Abstract
Pluripotent human embryonic stem cells (ESCs) can be differentiated in vitro into a variety of cells which hold promise for transplantation therapy. Human embryonal carcinoma cells (ECCs), stem cells of human teratocarcinomas, are considered a close but malignant counterpart to human ESCs. In this study, a comprehensive quantitative proteomic analysis of ESCs and ECCs was carried out using the iTRAQ method. Using two-dimensional liquid chromatography and tandem mass spectrometry analyses, we identified and quantitated ~1,800 proteins. Among these are proteins associated with pluripotency and development as well as tight junction signaling and TGF beta receptor pathway. Nearly ~200 proteins exhibit >2 fold difference in abundance between ESCs and ECCs. Examples of early developmental markers high in ESCs include beta-galactoside-binding lectin (LGALS1), undifferentiated embryonic cell transcription factor-1 (UTF1), DNA cytosine methyltransferase 3 isoform-B (DNMT3B), melanoma antigen family-A4 (MAGEA4), and interferon induced transmembrane protein-1 (IFITM1). In contrast, CD99-antigen (CD99), growth differentiation factor-3 (GDF3), cellular retinoic acid binding protein-2 (CRABP2), and developmental pluripotency associated-4 (DPPA4) were among the highly expressed proteins in ECCs. Several proteins that were highly expressed in ECCs such as heat shock 27 kDa protein-1 (HSPB1), mitogen-activated protein kinase kinase-1 (MAP3K1), nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor like-2 (NFKBIL2), and S100 calcium-binding protein-A4 (S100A4) have also been attributed to malignancy in other systems. Importantly, immunocytochemistry was used to validate the proteomic analyses for a subset of the proteins. In summary, this is the first large scale quantitative proteomic study of human ESCs and ECCs, which provides critical information about the regulators of these two closely related, but developmentally-distinct, stem cells.
Keywords: iTRAQ, mass spectrometry, quantitative proteomics, ESC, ECC
1. Introduction
Pluripotent cells are stem cells which can give rise to all cell types in the body. Pluripotent stem cells have been isolated from a variety of human sources as models for studying early human development as well as for in vitro differentiation into cardiocytes, motor neurons, hematopoetic cells and others for the purpose of transplantation therapy [1, 2]. Two of the most well-studied cell types include embryonic stem cells (ESCs) derived from the inner cell mass of blastocyst-staged embryos and embryonal carcinoma cells (ECCs), the stem cells of teratocarcinomas (mixed germ cell tumors) derived from progenitors of the germline [3]. Both of these cell types share the general properties of pluripotent stem cells in that they exhibit unlimited self-renewal and can give rise to derivatives of all three embryonic germ layers as demonstrated by embryoid bodies in cell culture and in the development of tumors after injection into adult mice. Thus, given these attributes, pluripotent stem cells can potentially provide sufficient numbers of differentiated cells to treat a wide variety of human conditions, including heart disease, diabetes, and many neurological disorders.
However, several major hurdles remain to be overcome if such cells are to be used clinically. Most importantly, these cells must be easily and reproducibly cultured and manipulated so that they possess the necessary characteristics for successful differentiation, transplantation and engraftment. For this purpose, identifying the factors involved in stem cell survival, proliferation and pluripotency is critical. Another critical factor lies with their chromosomal stability. For instance, most ECC lines are heteroploid, and those that are diploid exhibit alterations in their genomes as revealed through comparative genomic hybridization [4]. Nonetheless, to date the only clinical trial reported on the use of a pluripotent stem cell-derived source in humans are human ECC-derived postmitotic neurons implanted in regions of the brain damaged by stroke [5–7]. Although the outcome has been promising, the safety concerns regarding the use of a karyotypically unstable cell line will require further monitoring. In contrast, ESC lines are routinely maintained as normal diploids, except over long extended cultures which some lines have shown chromosomal abnormalities similar to those seen in ECCs [8]. Like karyotypic instability, the expression of factors associated with oncogenesis inherent in embryonic stem cells also raises concerns for their use in transplantation. Altered expression of many factors has now been associated with some cancers even though their role is unknown or is a secondary effect downstream of the cause of the tumorigenicity. Therefore, it is essential to determine those factors which turn on the oncogenic state versus those that enhance proliferation and self-renewal without inferring aberrant cell cycles and genomic instability. These factors can then be controlled and screened in cells before transplantation to minimize the risk of potential carcinogenic outcomes. The need for this information is highlighted by the recent approval by the FDA for the first human clinical trial utilizing human embryonic stem cells. This trial involves treating patients with spinal cord injury with hESC-derived oligodendrocyte neural progenitors [9]. The significance of identifying factors associated with pluripotency while avoiding those associated with tumorigenesis has also been highlighted by a series of studies that have shown the conversion of adult fibroblast cells into pluripotent-like stem cells by inserting four genes [10–13]. The resulting cells designated as induced pluripotent stem (iPS) cells express two pluripotent genes, OCT4 and SOX2, and two genes c-Myc and KLF4 which are frequently upregulated in tumors. Although this combination of genes successfully produced ES-like colonies that could generate chimeric animals including germline transmission, nearly 20% of the iPS-derived chimeric offspring developed tumors [2]. In addition, Maherali et al. [10] demonstrated that the expression of OCT4 was no longer required for iPS cell survival. Thus, while these types of studies provide hope for reprogramming adult cells for therapeutic uses, it further reiterates the necessity of finding genes associated with pluripotency while avoiding those associated with oncogenesis.
To define such genes, many attempts have been made to study the global stem cell genome as well as its chromatin state[14]. While these studies provide critical information for finding the factors associated with pluripotency, investigation into the protein levels in these cells are also required as levels of protein expression do not always directly correlate to transcriptomic changes. Indeed, with current developments in proteome-wide approaches, the characterization of the proteome of these cells has just begun. Some of the these proteomic studies which include analysis of mouse ESCs [15], human ESCs [16] and human embryonal carcinoma cells (ECCs) [17] involve non-quantitative analyses, which while useful, do not allow for differential analyses among these populations.
To date, a few proteomics studies and several transcriptomics studies have been reported comparing ESCs and ECCs [18–20]. Membrane proteomic approaches have also been reported recently using a label-free method of quantitation after extensive membrane fractionation [17]. More recent developments in quantitative methods to study proteomics have been employed to study ESCs in mice [21] but have not yet been applied to study human pluripotent stem cells. For this purpose, isobaric tagged for relative and absolute quantitation labeling (iTRAQ) is an effective method for comparing the expression level of even low abundance proteins. Alternatively, stable isotope labeling with amino acids in cell culture (SILAC) is another straightforward and simple approach for labeling proteins for mass spectrometry based analysis. This approach has been recently used for quantitative comparison of the membrane proteomes in human embryonic stem cells and their differentiation after their adaptation to SILAC media [22].
