Abstract
Synthesis of well-defined homodimeric protein-polymer conjugates using RAFT polymerization is described.
In nature, proteins often exist as dimers or multimers, or they self assemble into higher order oligomers in the active state.1 In many cases, this type of multimerization is critical for induction and regulation of biological processes.2 For example, approximately 70% (311) out of the total human enzymes (452) are multimers, and many of the multimers are homodimers (125).3 Cytokines, an important class of cell signaling molecules, often dimerize in the active state, and this association is crucial for tyrosine kinase receptor activation. Thus, by locking proteins into conjugate designs that present multiple copies of the biomolecules, one can study multivalent interactions in biological processes such as signal transduction and prepare therapeutics with higher activity than monovalent counterparts.1, 4, 5 For example, epidermal growth factor (EGF) is a well studied cytokine that regulates cell adhesion, proliferation, differentiation, and invasion. It is a monomeric protein that dimerizes in the active site. This dimerization causes two EGF receptors (EGFRs) to associate resulting in phosphorylation and cell signaling.6, 7 This suggests that tethering together two copies of EGF would provide a material with superior properties. Indeed, a dendrimer modified with an average of two EGFs exhibited significantly higher affinity to cells that over express EGFR than did free EGF.8 In another example, cyclic guanosine monophsophate (GMP) polymer dimers were able to bind to cyclic-nucleotide-gated channels with significantly higher affinity and were up to a thousand times more potent than the ligand alone.9 Yet despite these advantages, the majority of protein-polymer conjugates used in therapeutics and other applications consist of one protein bound to one or more polymers. This is likely due to the lack of available strategies to synthesize the polymers, particularly those that react with proteins to produce conjugates of defined stoichiometry. Herein we describe the efficient synthesis of protein reactive telechelic polymers by reversible addition-fragmentation chain transfer (RAFT) polymerization and their use to produce homodimeric protein-polymer conjugates.
RAFT polymerization was chosen because it enables predictable molecular weights10 and reactive chain ends that are susceptible to reducing or radical coupling reactions.11, 12 The ability to target molecular weight is important because biological activities of dimers can be significantly affected by the polymer spacer length.9 The chain end reactivity provides a convenient way to install a protein reactive group. These post-polymerization modification techniques have been recently used to synthesize heterodimeric protein-polymer conjugates by us and others.13, 14 But to our knowledge, homodimeric conjugates have not been synthesized by any controlled radical polymerization technique.
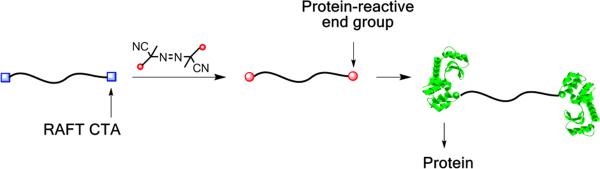
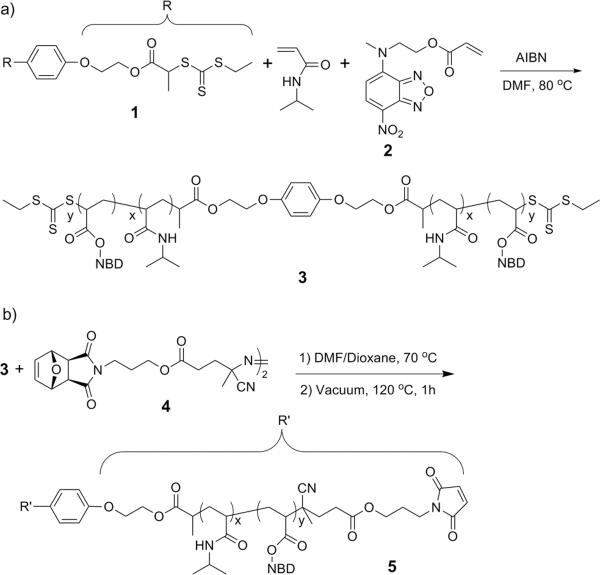
The strategy we employed was polymerization from a bistrithiocarbonate chain transfer agent (CTA), radical exchange of the trithiocarbonates with a protein reactive azo-initiator, followed by conjugation of the protein (Scheme 1). Specifically, a difunctional polyNIPAAm (pNIPAAm) was synthesized by RAFT polymerization (Scheme 2a) using a bisfunctionalized CTA (1). PNIPAAm was chosen because it responds to changes in temperature, and thus is utilized in numerous biotechnology applications.15 A small percentage (0.20 % of total monomer) of a monomer containing nitrobenz-2-oxa-1,3-diazole (NBD, 2) was incorporated because it provides a way to verify conjugate formation.16 Polymerization (initial ratios of [NIPAAm+NBD]:[1]:[AIBN] = 100:1:0.1) was carried out to 82% conversion under an argon atmosphere at 80 °C and the resultant difunctional pNIPAAm (3) was isolated after extensive purification by dialysis. The polymer had a low polydispersity index (PDI) of 1.06 and a number average molecular weight (Mn) of 15,500 Da by gel permeation chromatography (GPC) (Figure S2, ESI). The Mn was higher than the expected molecular weight of 9,800 Da because the value was determined by calibration with poly(methyl methacrylate) standards. To more accurately determine the molecular weight, the samples were subjected to GPC with laser light scattering (LLS) detection (Figure S4a), 1H NMR spectroscopy (Figure S5a, ESI), and matrix-assisted laser desorption/ionization time of flight (MALDITOF) mass spectrometry (Figure S3, ESI) resulting in values of 11,500, 10,500, and 8,990 Da, respectively. These molecular weights were similar to what was predicted, suggesting that the purified polymer had the designed symmetric structure. Utilizing the molecular weight obtained by MALDI-TOF MS, the end group retention was 91%, indicating that the trithiocarbonate groups were available for further elaboration.
