Abstract
Avian influenza A viruses of the H7 subtype have resulted in more than 100 cases of human infection since 2002. Highly pathogenic avian influenza (HPAI) H7 viruses have the capacity to cause severe respiratory disease and even death; however, the induction of the human innate immune response to H7 virus infection has not been well characterized. To better understand H7 virus pathogenesis in the human respiratory tract, we employed a polarized human bronchial epithelial cell model and primary human monocyte-derived macrophages. Here, we show that infection with HPAI H7 viruses resulted in a delayed and weakened production of cytokines, including the type I interferon response, compared with infections of other influenza A subtypes, including H7 viruses of low pathogenicity. These studies revealed that H7 viruses vary greatly in their ability to activate host innate responses and may contribute to the virulence of these viruses observed in humans.
Avian influenza A viruses of the H7 subtype are classified as highly pathogenic avian influenza (HPAI) and low pathogenic avian influenza (LPAI) viruses primarily based on mortality rates following chicken pathogenicity testing. Both H7 virus pathogenic phenotypes can cause outbreaks in poultry and have been transmitted directly to humans in recent years. Since 2002, H7 subtype viruses have caused more than 100 cases of human infection in Europe and North America, resulting in both ocular and respiratory illness [1]. A single fatal case of H7N7 infection in the Netherlands in 2003 due to acute respiratory distress syndrome resembled the severe respiratory infection that frequently follows HPAI H5N1 infection in humans [2, 3]. Respiratory disease symptoms following H7 virus infection have further been documented in human cases from New York (H7N2, 2003), Canada (H7N3, 2004), and the United Kingdom (H7N2, 2007) [4–6]. Recent studies demonstrating the transmissibility of selected H7 viruses in the ferret model underscore the potential public health risk associated with this subtype and the need to better understand the behavior of this subtype in human respiratory cells [7, 8].
Our current knowledge of the host response to HPAI viruses is based largely on studies of H5N1 virus infection. High virus load and hypercytokinemia have been associated with HPAI H5N1 virus–infected human cases [9, 10], with this severe disease generally recapitulated in laboratory animal models [11–14]. Similarly, HPAI H7N7 viruses have been shown to replicate efficiently in the respiratory tract of mice and induce high levels of proinflammatory cytokines in the lungs at the height of infection (days 3–6 postinoculation) [15–17]. However, induction of the host innate response early after H7 virus infection in humans has not been well characterized. Studying the type I interferon (IFN) response, activated following infection with human and avian influenza A viruses, is of particular importance given its central role in the establishment of an antiviral state that limits spread of progeny viruses to uninfected cells [18, 19].
The airway epithelium is the primary site of influenza virus replication in humans. Accordingly, the study of host responses following virus infection has been facilitated by the use of human respiratory epithelial cell models. Grown on transwell inserts, the human bronchial epithelial cell line Calu-3 forms polarized monolayers that resemble the in vivo airway epithelium, and has been used previously as an in vitro model for human and avian influenza viruses [20–22]. Human monocyte-derived macrophages also have been used previously to study influenza virus infection and represent a second major cell type that elicits immunomodulatory products upon infection with influenza [10, 23]. However, the permissiveness of these cell types to H7 virus infection, and the induction of innate immune responses in lung epithelial cells and macrophages after H7 virus exposure, was not known. We found that infection with HPAI H7 viruses in two cell types present in the human lung resulted in a weaker and delayed induction of innate immune responses, including the type I IFN response, compared with H7N2 viruses of low pathogenicity and other influenza A subtypes.
MATERIALS AND METHODS
Viruses
Influenza A virus stocks were grown in the allantoic cavity of 10-day-old embryonated hens’ eggs and clarified by centrifugation as described previously [13, 16, 20]. All experiments with HPAI viruses were conducted under biosafety level 3 containment, including enhancements required by the U.S. Department of Agriculture and the Select Agent Program [24].
Infection of Calu-3 Cells
Calu-3 cells (American Type Culture Collection) were cultured and infected on membrane inserts as previously described [20]. Virus was added to the apical surface of cells at a multiplicity of infection (MOI) of .01 (replication kinetics), 1 (real-time PCR), or 2 (cytokine quantification) for 1 h. Apical culture supernatants were harvested at various times postinfection (p.i.). At 11 h p.i. (MOI = 1), detection of influenza A virus nucleoprotein (NP) antigen in infected cells was performed as described [20]. Significance from these and all experiments presented in this study was assessed by Student t test.