Here, we report the use of an iTRAQ coupled to two-dimensional liquid chromatography and tandem mass spectrometry to compare the protein expression between two distinct, but phenotypically related, pluripotent populations - human ESCs and human ECCs. Our goal was to study the proteomic differences between ESCs and ECCs to identify potential candidates that might explain regulation of pluripotency and malignancy. This approach generated an initial high quality reference proteins of ~1,800 proteins, which include low abundance protein classes such as transcription factors and kinases that were not previously described in stem cells as well as previously documented stem cell markers. We also examined compartmental distribution of nuclear, cytoplasmic, and membrane proteins. Bioinformatics analysis of ESCs and ECCs revealed shared features of their pluripotent nature as well as distinguish the expression of key factors which may be related to the oncogenetic nature of ECCs.
2 Materials and Methods
2.1 Embryonic stem cell and carcinoma cell culture
Human ESCs (H1; WA01) were obtained from Wicell (Wisconsin) and cultured on Matrigel (BD Bioscience) coated dishes in the presence of conditioned medium derived from mouse embryonic fibroblasts (MEFs). MEFs (Millipore) were plated on gelatin (1%) -coated 10 cm dishes and cultured in DMEM, supplemented with 20% serum 3.5μl BME, 2 mM glutamax, 4 ng/mL basic fibroblast growth factor (bFGF, BD Bioscience) for 2 days. To generate conditioned media, MEFs were cultured in DMEM/F12 supplemented with 15% knock out serum, 3.5ul BME 2 mM glutamax and 2 mM NEAA, 4 ng/ml bFGF. Conditioned media is then collected every 24 hrs for 10 days. ESCs were passaged upon 80% confluence using 0.05% trypsin/ EDTA for 5 minutes at 37° C then neutralized using trypsin neutralizing solution (Lorenzo). ESCs were grown in DMEM/F12 supplemented with 2 mM glutamax, 10% FBS and passaged using trypsin similar to ESCs. The human embryonal carcinoma line, NTERA-2 cl.D1 was acquired through American Type Culture Collection (Virginia) and cultured on matrigel-coated plates under conditions described previously for this cell line [23]. Human ESCs and ECCs were constantly monitored for any differentiation events by immunocytochemistry. In order to completely remove traces of feeder cells ESCs cultured on MEFs were subsequently passaged on matrigel coated plates for five subcultures in conditioned media. Conditioned media was filtered using a 0.2 μM filter. Before lysis, ESCs and ECCs were washed 6 times with cold PBS to remove traces of contamination from serum. Karyotypic analysis of pluripotent cells is explained in Supplementary Methods.
2.2 Cell lysates, in-solution digestion and iTRAQ labeling
For the whole cell proteomic analysis, ESCs and ECCs were collected in serum-free media by washing them in ice cold PBS 3 times. The cells were lysed in 0.5% SDS and subsequently sonicated for 3 min on ice (Duty cycle 30%, output control at 3, on Sonifier 250, Branson). For the preparation of cytosolic and non-cytosolic fractions of cells, cells were washed in ice cold PBS for removal of serum. The cells were sheared by Dounce homogenizing 150 strokes in buffer containing 5 mM HEPES pH 7.4, 0.5 mM EDTA, 250 mM sucrose and freshly prepared 1 mM phenylmethylsulfonyl fluoride. Non-cytosolic fraction (pellet) was separated from cytoplasmic fraction by centrifugation at 100,000 g for 10 min at 4 °C. ESC and ECC samples from whole cell lysate, cytosolic and non-cytosolic preparations were normalized based on protein concentration and used for iTRAQ labeling.
Peptides from ESCs and ECCs were differentially labeled using iTRAQ reagent according to manufacturer’s instructions (Applied Biosystems). Briefly, 40 μg of each sample from duplicate ESCs and ECCs were treated with 2μl of reducing agent (tris(2-carboxyethyl) phosphine (TCEP)) at 60°C for 1 hr and alkylated with 1 μl of cysteine blocking reagent, methyl methanethiosulfonate (MMTS) for 10 minutes at room temperature. Protein samples were digested using sequencing grade trypsin (Promega) (1:15) for 12 hr at 37°C. Peptides from each sample in a final volume of 40μl were labeled with one of the four iTRAQ reagents in 70μl of ethanol at room temperature. After 2 hrs, iTRAQ labeling reactions were terminated by adding 100μl water to each sample and then samples are subsequently combined and organic solvent evaporated using a Speedvac. pH was adjusted to 3.0 using 100 mM phosphoric acid and then diluted to 1 ml in SCX solvent A (10 mM potassium phosphate buffer (pH 2.85) 25% acetonitrile). Combined mixtures of iTRAQ labeled tryptic digests from ESCs and ECCs were fractionated using strong cation exchange chromatography on a Polysulfoethyl A column (PolyLC, Columbia, MD) (300A, 5μm, 100 × 2.1mm) using an Agilent 1100 HPLC system containing a binary pump, UV detector and a fraction collector. Fractionation of peptides (0.2 ml fraction) were carried out by a linear gradient between solvent A and solvent B (solvent A, 350 mM KCl, pH 2.85). Three SCX fractionations were carried out for whole cell lysate, cytosolic and non-cytosolic preparations. The fractions were completely dried and reconstituted in 40μl of 0.2% formic acid and stored at −80°C until LC-MS/MS analysis.
2.3 Liquid chromatography and tandem mass spectrometry (LC-MS/MS)
Tandem mass spectrometry analysis of iTRAQ labeled peptides was carried out on a quadrupole time-of-flight mass spectrometer (QSTAR/pulsar, Applied Biosystems). Peptide fractions from SCX chromatography were further separated on reversed-phase liquid chromatography (RP-LC) system (Agilent 1100 system) interfaced with a mass spectrometer. The RP-LC system consisted of a desalting column (75μm × 3 cm, C18 material 5–10μm, 120Å) and an analytical column (75μm × 10 cm, C18 material 5μm, 120 Å) with a nanoflow solvent delivery. Electrospray source is fitted with an emitter tip 8μm (New Objective, Woburn, MA) and maintained at 900 v ion spray voltage. Peptide samples (40μl) were loaded onto a trap column in 0.1% formic acid, 5% acetonitrile for 15 min and LC-MS/MS data were acquired by online analysis of peptides eluted in an acetonitrile in 0.1% formic acid (5–40%) gradient for 30 min with a flow rate of 300 nl/min. Using Analyst v 1.1 (Applied Biosystems), MS/MS data were acquired by targeting three most abundance ions in the scan range of m/z 350 to 1200 Da and those ions selected were excluded from MS/MS for 45s. Unlike non-labeled peptides, twenty percent higher collision energy was applied during MS/MS scan of iTRAQ labeled peptides.