Scheme 1.
Straightforward synthesis of dimeric protein conjugates.
Scheme 2.
Synthesis of telechelic maleimide pNIPAAm.
Verification of the dimeric structure was carried out by aminolysis of the polymer with butylamine in order to cleave the middle ester bonds, followed by MALDI-TOF MS analysis (Figure S3, ESI). The molecular weight of the arm was 4,480 Da and the expected mass was 4,470 Da. This result further confirmed that the CTA was effective in producing the desired dimeric polymer.
Radical addition of a functional azo-initiator (4) to the polymer chain ends was chosen as a convenient method to introduce the cysteine-reactive functionality. To avoid any possible cross-linking reactions during radical coupling, the maleimide was protected as the Diels-Alder adduct with furan. Polymer 3 was heated at 70 °C in the presence of 4 in order to form the furan-protected polymer (Scheme 2b). We found that a 4 hour reaction time was optimal for modification of the chain ends without thermal removal of the protecting group. Comparison of the 1H NMR spectra before and after confirmed that the reaction had occurred (Figure S5a, b, ESI). The peak at 3.34 ppm corresponding to the methylene group of the chain-end CTA disappeared while new peaks appeared at 6.51, 5.25, and 2.85 ppm corresponding to the furan-protected maleimide. Additional analysis by UV-Vis at 306 nm gave a zero absorbance, which indicated that all of the trithiocarbonate chain ends had reacted. We were not able to obtain the molecular weight by MALDI-TOF MS. However, we were able to determine the values by NMR and GPC. Comparison of the integral of the peak for the main chain CH of pNIPAAm with those from the end group gave a Mn by 1H NMR of 11,600 Da. This result was the same as what was calculated prior to modification (11,500 Da) suggesting a high end group conversion. Analysis by LLS provided an Mn of 12,700. There was only a slight broadening of the polydispersity index to 1.10 (Figure S4b, ESI). These data further confirmed that the chain end modification reaction had occurred.
Deprotection of the end groups was facile. Upon heating the polymer at 120 °C under vacuum for 1 h, the retro Diels-Alder reaction occurred exposing the maleimides (5, Scheme 2b). During this process, the furan byproduct was removed, and no further purification of the polymer was necessary. The 1H NMR spectrum (Figure S5c, ESI) indicated that the deprotection had occurred as evident by the disappearance of the peaks at 6.51, 5.25, and 2.85 ppm and appearance of a new peak at 6.72 ppm. The polydispersity index remained unchanged (PDI = 1.09, Figure S4c, ESI). These data indicated that side reactions were minimal and that the telechelic maleimide polymer was formed.
Under neutral or slightly acidic pH, maleimides are known to react with thiols approximately 1000 times faster than with amines.17 Therefore, we investigated bioconjugation with a model protein that contains one free cysteine in order to create a site specific protein-polymer conjugate. A V131C mutant T4 lysozyme (T4L) was chosen as the protein. The bioconjugation reaction was carried out at 4 °C in phosphate buffered saline (PBS) in the presence of 10 mM of tris(2-carboxyethyl)phosphine hydrochloride (TCEP) and 10 mM of ethylenediaminetetraacetic acid (EDTA) for 16 h. The salts were removed by centrifugal filtration (MWCO 10,000).