Isolation and Infection of Human Macrophages
Primary human monocyte-derived macrophages were isolated from whole blood collected in ACD Vacutainer tubes (BD Biosciences) from healthy adult donors (18–30 years of age) in the Transfusion Medicine Program of Emory University Hospital who had not received a seasonal influenza vaccination within the year prior to blood collection (2008–2009). Peripheral blood mononuclear cells (PBMCs) were separated from buffy coats by centrifugation on a Histopaque-1077 density gradient (Sigma-Aldrich), and monocytes were isolated from PBMCs by negative selection using a Monocyte Isolate Kit II (Miltenyi Biotech). Monocytes were seeded at 3 × 105 cells per well in 24-well plates, and cultured with RPMI 1640 media (Gibco) supplemented with 5% heat-inactivated autologous plasma as described previously [23]. Monocytes were further purified by plastic adherence, and allowed to differentiate in vitro for 14 days. Cells were infected at an MOI of .01 (replication kinetics) or 2 (cytokine quantification) for 45 minutes, then washed and incubated in macrophage serum-free medium (Gibco), supplemented with 100 U/mL Pen/Strep and 1 mg/L N-p-tosyl-L-phenylalanine chloromethyl ketone (TPCK)–treated trypsin (Sigma-Aldrich).
Cytokine, Chemokine, and Cell Death Quantification
Culture supernatants were analyzed by enzyme-linked immunosorbent assay (ELISA) for the presence of human Interferon gamma-induced protein 10KDa (IP-10), Tumor Necrosis Factor-alpha (TNFα), Interleukin (IL)-6, IL-8 (BD Biosciences), Regulated upon Activation Normal T-cell Expressed (RANTES) (R&D Systems), Interferonα (specific for IFN-A, α2, αA/D, αD, αK, and α4b) or IFNβ (PBL Bimedical Laboratories). Quantification of nucleosomes 24 h following virus infection (MOI = 1) of Calu-3 cells was performed with the Cell Death Detection ELISAPLUS assay (Roche Applied Science). A minimum of 2 independent samples were collected and tested in duplicate or triplicate for each condition.
Real-Time Quantitative Reverse Transcription PCR (RT-PCR)
Total RNA was extracted from normal, mock-infected, or virus-infected Calu-3 cells, reverse transcribed, and used for real-time PCR with SYBR green (Applied Biosciences) as described [20] with all reaction performed in duplicate or triplicate. All quantifications (threshold cycle [CT] values) were normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and analyzed to determine the relative level of gene expression [20]. Primer sequences were designed with PrimerExpress (Applied Biosystems) and have been published previously [20]. The RT2 Profiler PCR Array (SABiosciences) for Human Interferons and Receptors was performed with total RNA extracted 20 h p.i. and analyzed per manufacturer's instructions.
RESULTS
H7 Viruses Replicate Productively in Both Human Monocyte-Derived Macrophages and Bronchial Epithelial Cells (Calu-3)
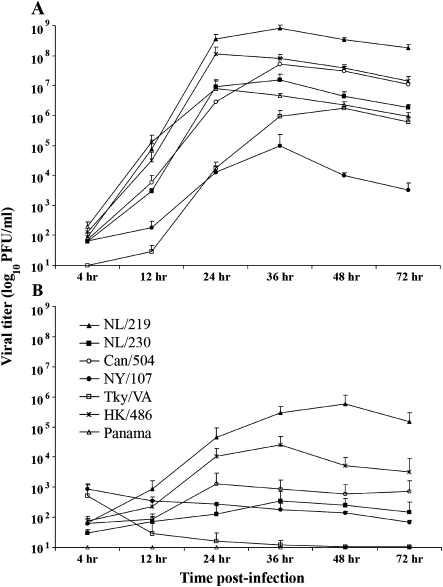
To determine if the kinetics of replication differed between viruses within the H7 subtype, we utilized 2 human cell types: Calu-3 and primary monocyte-derived macrophages, previously shown to support both avian and human influenza virus replication [20, 25]. α2-3 and α2-6 linked sialic acids, the preferred receptors for avian and human influenza viruses, respectively, were present on both cell types in relatively equal proportions ([20] and data not shown). We infected these cells with selected H7 influenza viruses associated with disease in humans as well as an HPAI H5N1 virus and a seasonal H3N2 virus (Table 1). The percentage of infected cells was monitored by immunofluorescence staining of viral NP and was found to be similar following infection (MOI = 1, measured at 8 h p.i.) with all human and avian influenza viruses, with an average of 20%–30% and >90% of Calu-3 cells and macrophages expressing NP, respectively (data not shown), demonstrating similar infectivity as shown in previous studies [20, 25]. The HPAI H7N7 viruses NL/219 and NL/230 and the HPAI H7N3 virus Can/504, in addition to the H5N1 HK/486 and H3N2 Panama viruses, replicated in Calu-3 cells to significantly higher titers (P < .005) compared with the LPAI H7N2 viruses NY/107 and Tky/VA through 36 h p.i. (Figure 1A). In contrast, only NL/219 and HK/486 viruses replicated to titers over 100-fold above baseline by 36 h p.i. in monocyte-derived macrophages (Figure 1B), whereas LPAI H7N2 and H3N2 viruses did not rise above baseline titer through 72 h p.i. The NL/219 virus, which replicated to highest titer in Calu-3 cells, also replicated to highest titer in macrophages.