2.4 Mass spectrometry data analysis
Peptide and protein identification was carried out in compliance with Molecular and Cellular Proteomics guidelines. ProteinPilot software V3.0 (Applied Biosystems) was used for database search and quantitation, which uses Paragon algorithm for protein identification and quantitation. Estimates of both local and global FDR are given in Supplementary Table 1. ProGroup algorithm further process these data to determine minimal set of justifiable identified proteins. Instrument raw files were uploaded from three sets of experiment separately (Whole cell, cytosolic and non-cytosolic) and searched against human RefSeq database version 35 containing 33,888 proteins. Search parameters included iTRAQ labeling at N-terminus and lysine residues, cysteine modification by methyl methanethiosulfonate (MMTS), methionine oxidation and digestion by trypsin. We carried out the data analysis using ProteinPilot 3.0, which gives both global and local FDRs. The list of proteins shows the estimate of proteins at 1% and 5% FDR levels. Since in ProteinPilot, we used >95% confidence score cutoff (>1.3 unused score) for protein identification before FDR analysis, we included proteins identified up to 5% FDR
Relative abundance of proteins calculated based on individual peptide ratios. Shared peptides were not included for quantitation except for first hit protein among the other proteins and isoform specific identification of protein was carried out by selecting peptides distinct to each form. The ion count threshold value for considering reporter ions for fold calculation was set at 7. When the same protein was identified in more than one experiment, the quantitation ratio is selected from the experiment with the best p-values. ProteinPilot software quantitates protein ratios for those identified with at least two peptides considering the error factor and p-value, both are estimation of confident interval indicating the likelihood that protein is differentially expressed. In addition, we have included background noise reduction and bias correction feature of ProteinPilot.
2.5 Functional Analysis
For the functional analysis we used Ingenuity Pathways Analysis (IPA) software version 7.1 (Ingenuity Systems, Mountain View, CA) (<http://www.ingenuity.com/products/pathways_analysis.html>). We uploaded the Entrez gene symbols corresponding to all proteins quantitated from both ESCs and ECCs. Proteins with least p-value for iTRAQ ratio were selected from three experiments which represent the aggregate of whole cell lysate, cytosolic and non-cytosolic fractions. IPA software was used to overlay the proteins identified in ESCs and ECCs in different canonical pathways and networks along with their expression level values (p-value <0.05). For cellular localization annotation, all gi accession numbers were mapped to HPRD accession and clustered according to primary localization (nucleus, plasma membrane, cytoplasm, extracellular matrix and unknown category) to understand the proteomic coverage attained by our method.
2.6 Immunocytochemical staining
Antibodies and the concentrations used for immunocytochemical validation are summarized in Supplementary Methods. ESCs and ECCs were fixed in 4% paraformaldehyde for 15 min and antibodies diluted in Dulbecco’s PBS (DPBS) containing 15% goat serum and incubated with the fixed cells for an hr at 25°C. Fluorescently-labeled secondary antibodies (1:200 dilution; Molecular Probes) diluted in DPBS in 15% goat serum were used for detection. Nuclei were stained using DAPI (Sigma) and controls were performed with secondary antibodies alone. Fluorescent images were visualized using a Nikon Eclipse E800 microscope (Nikon, Inc., Melville, NY) and were captured with a Photometrics 20 MHz cooled interlined CCD camera. Alexa Fluor 488 (cyan-green color) was detected using a FITC excitation filter, a 505 nm dichroic mirror and a barrier filter (Chroma, Inc., Burlington, VT)with a band width of 515–555 nm. Alexa Fluor 594 (orange-red) fluorescence was detected using a G2ERHOD 541–551 nm excitation filter, a 575 nm dichroic mirror and a barrier filter with a band width of 590. DAPI was detected using a standard DAPI/Hoechst filter set, UV 2E/C 340380 nm excitation filter, 400 nm dichroic mirror, and a barrier filter with a band width of 435–485 nm. The images were processed using Metamorph software, v.6.2 (Universal Imaging Corp). Importantly, to confirm differences in the relative expression between cell lines, images were captured with the same exposure time for each treatment.