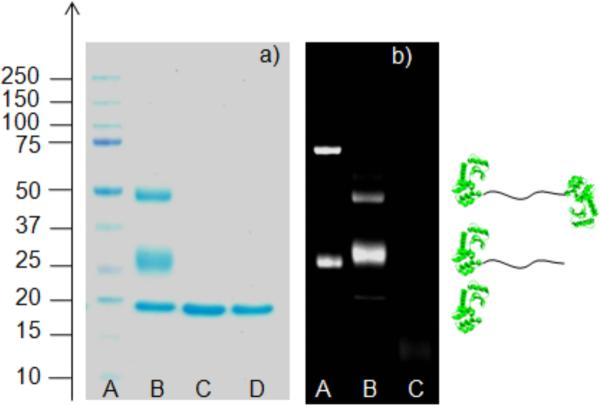
SDS-PAGE analysis indicated that T4L-polymer conjugates formed (Figure 1). The gel with coomassie staining had two new higher molecular weight spots at ~26 kDa and ~48 kDa compared to unmodified T4L at ~18 kDa (Lanes B compared to D). These new bands were attributed to the monomer and dimer protein-polymer conjugate adducts which amounted to 79% and 21% based on fluorescnece quanitification, respectively. The spots were not visible when the control furan protected maleimide polymer was incubated with the protein (lane C). Liquid chromatography mass spectrometry (LC-MS/MS) of the trypsin digests showed that T4L was present in the bands attributed to the monomer and dimer conjugates (see ESI for details). The polymer contained NDB units; thus the gel was also exposed to UV light for visualization. The spots attributed to the monomer and dimer conjugate were observed (Figure 1b, lane B), confirming that polymer was attached to the proteins in these cases. For the control conjugation, again no conjugate was observed (lane C).
Figure 1.
SDS-PAGE of the protein-polymer conjugates a) stained with Coomassie blue, b) visualized with UV light.
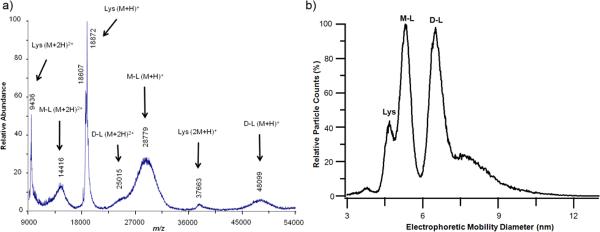
To confirm the PAGE results, analysis by mass spectrometry was undertaken. The theoretical molecular weights of the protein-polymer conjugate and protein-polymer-protein conjugate calculated using the Mn determined by mass spectrometry of the polymer were 27.8 kDa and 46.6 kDa, respectively. The MALDI-TOF spectrum of the conjugate (Figure 2a) displayed peaks at 18.8 kDa, 28.8 kDa, and 48.1 kDa corresponding to unmodified protein, monomeric and dimeric conjugate respectively.
Figure 2.
a) MALDI-TOF MS, b) ESI-GEMMA of the protein-polymer conjugates; relative particle counts axis up to 100 only. Lys: T4L, M-L: monomer linked conjugate (T4L-pNIPAAm), and D-L: dimer linked conjugate (T4L-pNIPAAm-T4L).
Electrospray ionization – gas-phase electrophoretic-mobility macromolecule analysis (ESI-GEMMA) was also performed (Figure 2b).18 ESI-GEMMA separates macromolecules based on their electrophoretic mobility in air. ESI-GEMMA has a higher sensitivity than MALDI-TOF for high mass proteins and polymers. ESI produced, multiply charged particles are subject to charge reduction such that particles detected are mostly singly charged. Thus, the observation of complicated ESI mass spectra of heterogeneous samples and large particles is minimized using the ESI-GEMMA method. The electrophoretic mobility diameter (EMD) of the peaks identified in the ESI-GEMMA spectra and the calculated molecular weights were 4.6 nm and 18 kDa for T4L, 5.3 nm and 27 kDa for the polymer with one protein, and 6.5 nm and 50 kDa for the polymer with two proteins. Based on the standard deviation of the density used to calculate the molecular weights from the EMD values (see ESI), the molecular weights were expected to be within ± 10% of the reported values. Combining the gel electrophoresis, MALDI-TOF and ESI-GEMMA data, it was concluded that the desired protein dimer polymer conjugate formed.