Table 1.
Influenza A Viruses Used in This Study
| Virus | Name in Study | Subtype | IVPIa | Patient Symptomsb | HA Phylogenetic Lineage | SA Binding Preferencec |
| A/Panama/2007/99 | Panama | H3N2 | NAd | Respiratory | H3 North American | α2-6 |
| A/New York/107/03 | NY/107 | H7N2 | LPAI | Respiratory | H7 North American | α2-3, α2-6 |
| A/Tky/Virginia/4529/02 | Tky/VA | H7N2 | LPAI | NA | H7 North American | α2-3, α2-6 |
| A/Hong Kong/486/97 | HK/486 | H5N1 | HPAI | Respiratory | H5 Eurasian | α2-3 |
| A/Netherlands/219/03 | NL/219 | H7N7 | HPAI | Respiratory, fatal | H7 Eurasian | α2-3 |
| A/Netherlands/230/03 | NL/230 | H7N7 | HPAI | Conjunctivitis | H7 Eurasian | α2-3 |
| A/Canada/504/04 | Can/504 | H7N3 | HPAI | Conjunctivitis, respiratory | H7 North American | α2-3, α2-6 |
Figure 1.
Replication kinetics of H7 influenza viruses in human bronchial epithelial cells and macrophages. Calu-3 cells (A) or primary monocyte-derived macrophages (B) were cultured as described previously [20, 23] and infected with human or avian influenza viruses of multiple subtypes at a multiplicity of infection (MOI) of .01. Supernatants were collected at indicated times postinfection (p.i.) and titered by standard plaque assay for the presence of infectious virus.
HPAI H7 Viruses Exhibit Decreased Cytokine Production Compared With HPAI H5N1 and LPAI H7N2 Viruses
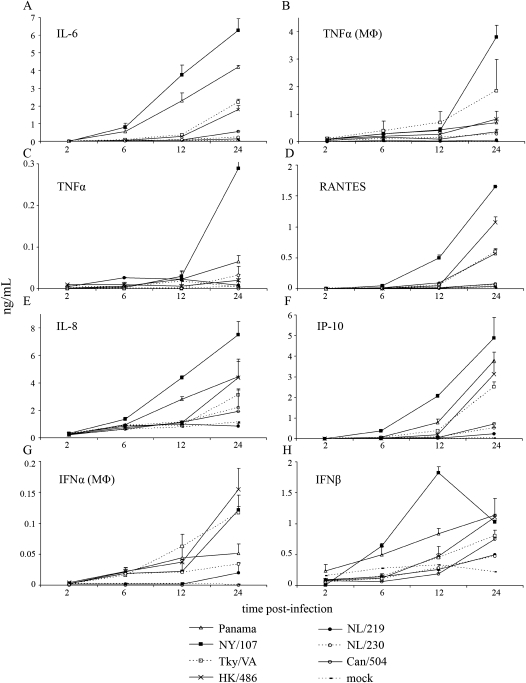
Cytokine dysregulation has been proposed to contribute to the high pathogenicity observed following HPAI virus infection in humans, with differential host immune responses identified between avian H5N1 and seasonal human influenza viruses [9, 20]. We next assessed the production of cytokines in Calu-3 cells or monocyte-derived macrophages infected with HPAI and LPAI H7 viruses in comparison to infection with an HPAI H5N1 or human H3N2 virus. Calu-3 cells infected with NY/107 or Panama viruses secreted the highest levels of the proinflammatory cytokine IL-6 (Figure 2A). In contrast, HPAI H7 virus infection produced significantly lower levels of IL-6 compared with levels induced by the HPAI H5N1 virus, the LPAI H7N2 virus, or H3N2 virus infection at 12 and 24 h p.i. (P < .005). Similar to trends observed with IL-6 production, infection with the HPAI H7 viruses NL/219 and Can/504 produced significantly less TNFα (P < .05) compared with all other viruses tested at 12 h p.i. in Calu-3 cells, a trend also observed in primary human macrophages (Figure 2B–C). Additionally, infection of Calu-3 cells with the HPAI H7 viruses NL/219, NL/230, and Can/504 resulted in significantly less production (P < .05) of the chemokines IP-10 (CXCL10), RANTES, and IL-8 by 24 h p.i. as compared with the H5N1, H7N2, or H3N2 viruses (Figure 2D–F).
Figure 2.
Cytokine production following influenza virus infection in Calu-3 cells and human monocyte-derived macrophages. Calu-3 cells (A, C–F, H) or human primary monocyte-derived macrophages (MΦ) (B, G) were infected at a multiplicity of infection (MOI) of 2. Supernatants were taken at indicated times postinfection (p.i.) and levels of each analyte were quantified by ELISA.