3 Results
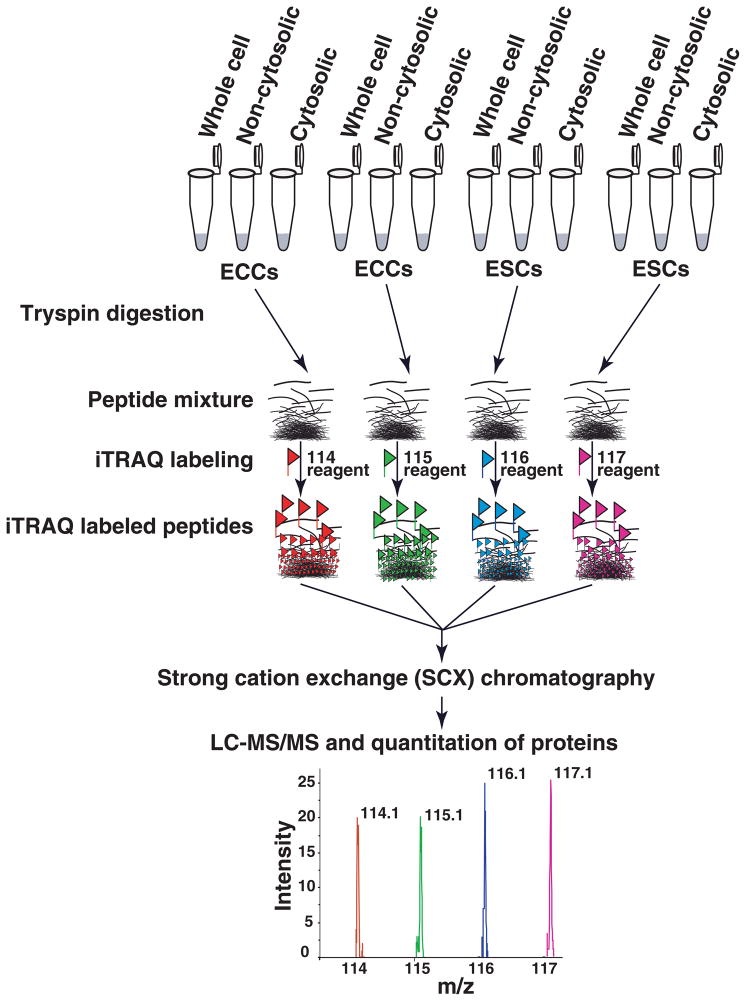
3.1 LC-MS/MS analysis of iTRAQ labeled peptides and mass spectrometry data analysis
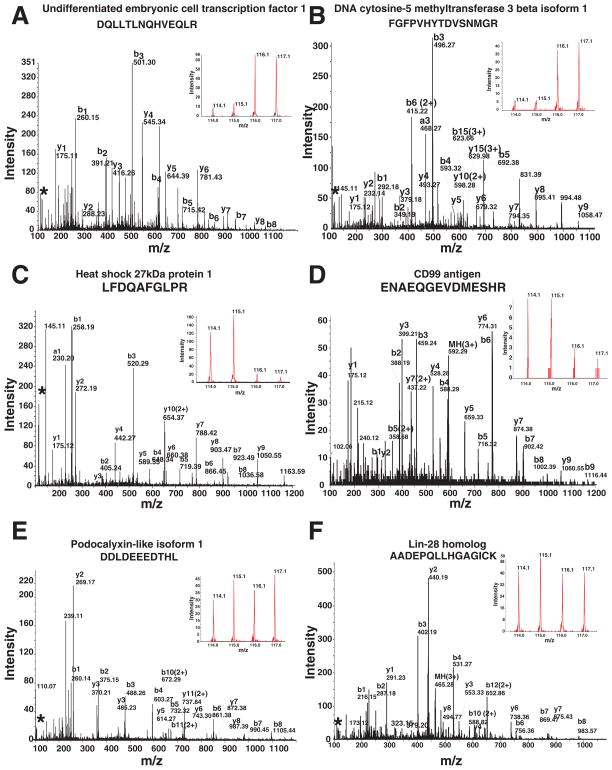
The integrity of human ESCs and ECCs isolated for quantitative proteomic analysis was verified with well-established markers of pluripotency, POU class 5 homeobox 1 transcription factor, (OCT4), tumor rejection antigen 1–81 (TRA-1-81) and stage specific antigen-4 (SSEA4) (Supplementary Fig. 1). Whole cell lysate, cytosolic and non-cytosolic fractions of ESCs and ECCs were compared with non-cytosolic fractions containing membrane, nuclear and other organelle proteomes. Importantly, technical replicates were performed for each experiment dividing the same lysate into two aliquots. Peptides from ECCs were labeled with reagents containing 114 and 115 iTRAQ reporters while peptides from ESCs were labeled with reagents containing 116 and 117 iTRAQ reporters (Fig. 1). LC-MS/MS analysis of 70 SCX fractions from whole cell lysates, cytosolic and non-cytosolic preparations generated a total of >100,000 MS/MS spectra. Using confidence cutoff score ProtScore value >1.3 (95 % confidence), a total of ~1,800 proteins were identified from 36,967 distinct peptides. MS/MS and iTRAQ reporter ion spectra of representative peptides from proteins with different expression levels in ESCs and ECCs are shown in Fig. 2. Panels A and B show the MS/MS spectra of peptides from undifferentiated embryonic cell transcription factor 1 (UTF1) and DNA cytosine-5 methyltransferase 3 beta isoform 1 (DNMT3B), which were highly expressed in ESCs. Panel C and D show the MS/MS spectra of peptides from heat shock 27kDa protein 1 (HSPB1), and CD99 antigen (CD99), which were highly expressed in ECCs. Panel E and F show the MS/MS spectra of peptides from podocalyxin-like isoform 1 (PODXL) and LIN28 homolog (LIN28), which showed no significant change in expression in whole cell analysis. Supplementary Fig. 2 shows additional MS/MS spectra and iTRAQ ratios of 8 peptides from proteins 1) highly expressed in ESCs: Beta-galactoside-binding lectin (LGALS1), biglycan (BGN), gelsolin (GSN), 2) highly expressed in ECCs: developmental pluripotency associated 4 (DPPA4), cellular retinoic acid binding protein 2 (CRABP2), and nucleolar protein 1, 120kDa (NOP2) and 3) protein (talin 1) which show similar level of expression. HELLS1 showed slight higher level of expression in ESCs compared to ECCs as shown by MS/MS spectrum and immunocytochemical staining. The complete list of these proteins along with iTRAQ ratios and FDR values can be found in Supplementary Table 1. Importantly, quantitation data is supported by p-values wherever more than two peptides are used for quantitations, each with technical replicates. Error factor and number of peptides (>95% confidence) used for quantitation are included. The error factor is similar to standard deviation and it gives a measure of the certainty of the average ratio. ProteinPilot calculates Error factor, = 1095%Confidence error.
Fig. 1. Outline of the quantitative proteomic strategy using 4-plex iTRAQ reagents.
iTRAQ labeling was carried out separately using whole cell lysate, cytosolic or non-cytosolic fractions. Samples were digested using trypsin in duplicate and labeled using iTRAQ reagents. Peptides from ECCs were labeled with iTRAQ reagent having 114 and 115 reporters and peptides from ESCs were labeled with iTRAQ reagent having 116 and 117 reporters. After labeling, peptides from all four samples were combined and fractionated by strong cation exchange (SCX) chromatography. Each fraction was then analyzed by LC-MS/MS on a quadrupole time of flight mass spectrometer.
Fig. 2. MS/MS spectra of iTRAQ labeled peptides from selected proteins.
Panels A to F show the MS/MS spectra of peptides from undifferentiated embryonic cell transcription factor 1 (UTF1) and DNA cytosine-5 methyltransferase 3 beta isoform 1 (DNMT3B), heat shock 27kDa protein 1 (HSPB1), CD99 antigen (CD99), podocalyxin-like isoform 1 (PODXL) and LIN28 homolog (LIN28), respectively. The reporter ions in the inset show the examples of high, low and equal expression of proteins in ESCs and ECCs.
