:899–909. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ting AY, Kain KH, Klemke RL, Tsien RY. Proc Natl Acad Sci U S A. 2001;98:15003–15008. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, Svoboda K. Proc Natl Acad Sci U S A. 2008;105:19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M, Ozawa T, Inukai K, Asano T, Umezawa Y. Nat Biotechnol. 2002;20:287–294. doi: 10.1038/nbt0302-287. [DOI] [PubMed] [Google Scholar]
- 9.Kurokawa K, Mochizuki N, Ohba Y, Mizuno H, Miyawaki A, Matsuda M. J Biol Chem. 2001;276:31305–31310. doi: 10.1074/jbc.M104341200. [DOI] [PubMed] [Google Scholar]
- 10.Allen MD, DiPilato LM, Rahdar M, Ren YR, Chong C, Liu JO, Zhang J. ACS Chem Biol. 2006;1:371–376. doi: 10.1021/cb600202f. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 12.Zondlo SC, Gao F, Zondlo NJ. J Am Chem Soc. 2010;132:5619–5621. doi: 10.1021/ja100862u. [DOI] [PubMed] [Google Scholar]
- 13.Ray-Saha S, Schepartz A. Chembiochem. 2010;11:2089–2091. doi: 10.1002/cbic.201000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Allen MD. Mol Biosyst. 2007;3:759–765. doi: 10.1039/b706628g. [DOI] [PubMed] [Google Scholar]
- 15.Kerppola TK. Nat Methods. 2006;3:969–971. doi: 10.1038/nmeth1206-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodgson L, Shen F, Hahn K. Curr Protoc Cell Biol. 2010;Chapter 14(Unit 14.11.1–26) doi: 10.1002/0471143030.cb1411s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni Q, Ganesan A, Aye-Han NN, Gao X, Allen MD, Levchenko A, Zhang J. Nat Chem Biol. 2011;7:34–40. doi: 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boute N, Jockers R, Issad T. Trends Pharmacol Sci. 2002;23:351–354. doi: 10.1016/s0165-6147(02)02062-x. [DOI] [PubMed] [Google Scholar]
- 19.Willoughby D, Cooper DM. Nat Methods. 2008;5:29–36. doi: 10.1038/nmeth1135. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Lee KC, Bhojani MS, Khan AP, Shilman A, Holland EC, Ross BD, Rehemtulla A. Nat Med. 2007;13:1114–1119. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- 21.Nyati S, Ranga R, Ross BD, Rehemtulla A, Bhojani MS. Anal Biochem. 2010;405:246–254. doi: 10.1016/j.ab.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan CT, Paulmurugan R, Reeves RE, Solow-Cordero D, Gambhir SS. Mol Imaging Biol. 2009;11:144–158. doi: 10.1007/s11307-008-0187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen MD, Zhang J. Angew Chem Int Ed Engl. 2008;47:500–502. doi: 10.1002/anie.200703493. [DOI] [PubMed] [Google Scholar]
- 24.Allen MD, Zhang J. Biochem Biophys Res Commun. 2006;348:716–721. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen AW, Daugherty PS. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 26.Rizzo MA, Springer GH, Granada B, Piston DW. Nat Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Hupfeld CJ, Taylor SS, Olefsky JM, Tsien RY. Nature. 2005;437:569–573. doi: 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- 28.Depry C, Allen MD, Zhang J. Mol Biosyst. 2011;7:52–58. doi: 10.1039/c0mb00079e. [DOI] [PubMed] [Google Scholar]
- 29.Stefan E, Aquin S, Berger N, Landry CR, Nyfeler B, Bouvier M, Michnick SW. Proc Natl Acad Sci U S A. 2007;104:16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- 31.Ni Q, Titov DV, Zhang J. Methods. 2006;40:279–286. doi: 10.1016/j.ymeth.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Loening AM, Fenn TD, Wu AM, Gambhir SS. Protein Eng Des Sel. 2006;19:391–400. doi: 10.1093/protein/gzl023. [DOI] [PubMed] [Google Scholar]
- 33.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 34.DiPilato LM, Cheng X, Zhang J. Proc Natl Acad Sci U S A. 2004;101:16513–16518. doi: 10.1073/pnas.0405973101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Connor KA, Roth BL. Nat Rev Drug Discov. 2005;4:1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.