Next, we examined the kinetics of IFNβ production as this cytokine has been shown to mediate protective responses during influenza virus infection [27]. Infection of Calu-3 cells with NY/107 and Panama viruses elicited the most rapid production of IFNβ, with significant levels over mock-infected cells detected at 12 h p.i. (P < .05) (Figure 2H). In contrast, levels of IFNβ induced by infection with the HPAI H7 viruses NL/219, NL/230, and Can/504 were significantly lower compared with H7N2, H5N1, and H3N2 viruses (P < .001) at 24 h p.i. As Calu-3 cells do not produce substantial levels of IFNα following influenza virus infection (data not shown), we utilized human monocyte-derived macrophages to measure the production of IFNα [20]. Similar to the reduced levels of IFNβ detected in Calu-3 cells, HPAI H7 virus–infected cells produced significantly lower levels of IFNα at 12 and 24 h p.i. (P < .001) compared with all other viruses tested (Figure 2G). These findings suggest that infection with HPAI H7N7 and H7N3 viruses results in reduced early induction of type I interferons, cytokines, and chemokines in human cells.
HPAI H7 Viruses Elicit Reduced Levels of Cytokine, IFN, and IFN-Stimulatory Genes
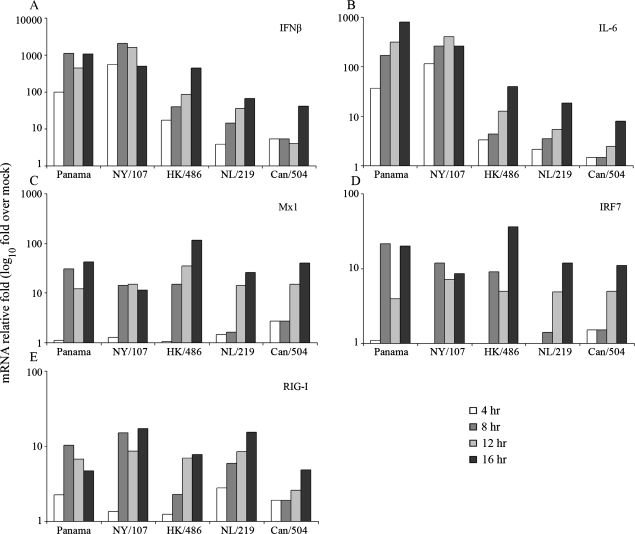
To ascertain if the reduction in cytokine production following HPAI H7 virus infection was found at the transcriptional level, we examined the kinetics of mRNA production of selected cytokines following virus infection in Calu-3 cells. In agreement with IFNβ protein production, NY/107 and Panama viruses induced IFNβ transcription at levels 100-fold above mock-infected cells as early as 4 h p.i. (Figure 3A). In contrast, infection with the HPAI viruses NL/219, Can/504, and HK/486 resulted in approximately 10-fold lower levels of IFNβ mRNA compared with LPAI H7N2 and H3N2 viruses tested through 8 h p.i. Similarly, NY/107 and Panama viruses possessed the highest levels of IL-6 mRNA through 16 h p.i. compared with all other viruses tested (Figure 3B). At the mRNA level, infection with the HPAI viruses NL/219, Can/504, and HK/486 consistently resulted in reduced levels (≤24-fold) of IL-6 mRNA at 8 and 12 h p.i. compared with LPAI viruses and the human H3N2 virus.
Figure 3.
HPAI H7 viruses induce a delayed and weaker cytokine and IFN-stimulated gene (ISG) response compared with LPAI H7 viruses or HPAI H5N1 virus. Calu-3 cells were infected at a multiplicity of infection (MOI) of 1. Total RNA was isolated from cells at indicated times postinfection (p.i.) and examined by real-time RT-PCR. The data are presented as the relative fold gene induction above mock-infected cells and represent the mean of duplicate experiments.
The reduction in IFNα/β production observed following infection with HPAI H7 viruses prompted us to examine the concurrent expression of the IFN-stimulated genes (ISGs) myxovirus resistance 1 (Mx1), interferon response factor 7 (IRF7), and retinoic acid inducible gene I (RIG-I) in Calu-3 cells. NL/219 and Can/504 virus-infected cells produced 2.5–9-fold reduced mRNA levels of these ISGs compared with all other viruses tested at 8 h p.i. (Figure 3C–E), consistent with the diminished type I IFN responses observed with these HPAI H7 viruses. Transcription levels for all genes were generally similar for all influenza viruses tested by 16 h p.i. Taken together, these results indicate that infection with HPAI H7 viruses elicits a significantly delayed and weakened type I IFN response compared with an HPAI H5N1 virus, LPAI H7N2 viruses, or the H3N2 virus.