3.2 Proteins differentially expressed in ESCs and ECCs
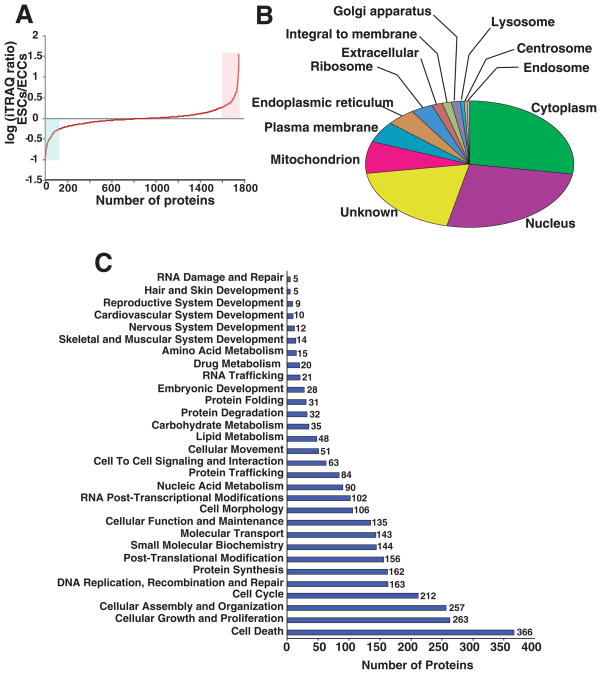
Fig. 3A shows iTRAQ fold changes for all proteins and differential expression of a small subset of proteins from ESCs and ECCs. Nearly, 213 ESC proteins showed >2 fold changes in expression levels while ~208 proteins were found to be expressed more in ECCs. Table 1 shows the partial list of proteins (top 55) along with their iTRAQ ratio that were overexpressed in ESCs when compared to ECCs. The transcription factors, UTF1 and general transcription factor IIIC, polypeptide 4 (GTF3C4) were highly expressed in ESCs (14 and 2.4 fold respectively). UTF1 is a known pluripotency marker which decreases during the onset of differentiation of stem cells [24]. Highly expressed ESCs membrane protein include annexin 1 (ANXA1) caspase recruitment domain family, member 11 (CARD11) (4.0 fold) and cadherin EGF LAG seven-pass G-type receptor 3 (CELSR3) (9.2) fold, catenin (cadherin-associated protein) beta 1(CTNNB1) (2.6 fold), interferon induced transmembrane protein 1 (IFITM1) (2.7 fold) and zyxin (ZYX) 5.3 fold. In contrast, Table 2 shows the partial list of proteins (top 55) identified in this study that were highly expressed in ECCs compared to ESCs. Among them, growth differentiation factor 3 (GDF3), DPPA4, MFGE8 and HSPB1 were identified.
Fig. 3. Localization and functional annotation of proteins identified from ESCs and ECCs.
Panel A shows the distribution of iTRAQ fold changes (proteins expression levels) observed between ESCs and ECCs. Panel B shows the gene ontology analysis for cellular localization of all the proteins identified. Primary and alternate localization data was downloaded from human protein reference database (www.hprd.org) [25] and Panel C shows functional classification of all the proteins quantitated in this study. Using Ingenuity pathway analysis tool, proteins justifying specific biological function significantly (p <0.05) are listed.
Table 1.
A partial list of proteins expressed at higher levels in ESCs as compared to ECCs
| Gene Symbol | Accession | Protein name | Peptides¶ | ESC/ ECC iTRAQ ratio | Sample |
|---|---|---|---|---|---|
| SERBP1 | gi|66346683 | SERPINE1 mRNA binding protein 1 isoform 3 | 8 | 8* | Whole cell |
| CALD1 | gi|15149465 | caldesmon 1 isoform 5 | 8 | 9.3* | Whole cell |
| TRIM28 | gi|5032179 | tripartite motif-containing 28 protein | 5 | 3.9* | Non-cytosolic |
| NES | gi|38176300 | nestin | 9 | 5.3* | Whole cell |
| MAGEA4 | gi|58530871 | melanoma antigen family A, 4 | 3 | 35.3* | Cytosolic |
| LAMA1 | gi|38788416 | laminin, alpha 1 precursor | 6 | 12* | Non-cytosolic |
| RUVBL1 | gi|4506753 | RuvB-like 1 | 6 | 5.5* | Whole cell |
| CALM1 | gi|5901912 | calmodulin 1 | 10 | 7.2* | Whole cell |
| L1TD1 | gi|31542663 | LINE-1 type transposase domain containing 1 | 5 | 4.6* | Whole cell |
| TAGLN | gi|48255907 | transgelin | 4 | 3.7* | Cytosolic |
| UGP2 | gi|48255966 | UDP-glucose pyrophosphorylase 2 isoform a | 4 | 5.3* | Whole cell |
| YBX1 | gi|34098946 | nuclease sensitive element binding protein 1 | 12 | 18.5* | Whole cell |
| PSIP1 | gi|19923653 | PC4 and SFRS1 interacting protein 1 isoform 2 | 3 | 18.4* | Whole cell |
| EEF2 | gi|4503483 | eukaryotic translation elongation factor 2 | 40 | 13.8* | Whole cell |
| COPA | gi|4758030 | coatomer protein complex, subunit alpha | 6 | 3.7* | Non-cytosolic |
| KPNA2 | gi|4504897 | karyopherin alpha 2 | 14 | 2.8** | Whole cell |
| SERPINB9 | gi|4758906 | Protease inhibitor 9 | 3 | 5** | Cytosolic |
| DNMT3B | gi|5901940 | DNA cytosine-5 methyltransferase 3 beta isoform 1 | 4 | 5.6** | Non-cytosolic |
| EZR | gi|21614499 | villin 2 | 8 | 4.3** | Cytosolic |
| VCL | gi|7669550 | vinculin isoform meta-VCL | 19 | 10.2** | Whole cell |
| UTF1 | gi|71043876 | undifferentiated embryonic cell transcription factor 1 | 2 | 14.7** | Whole cell |
| YWHAQ | gi|5803227 | 14-3-3 theta | 11 | 7.2** | Whole cell |
| BASP1 | gi|30795231 | brain abundant, membrane attached signal protein 1 | 3 | 10.5** | Whole cell |
| CASP3 | gi|14790119 | caspase 3 preproprotein | 3 | 5.4** | Cytosolic |
| FLNB | gi|105990514 | filaminB, beta (actin binding protein 278) | 21 | 3** | Whole cell |
| WDR3 | gi|5803221 | WD repeat-containing protein 3 | 1 | 18.4** | Whole cell |
| DDX1 | gi|4826686 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 | 4 | 3.3** | Whole cell |
| HMGA1 | gi|22208977 | high mobility group AT-hook 1 isoform a | 5 | 5** | Whole cell |
| SR140 | gi|122937227 | U2-associated SR140 protein | 1 | 5.4** | Whole cell |
| PSMD3 | gi|25777612 | proteasome 26S non-ATPase subunit 3 | 3 | 3.2** | Whole cell |
| TJP1 | gi|116875767 | tight junction protein 1 isoform a | 3 | 4.9*** | Whole cell |