It was not unexpected that monomeric conjugate was observed along with the desired dimer. It is known that macromolecular reaction dynamics can hinder reactions that would be efficient for small molecules.19 To remove the monomer adduct, a standard cation exchange resin was employed and the product eluted with increasing salt concentrations. Analysis by SDS PAGE (Figure S6, ESI) indicated that with 0.14 M NaCl, the monomeric conjugate eluted. The dimer was retained on the column until a concentration of 0.18 M salt was introduced, providing a way to effectively isolate the desired conjugate.
Taken together these data clearly demonstrate that RAFT polymerization, followed by radical addition of a protein-reactive azo-initiator is an effective method to synthesize polymers that produce dimeric protein conjugates. We predict that this straightforward strategy will provide access to dimeric protein polymer conjugates resulting in materials with superior biological activities compared to the monomeric conjugates or unmodified proteins. We are currently exploring these possibilities.
Supplementary Material
Acknowledgments
This work was supported by the NSF (CHE-0809832). HDM thanks Amgen (New Faculty Award) and the Alfred P. Sloan Foundation for further support. The UCLA Functional Proteomics Center was established and equipped by a grant from the W. M. Keck Foundation. JAL acknowledges support from the NIH (RR20004). CSK thanks the Chancellor's Dissertation Year Fellowship and GNG thanks the Christopher S. Foote Graduate Fellowship.
Footnotes
Electronic Supplementary Information (ESI) available: [experimental procedures and data not provided in the text]. See DOI: 10.1039/b000000x/
References
- 1.Mammen M, Choi SK, Whitesides GM. Angew. Chem. Int. Ed. 1998;37:2755–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Thornton JM. Prog. Biophys. Molec. Biol. 1995;63:31–65. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 3.Marianayagam NJ, Sunde M, Matthews JM. Trends Biochem. Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Kiessling LL, Gestwicki JE, Strong LE. Angew. Chem. Int. Ed. 2006;45:2348–2368. doi: 10.1002/anie.200502794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Sampson NS. Current Opin. Struc. Biol. 2006;16:544–550. doi: 10.1016/j.sbi.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Schreiber AB, Libermann TA, Lax I, Yarden Y, Schlessinger J. J. Biol. Chem. 1983;258:846–853. [PubMed] [Google Scholar]
- 7.Zhang XW, Gureasko J, Shen K, Cole PA, Kuriyan J. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Thomas TP, Shukla R, Kotlyar A, Liang B, Ye JY, Norris TB, Baker JR. Biomacromolecules. 2008;9:603–609. doi: 10.1021/bm701185p. [DOI] [PubMed] [Google Scholar]
- 9.Kramer RH, Karpen JW. Nature. 1998;395:710–713. doi: 10.1038/27227. [DOI] [PubMed] [Google Scholar]
- 10.Moad G, Rizzardo E, Thang SH. Australian J. Chem. 2005;58:379–410. [Google Scholar]
- 11.Kulkarni S, Schilli C, Muller AHE, Hoffman AS, Stayton PS. Bioconjugate Chem. 2004;15:747–753. doi: 10.1021/bc034215k. [DOI] [PubMed] [Google Scholar]
- 12.Perrier S, Takolpuckdee P, Mars CA. Macromolecules. 2005;38:2033–2036. [Google Scholar]
- 13.Boyer C, Liu J, Bulmus V, Davis TP, Barner-Kowollik C, Stenzel MH. Macromolecules. 2008;41:5641–5650. [Google Scholar]
- 14.Heredia KL, Grover GN, Tao L, Maynard HD. Macromolecules. doi: 10.1021/ma8022712. DOI: 10.1021/ma8022712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman AS, Stayton PS. Macromolec. Symp. 2004;207:139–151. [Google Scholar]
- 16.Chen GJ, Tao L, Mantovani G, Ladmiral V, Burt DP, Macpherson JV, Haddleton DM. Soft Matter. 2007;3:732–739. doi: 10.1039/b618325e. [DOI] [PubMed] [Google Scholar]
- 17.Hermanson GT, Hermanson GT. Bioconjugate techniques. 1996 [Google Scholar]
- 18.Kaddis CS, Lomeli SH, Yin S, Berhane B, Apostol MI, Kickhoefer VA, Rome LH, Loo JA. J. Am. Soc. Mass Spec. 2007;18:1206–1216. doi: 10.1016/j.jasms.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natarajan A, Du WJ, Xiong CY, DeNardo GL, DeNardo SJ, Gervay-Hague J. Chem. Commun. 2007:695–697. doi: 10.1039/b611636a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.