NL/219 Virus Infection Consistently Resulted in the Lowest Upregulation of IFN and Related Genes Compared With Other Influenza Viruses Tested
To better establish the breadth of host cell responses affected by H7 virus infection, the expression of a panel of genes controlled by or involved in cell signaling mediated by human IFNs and receptors was determined by real-time PCR array. Genes significantly upregulated following infection of Calu-3 cells with representative human and avian viruses are presented as fold change over mock-infected controls (Table 2). As predicted by ELISA and RT-PCR results shown in Figures 2 and 3, the H7N2 virus NY/107 consistently induced the highest levels of IFN and related genes compared with all other viruses tested (Table 2). IL-28A and IL-29, type III interferons with antiviral activity and previously shown to be secreted by alveolar epithelial cells following influenza virus infection, were most dramatically upregulated by the LPAI H7N2 virus [28, 29]. Concurrently, NL/219 virus infection consistently resulted in the lowest upregulation of these IFN and related genes compared with all other influenza viruses tested.
Table 2.
Fold Regulation of Human IFNs and Receptors Following Avian and Human Influenza Virus Infection in Calu-3 Cells.
| Virus | |||||
| Name | Panama | NY/107 | HK/486 | NL/219 | |
| IFN and other genes related to IFNs | |||||
| IFNA2 | 95.5 | 753.1 | 195.2 | 81.1 | |
| IFNA5 | 56.4 | 667.8 | 175.5 | 159.6 | |
| IFNA6 | 427.8 | 1454.9 | 408.8 | 318.5 | |
| IFNA8 | 17.5 | 236.1 | 59.9 | 20.9 | |
| IFNB1 | 7167.9 | 7115.1 | 2975.0 | 1049.4 | |
| IFNG | 1.8 | 77.2 | 3.9 | 13.5 | |
| IL6 | 982.7 | 4661.8 | 1051.8 | 175.1 | |
| IL28A | 21429.8 | 36023.9 | 17064.0 | 5963.7 | |
| IL29 | 3575.7 | 11216.5 | 6898.2 | 1111.8 | |
| IFN-class cytokine receptors | |||||
| IL2RB | 2.0 | 14.3 | 2.9 | 4.8 | |
| IL3RA | 3.5 | 13.1 | 3.9 | 5.2 | |
| IL5RA | 8.7 | 39.9 | 12.2 | 81.5 | |
| IL9R | 3.7 | 9.5 | 7.5 | 40.0 | |
| IL10RA | 27.9 | 57.5 | 17.3 | 8.6 | |
| IL12B | 21.9 | 204.6 | 56.3 | 40.2 | |
| IL21R | 9.5 | 78.2 | 22.9 | 46.1 | |
| IFN-inducible proteins | |||||
| IP-10 | 2185.9 | 2195.0 | 2187.9 | 283.1 | |
| IFI6 | 56.0 | 36.0 | 38.8 | 11.9 | |
| IFI27 | 22.7 | 25.9 | 27.0 | 13.9 | |
| IFIT1L | 144.1 | 190.5 | 47.4 | 254.6 | |
| IFITM1 | 91.8 | 74.2 | 114.7 | 51.6 | |
| IRGM | 14.9 | 244.4 | 18.5 | 12.5 | |
| ISG15 | 79.6 | 64.6 | 94.7 | 23.6 | |
| MX1 | 113.3 | 53.3 | 99.4 | 31.7 | |
| OAS1 | 27.6 | 21.8 | 29.5 | 11.1 | |
NOTE. Values in bold and underlined represent the highest and lowest fold induction over mock-infected cells for each analyte, respectively.
In addition to production of IFNs, engagement of the type I IFN system induces the expression of a wide variety of gene products, including IFN-class cytokine receptors and IFN-inducible proteins, which contribute toward the overall IFN-mediated response to influenza viruses [27]. NY/107 virus infection resulted in the highest detected levels of type I cytokine receptors, with the exception of IL-5RA and IL9R, which were detected at highest magnitude following NL/219 virus infection (Table 2). The H3N2 human virus Panama consistently resulted in the lowest induction of type I cytokine receptors. The H5N1 virus HK/486 induced high levels of several IFN-induced proteins, including Mx1, 2′-5′-oligoadenylate synthetase1, and ISG15. NL/219 virus infection consistently elicited the lowest levels of IFN-inducible proteins compared with all other viruses, including the induction of IP-10, which was significantly reduced compared with all other viruses tested (P < .001). In summary, we observed a pronounced upregulation of IFN and IFN-related genes following NY/107 virus infection, and a concurrent diminished presence of these genes following NL/219 virus infection.