| LOC643752 | gi|88983788 | PREDICTED: similar to RAS related protein 1b | 2 | 3.4*** | Whole cell |
| AKAP12 | gi|21493022 | A-kinase anchor protein 12 isoform 1 | 2 | 2.5*** | Whole cell |
| TXN | gi|50592994 | thioredoxin | 4 | 4.4*** | Whole cell |
| ACTC1 | gi|4885049 | cardiac muscle alpha actin 1 proprotein | 54 | 33.4 | Whole cell |
| CELSR3 | gi|13325066 | cadherin EGF LAG seven-pass G-type receptor 3 | 1 | 9.2 | Whole cell |
| BGN | gi|4502403 | biglycan | 2 | 11.5 | Whole cell |
| CBR1 | gi|4502599 | carbonyl reductase 1 | 2 | 8.8 | Cytosolic |
| FABP5 | gi|4557581 | fatty acid binding protein 5 (psoriasis-associated) | 1 | 5.7 | Whole cell |
| CNPY2 | gi|7657176 | transmembrane protein 4 | 2 | 14.7 | Whole cell |
| TXNL1 | gi|4759274 | thioredoxin-like 1 | 1 | 5.1 | Whole cell |
| ETV1 | gi|31742534 | ets variant gene 1 | 1 | 14.2 | Non-cytosolic |
| LOC644755 | gi|89028687 | PREDICTED: hypothetical protein | 1 | 4.7 | Whole cell |
| TOP2B | gi|19913408 | DNA topoisomerase II, beta isozyme | 4 | 4.4 | Whole cell |
| SRP72 | gi|109638749 | signal recognition particle 72kDa | 0 | 4.4 | Non-cytosolic |
| HN1 | gi|7705877 | hematological and neurological expressed 1 isoform 1 | 2 | 4.3 | Whole cell |
| SPG20 | gi|21703346 | spartin | 1 | 3.8 | Whole cell |
| HMGB1 | gi|4504425 | high-mobility group box 1 | 3 | 4 | Cytosolic |
| NQO1 | gi|70995422 | NAD(P)H menadione oxidoreductase 1, dioxin-inducible | 1 | 3.4 | Cytosolic |
| MTHFD1L | gi|36796743 | methylenetetrahydrofolate dehydrogenase 1-like | 2 | 3.2 | Non-cytosolic |
| AKAP12 | gi|21493024 | A-kinase anchor protein 12 isoform 2 | 1 | 3.2 | Non-cytosolic |
| GJA1 | gi|4504001 | connexin 43 | 2 | 3 | Whole cell |
| FERMT2 | gi|29789006 | Kindlin 2 | 1 | 5.5 | Whole cell |
| LGALS1 | gi|4504981 | beta-galactoside-binding lectin precursor | 2 | 2.5 | Whole cell |
| ANXA1 | gi|4502101 | annexin 1 | 10 | 10 | Whole cell |
(p value
<0.001,
<0.01 and
<0.05, unused score >1.3,
Number of peptides with >95% confidence score used for quantitation)
Table 2.
A partial list of proteins expressed at higher levels in ECCs as compared to ESCs
| Gene Symbol | RefSeq Accession | Protein Name | Peptides ¶ | ESC/ ECC iTRAQ ratio | Sample |
|---|---|---|---|---|---|
| PARP1 | 4501955 | poly (ADP-ribose) polymerase family, member 1 | 25 | 0.3* | Non-cytosolic |
| PHGDH | 23308577 | phosphoglycerate dehydrogenase | 32 | 0.1* | Whole cell |
| TPI1 | 4507645 | triosephosphate isomerase 1 | 19 | 0.2* | Cytosolic |
| FKBP4 | 4503729 | FK506-binding protein 4 | 17 | 0.3* | Cytosolic |
| FLNC | 116805322 | gamma filamin | 15 | 0.3* | Cytosolic |
| GANAB | 38202257 | alpha glucosidase II alpha subunit isoform 2 | 17 | 0.5* | Non-cytosolic |
| LOC654188 | 113429184 | PREDICTED: similar to peptidylprolyl isomerase A isoform 1 | 5 | 0.2* | Non-cytosolic |
| AHCY | 9951915 | S-adenosylhomocysteine hydrolase | 10 | 0.1* | Whole cell |
| STRAP | 20149592 | serine/threonine kinase receptor associated protein | 9 | 0.3* | Cytosolic |
| DHX9 | 100913206 | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | 12 | 0.4* | Cytosolic |
| PRKDC | 126032350 | protein kinase, DNA-activated, catalytic polypeptide 2 | 17 | 0.2* | Whole cell |
| RAN | 5453555 | ras-related nuclear protein | 13 | 0.1* | Whole cell |
| SNRNP200 | 40217847 | activating signal cointegrator 1 complex subunit 3-like 1 | 9 | 0.4* | Whole cell |
| LRPPRC | 31621305 | leucine-rich PPR motif-containing protein | 10 | 0.3* | Whole cell |
| KHSRP | 4504865 | KH-type splicing regulatory protein (FUSE binding protein 2) | 12 | 0.2* | Whole cell |
| RPL7A | 4506661 | ribosomal protein L7a | 6 | 0.3* | Whole cell |
| KRT18 | 4557888 | keratin 18 | 22 | 0.2** | Whole cell |
| HSPB1 | 4504517 | heat shock 27kDa protein 1 | 10 | 0.1** | Whole cell |
| TCP1 | 57863257 | T-complex protein 1 isoform a | 7 | 0.2** | Whole cell |
| PCNA | 4505641 | proliferating cell nuclear antigen | 3 | 0.2** | Whole cell |
| DEK | 4503249 | DEK oncogene | 3 | 0.1** | Whole cell |
| PCMT1 | 4885539 | protein-L-isoaspartate (D-aspartate) O-methyltransferase | 4 | 0.1** | Whole cell |
| FSCN1 | 4507115 | fascin 1 | 4 | 0.4** | Non-cytosolic |
| PRDX2 | 32189392 | peroxiredoxin 2 isoform a | 4 | 0.4** | Cytosolic |
| NHP2L1 | 51317376 | NHP2 non-histone chromosome protein 2-like 1 | 3 | 0.2** | Whole cell |
| PSMB2 | 4506195 | proteasome beta 2 subunit | 3 | 0.1** | Whole cell |
| MAGOHB | 8922331 | mago-nashi homolog 2 | 5 | 0.3** | Cytosolic |
| PRDX4 | 5453549 | thioredoxin peroxidase | 7 | 0.2** | Whole cell |
| PA2G4 | 124494254 | ErbB3-binding protein 1 | 2 | 0.1** | Whole cell |
| SFRS4 | 21361282 | splicing factor, arginine/serine-rich 4 | 1 | 0.1** | Whole cell |