Apoptosis and Necrosis Following H7 Virus Infection
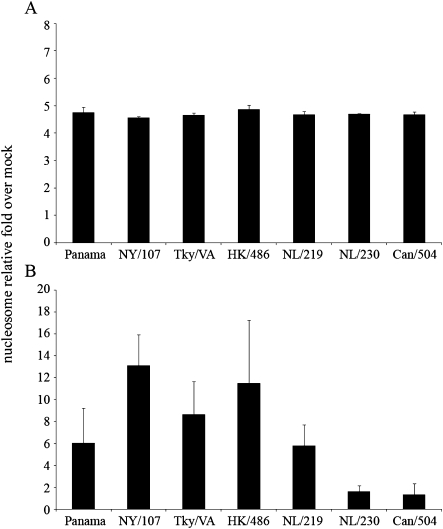
Previous studies have shown that the production of IFN following virus infection can lead to an antiviral state in the host that includes an increased sensitivity of infected cells to apoptosis [30, 31]. Expression of Mx protein in infected cells has further been linked with promotion of apoptotic pathways [32, 33]. To determine if there was an association between heightened expression of these factors and increased cell death, we quantified levels of cytoplasmic histone-associated DNA fragments (nucleosomes) in cell lysates (indicative of apoptosis) and culture supernatants (indicative of necrosis) after influenza virus infection. Levels of nucleosomes in cell lysates of Calu-3 cells infected with representative viruses at 24 h p.i. remained unchanged (Figure 4A). However, NY/107 and HK/486 virus infection resulted in the highest enrichment of nucleosomes in supernatants compared with all other viruses tested (>13- and 11-fold higher than mock-infected cells, respectively) (Figure 4B). Elevated nucleosome levels compared with mock were also detected following Panama, Tky/VA, and NL/219 virus infection; these levels were significantly higher compared with NL/230 and Can/504 viruses, which possessed <2-fold levels of nucleosomes compared with mock infection at this time p.i. (P < .05). In summary, we observed elevated levels of nucleosomes in the cytoplasm of Calu-3 cells following infection with viruses shown to elicit high levels of IFN and Mx protein, suggesting heightened necrosis of cells infected with viruses producing factors connected to the establishment of an antiviral state.
Figure 4.
Detection of nucleosomes in the cytoplasm of cells infected with human and avian influenza viruses. Calu-3 cells were infected at a multiplicity of infection (MOI) of 1. Cell lysates (A) or supernatants (B) were collected at 24 h postinfection (p.i.) and the presence of nucleosomes was detected by ELISA. The data are presented as the relative fold above mock-infected cells and represent the mean of duplicate experiments.
DISCUSSION
HPAI H7N7 and H5N1 viruses are both capable of causing severe disease and death following human infection, and similar molecular determinants of pathogenicity between these viruses suggest a shared mechanism of action following virus infection [3, 34–36]. However, while previous studies have shown that numerous cell types present in the human respiratory tract support H5N1 virus replication and contribute toward a detrimental “cytokine storm” in some infected hosts [20, 23, 25, 36], it was unknown if H7 viruses share this ability. Here, we provide extensive side-by-side data using two cell types found in the respiratory tract to more accurately compare the activation of human host responses following infection with avian influenza viruses of multiple subtypes and lineages. We found that HPAI H7 viruses from both North American and Eurasian lineages exhibited a delayed and diminished induction of innate immune responses in human cells compared with LPAI H7N2, HPAI H5N1, or human H3N2 viruses.
Differential cytokine production in human respiratory cells following infection with avian and human viruses that exhibit similar infectivity and replication kinetics has been shown previously [20, 23, 25, 37]. HPAI H5N1 viruses from 1997 have been shown to elicit hypercytokinemia following virus infection compared with human viruses [20, 23, 25], whereas an attenuated IFN response was observed following infection of human respiratory cells with H5N1 viruses isolated from 2004 [20, 23, 25]. In a separate study, infection of human monocyte-derived macrophages and dendritic cells with the reconstructed 1918 H1N1 virus or a 2009 H1N1 pandemic virus revealed diminished cytokine production and reduced induction of antiviral genes, demonstrating the ability of pandemic influenza viruses to efficiently attenuate host innate immune responses [37, 38]. As we found that HPAI H7 viruses from multiple lineages similarly escaped induction of host interferons and receptors following infection, the pandemic potential of this virus subtype should not be overlooked.
The relationships between IFN signaling, cytokine production, virus replication, and overall pathogenesis in a host following influenza virus infection are complex and not fully understood [27]. The dramatic upregulation of both type I and type III IFNs following NY/107 virus infection is striking in that these classes of IFN employ different receptors (IFN-αβR for type I, IFN-λR for type III) [39]. Both type I and type III IFNs use the JAK/STAT signaling pathway, suggesting a common virus-induced event during initial signaling events [39]. Further study is needed to ascertain if the heightened expression of IFN related genes following NY/107 virus infection contributed to the elevated levels of nucleosomes detected in culture supernatant from infected cells, or if the attenuated IFN response displayed by HPAI H7 viruses prevented induction of necrosis in this system [30, 40]. Despite this marked difference in IFN production, all viruses tested were equally sensitive to the antiviral effects of IFNβ as demonstrated by a delay in viral replication following pretreatment (data not shown).
Delayed transcription in HPAI H7–infected cells of IRF7 and RIG-I, genes that contribute to the induction and augmentation of the IFN response [41, 42], has been demonstrated following infection with select H5N1 viruses and may contribute to the poor initiation of the IFN response observed with these H7 viruses [20]. The reduction of host antiviral responses early after infection with HPAI H7 influenza viruses could additionally allow for the growth advantage of these viruses observed in 2 cell types [43]. Interestingly, the HPAI H7N7 and H7N3 viruses tested here, all of which poorly elicited cytokine and chemokine responses early following infection, markedly differed in their virulence in both humans and mammalian models [16, 17]. The efficient replication of the HPAI H7N7 virus NL/219 may in part be attributed to the E627K substitution in PB2, the only virus in this study to contain this substitution; recent work has demonstrated the necessity of 627K for the optimal replication of this virus in a human lung cell line [35, 36, 44]. Infection with LPAI H7N2 viruses, which possess an HA receptor binding specificity resembling that of human viruses (Table 1), resulted in production of cytokines and chemokines similar to the H3N2 virus [7]. Despite this similarity, titers of NY/107 virus were 2–3 logs lower than the H3N2 virus 12–24 h p.i. in Calu-3 cells, suggesting differing mechanisms between these viruses to overcome host responses. Further study characterizing the role of the NS1 protein of H7 viruses, in addition to examining differences between these viruses in the early activation of signaling pathways associated with cytokine production, will help elucidate those molecular determinants that contribute to differential induction of the early innate immune response following H7 virus infection.
The severity of HPAI H5N1 infection in humans is in part attributed to a high viral load and heightened production of proinflammatory cytokines [9]. In accord with other studies, this work suggests that the severe disease observed following infection with some HPAI viruses may in part be caused not by cytokine dysregulation but rather a suppression of the early innate immune response [45]. Future investigation of innate and adaptive immune responses to H7 viruses throughout the course of infection, especially in in vivo models, is warranted. Furthermore, given the ocular tropism displayed associated with H7 viruses, study of host responses within ocular cell types would further improve our understanding of this virus subtype. These studies allow for a better understanding of not only human infection with viruses within the H7 subtype but of the complex host response following infection with HPAI viruses.
Acknowledgments
We thank Ron Fouchier (Erasmus Medical Center) and Yan Li (Canadian Center for Human and Animal Health) for providing some of the viruses used in this study. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- 1.Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15:859–65. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chotpitayasunondh T, Ungchusak K, Hanshaoworakul W, et al. Human disease from influenza A (H5N1), Thailand, 2004. Emerg Infect Dis. 2005;11:201–9. doi: 10.3201/eid1102.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier RA, Schneeberger PM, Rozendaal FW, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci U S A. 2004;101:1356–61. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC) Update: influenza activity–United States and worldwide, 2003-04 season, and composition of the 2004-05 influenza vaccine. MMWR Morb Mortal Wkly Rep. 2004;53:547–52. [PubMed] [Google Scholar]
- 5.Tweed SA, Skowronski DM, David ST, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis. 2004;10:2196–9. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudley JP. Public health and epidemiological considerations for avian influenza risk mapping and risk assessment. Ecol Soc. 2008;13:21. [Google Scholar]
- 7.Belser JA, Blixt O, Chen LM, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci U S A. 2008;105:7558–63. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, Wan H, Araya Y, Perez DR. Partial direct contact transmission in ferrets of a mallard H7N3 influenza virus with typical avian-like receptor specificity. Virol J. 2009;6:126. doi: 10.1186/1743-422X-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peiris JS, Yu WC, Leung CW, et al. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–9. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao P, Watanabe S, Ito T, et al. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–9. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999;73:5903–11. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maines TR, Lu XH, Erb SM, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zitzow LA, Rowe T, Morken T, Shieh WJ, Zaki S, Katz JM. Pathogenesis of avian influenza A (H5N1) viruses in ferrets. J Virol. 2002;76:4420–9. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wit E, Munster VJ, Spronken MI, et al. Protection of mice against lethal infection with highly pathogenic H7N7 influenza A virus by using a recombinant low-pathogenicity vaccine strain. J Virol. 2005;79:12401–7. doi: 10.1128/JVI.79.19.12401-12407.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belser JA, Lu X, Maines TR, et al. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J Virol. 2007;81:11139–47. doi: 10.1128/JVI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph T, McAuliffe J, Lu B, Jin H, Kemble G, Subbarao K. Evaluation of replication and pathogenicity of avian influenza a H7 subtype viruses in a mouse model. J Virol. 