| HNRNPH1 | 5031753 | heterogeneous nuclear ribonucleoprotein H1 | 13 | 0.2** | Whole cell |
| CSTB | 4503117 | cystatin B | 3 | 0.1** | Whole cell |
| SLC25A5 | 4502099 | solute carrier family 25, member 5 | 11 | 0.1*** | Whole cell |
| DDOST | 20070197 | dolichyl-diphosphooligosaccharide-protein glycosyltransferase | 2 | 0.3*** | Whole cell |
| AHNAK | 61743954 | AHNAK nucleoprotein isoform 1 | 3 | 0.3*** | Non-cytosolic |
| CLTC | 4758012 | clathrin heavy chain 1 | 23 | 0.4*** | Whole cell |
| CALU | 4502551 | calumenin precursor | 3 | 0.2*** | Non-cytosolic |
| PSMC1 | 24430151 | proteasome 26S ATPase subunit 1 | 6 | 0.3*** | Whole cell |
| MLLT4 | 90819237 | Myeloid/lymphoid or mixed lineage leukemia, translocated to 4 | 1 | 0.4 | Non-cytosolic |
| PEBP1 | 4505621 | prostatic binding protein | 7 | 0.3 | Whole cell |
| MFGE8 | 5174557 | milk fat globule-EGF factor 8 protein | 2 | 0.4 | Whole cell |
| MIF | 4505185 | macrophage migration inhibitory factor | 4 | 0.1 | Whole cell |
| CD99 | 4505183 | CD99 antigen | 1 | 0.3 | Cytosolic |
| CTNNBL1 | 18644734 | beta catenin-like 1 | 1 | 0.3 | Whole cell |
| PODXL | 66277202 | podocalyxin-like precursor isoform 1 | 3 | 0.3 | Non-cytosolic |
| GDF3 | 10190670 | growth differentiation factor 3 precursor | 1 | 0.3 | Non-cytosolic |
| THY1 | 19923362 | Thy-1 cell surface antigen | 1 | 0.4 | Whole cell |
| PTPN9 | 4506301 | protein tyrosine phosphatase, non-receptor type 9 | 1 | 0.5 | Non-cytosolic |
| SPARC | 4507171 | secreted protein, acidic, cysteine-rich (osteonectin) | 1 | 0.5 | Cytosolic |
| MAPK1 | 66932916 | mitogen-activated protein kinase 1 | 1 | 0.5 | Whole cell |
| SART1 | 10863889 | squamous cell carcinoma antigen recognized by T cells 1 | 1 | 0.5 | Cytosolic |
| TEX264 | 7706708 | testis expressed sequence 264 | 1 | 0.5 | Whole cell |
| CSNK2A1 | 4503095 | casein kinase II alpha 1 subunit isoform a | 4 | 0.4 | Cytosolic |
| DDB1 | 13435359 | damage-specific DNA binding protein 1 | 8 | 0.3 | Whole cell |
| MGB2 | 11321591 | high-mobility group box 2 | 1 | 0.3 | Non-cytosolic |
(p value
<0.001,
<0.01 and
<0.05, unused score >1.3,
Number of peptides with >95% confidence score used for quantitation)
3.3 Categorization and functional annotation analysis of proteins quantitated in ESCs and ECCs
Functional annotations of the combined list of proteins from all three experiments are shown in Fig. 3B. Categories are based on primary localization with the total number of proteins in parenthesis, which include cytoplasm (493) nucleus (466), mitochondrion (146), endoplasmic reticulum (80), ribosome (61), extracellular (37), integral to membrane (25), golgi apparatus (25), lyzosome (10), centrosome (9), and endosome (7). In addition to the 97 proteins localized primarily to the plasma membrane, another 160 proteins were found in which plasma membrane was their alternate localization. The list of proteins with NCBI sequence identifier GI number and localization information derived from HPRD database [25] is given in Supplementary Table 2. Functional annotation of the protein dataset using the Ingenuity pathway analysis tool revealed identification of a large number of molecules from several canonical pathways (Fig. 3C). Supplementary Table 3 shows the list of ~480 proteins classified as cancer gene clusters using Ingenuity pathway analysis (IPA) tool. With a 2 fold change in expression as cutoff, we found ESCs and ECCs showed 15% and 11% of highly expressed proteins in respective cells. Cancer markers that were expressed at lower levels in ESCs when compared to ECCs, included p53 induced protein (0.5 fold in non-cytosolic fraction), NFKBIL2 (0.8), S100A4 (0.7) and HSPB1 (0.3) (p<0.05).
In contrast, when pathways associated with pluripotency were studied, the Wnt pathway demonstrated a significant number of proteins that could be detected in both cell lines. Specifically, 17 molecules associated with the Wnt pathway (www.netpath.org) such as ARRB1, LRP1, CTBP2, MAP1B, CSNK2B, CDC2, CSNK2A1, PPP2CA, RUVBL1, PIN1, SUMO1, SUMO2, MARK2 PPP2R5B, RHOA and RAC1 were identified in both ESCs and ECCs. These proteins were expressed in similar levels between both cell types while other members of this pathway such as ARRB1 (~1.5 fold), CTNNB1 (1.4 fold), PPP2CA (~1.7 fold) were high in ESCs and CDC2 (<0.6) was low in ESCs. Among the member of TGF beta receptor pathway, 19 molecules (ANAPC4, AP2B1, CAV1, CDC2, CDC27, CTNNB1, HDAC1, HSPA8, KPNB1, NUP153, NUP214, PPP2R2A, SNX2, SNX6, SPARC, STRAP, SUMO1, TRAP1 and XPO1) were identified in this study. Among them SNX2, CDC2, CDC27, and STRAP showed 0.5, 0.6, 0.7 and 0.7 fold changes in ESC when compared to ECCs respectively. Only SPARC was found to be high (2.0 fold) in ESCs.