2007;81:10558–66. doi: 10.1128/JVI.00970-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan MC, Cheung CY, Chui WH, et al. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir Res. 2005;6:135. doi: 10.1186/1465-9921-6-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayman A, Comely S, Lackenby A, et al. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology. 2006;347:52–64. doi: 10.1016/j.virol.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Zeng H, Goldsmith C, Thawatsupha P, et al. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J Virol. 2007;81:12439–12449. doi: 10.1128/JVI.01134-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florea BI, Cassara ML, Junginger HE, Borchard G. Drug transport and metabolism characteristics of the human airway epithelial cell line Calu-3. J Control Release. 2003;87:131–8. doi: 10.1016/s0168-3659(02)00356-5. [DOI] [PubMed] [Google Scholar]
- 22.Pekosz A, Newby C, Bose PS, Lutz A. Sialic acid recognition is a key determinant of influenza A virus tropism in murine trachea epithelial cell cultures. Virology. 2009;386:61–7. doi: 10.1016/j.virol.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheung CY, Poon LL, Lau AS, et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–7. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 24.Chosewood LC, Wilson DE. Biosafety in microbiological and biomedical laboratories. 5th ed. Atlanta, GA: Centers for Disease Control and Prevention; 2009. Laboratory biosafety level criteria. 30–59. [Google Scholar]
- 25.Zhou J, Law HK, Cheung CY, Ng IH, Peiris JS, Lau YL. Differential expression of chemokines and their receptors in adult and neonatal macrophages infected with human or avian influenza viruses. J Infect Dis. 2006;194:61–70. doi: 10.1086/504690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subbarao K, Klimov A, Katz J, et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science. 1998;279:393–6. doi: 10.1126/science.279.5349.393. [DOI] [PubMed] [Google Scholar]
- 27.Wolff T, Ludwig S. Influenza viruses control the vertebrate type I interferon system: factors, mechanisms, and consequences. J Interferon Cytokine Res. 2009;29:549–57. doi: 10.1089/jir.2009.0066. [DOI] [PubMed] [Google Scholar]
- 28.Sheppard P, Kindsvogel W, Xu W, et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63–8. doi: 10.1038/ni873. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Oberley-Deegan R, Wang S, et al. Differentiated human alveolar type II cells secrete antiviral IL-29 (IFN-lambda 1) in response to influenza A infection. J Immunol. 2009;182:1296–304. doi: 10.4049/jimmunol.182.3.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balachandran S, Roberts PC, Kipperman T, et al. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. J Virol. 2000;74:1513–23. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka N, Sato M, Lamphier MS, et al. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells. 1998;3:29–37. doi: 10.1046/j.1365-2443.1998.00164.x. [DOI] [PubMed] [Google Scholar]
- 32.Mibayashi M, Nakad K, Nagata K. Promoted cell death of cells expressing human MxA by influenza virus infection. Microbiol Immunol. 2002;46:29–36. doi: 10.1111/j.1348-0421.2002.tb02673.x. [DOI] [PubMed] [Google Scholar]
- 33.Numajiri A, Mibayashi M, Nagata K. Stimulus-dependent and domain-dependent cell death acceleration by an IFN-inducible protein, human MxA. J Interferon Cytokine Res. 2006;26:214–9. doi: 10.1089/jir.2006.26.214. [DOI] [PubMed] [Google Scholar]
- 34.Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–73. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- 35.Munster VJ, de Wit E, van Riel D, et al. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J Infect Dis. 2007;196:258–65. doi: 10.1086/518792. [DOI] [PubMed] [Google Scholar]
- 36.de Wit E, Munster VJ, van Riel D, et al. Molecular determinants of adaptation of HPAI H7N7 viruses to efficient replication in the human host. J Virol. 2010;84(3):1597–606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrone LA, Plowden JK, Garcia-Sastre A, Katz JM, Tumpey TM. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 2008;4:e1000115. doi: 10.1371/journal.ppat.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osterlund P, Pirhonen J, Ikonen N, et al. Pandemic H1N1 2009 influenza A virus induces weak cytokine response in human macrophages and dendritic cells and is highly sensitive to antiviral actions of interferons. J Virol. 2010;89(3):1414–22. doi: 10.1128/JVI.01619-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 40.Hinshaw VS, Olsen CW, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol. 1994;68:3667–73. doi: 10.1128/jvi.68.6.3667-3673.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marie I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–9. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoneyama M, Kikuchi M, Natsukawa T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 43.Veckman V, Osterlund P, Fagerlund R, Melen K, Matikainen S, Julkunen I. TNF-alpha and IFN-alpha enhance influenza-A-virus-induced chemokine gene expression in human A549 lung epithelial cells. Virology. 2006;345:96–104. doi: 10.1016/j.virol.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 44.Shinya K, Hamm S, Hatta M, Ito H, Ito T, Kawaoka Y. PB2 amino acid at position 627 affects replicative efficiency, but not cell tropism, of Hong Kong H5N1 influenza A viruses in mice. Virology. 2004;320:258–66. doi: 10.1016/j.virol.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 45.Kobasa D, Jones SM, Shinya K, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–23. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]