Among the proteins associated with kit receptor pathway, nine proteins (CLTC, CRKL, GRB2, MAPK1, PLCG1, PTPN11, RPS6KA1, STAT1 and VAV2) were detected in our study. MAPK1 was expressed in low level in ESCs (0.54 fold respectively). Under the Notch pathway, APP, HDAC1, HDAC2, MAPK1, SIN3A and WDR12 were identified and specifically cell proliferation regulatory protein WDR12 was ~2.1 fold highly expressed in ESCs when compared to ECCs. Twenty two proteins were identified from MAPK signaling pathway including ARRB1, CASP3, CDC42, CRKL, DUSP5, FLNA, FLNB, FLNC, GRB2, HSPA1A, HSPA8, HSPB1, MAP2K2, MAPK1, PAK1, PAK2, PPM1A, PPP5C, RAC1, RPS6KA1 and RRAS2. Both CASP3 showed 5.4 fold while PAK1 and PAK2 showed 2.3 fold changes in expression level in ESCs when compared to ECCs. MAPK1, RRAS2 and HSPB1 showed less than 0.5 fold expression levels in ESCs. Among the large number of molecules associated with EGFR pathway, molecules such as ARF4, KRT18, KRT8, MAPK1 and NDUFA13, were low in ESCs when compared to ECCs. In contrast, VAV2, connexin 43 and PAK1 levels were high in ESCs when compared to ECCs (Table 2). GRB2 involved in receptor tyrosine kinase signaling showed slightly higher expression (1.3 fold, p<0.03) in ESCs compared to ECCs.
In addition to identifying specific pathways, our data also classified proteins by cellular function including embryonic stem cells survival and cell death, cellular growth and proliferation, cellular assembly and organization, cell cycle, DNA replication, and recombination and repair (Fig. 3C). Interestingly, many proteins involved in early embryonic development were also detected in our analyses. These included DNMT3A, DNMT3B, MAGEA4, HELLS, GDF3, UTF1 and CTNNB1. Hence this proteomic dataset is a valuable resource to investigate subset of proteins in specific pathways.
3.4 Immunocytochemical validation of differentially-expressed proteins
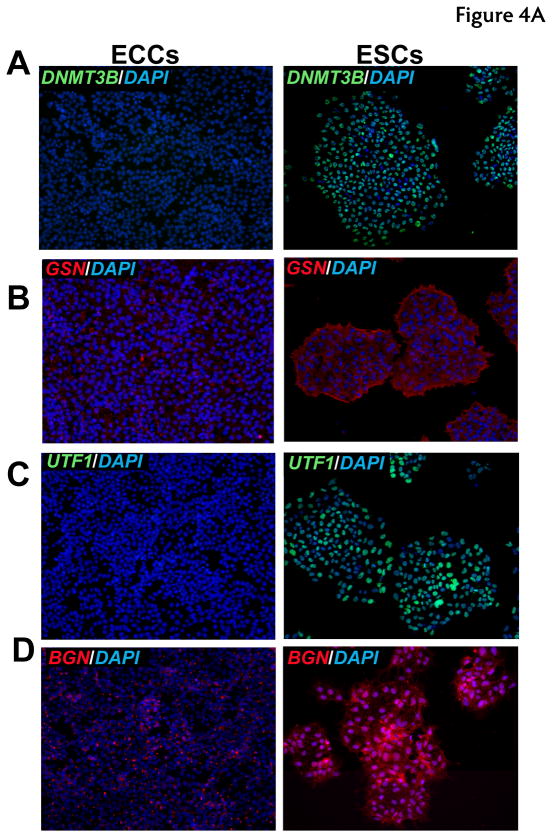
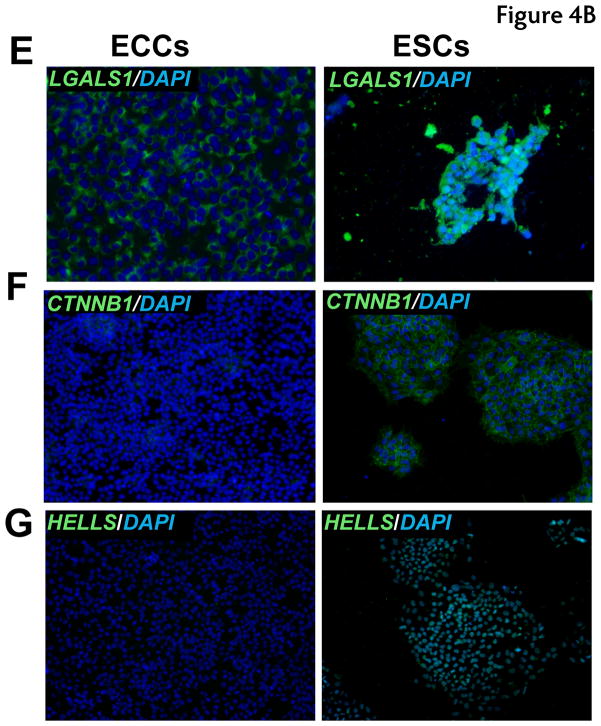
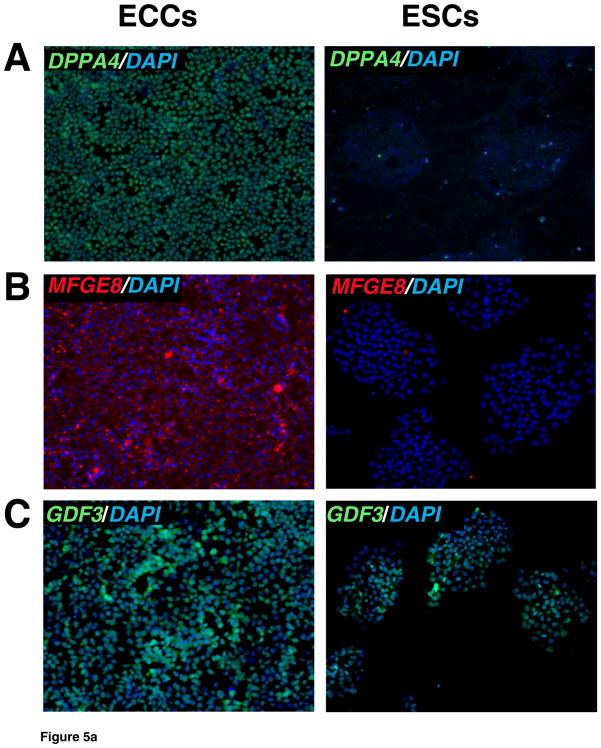
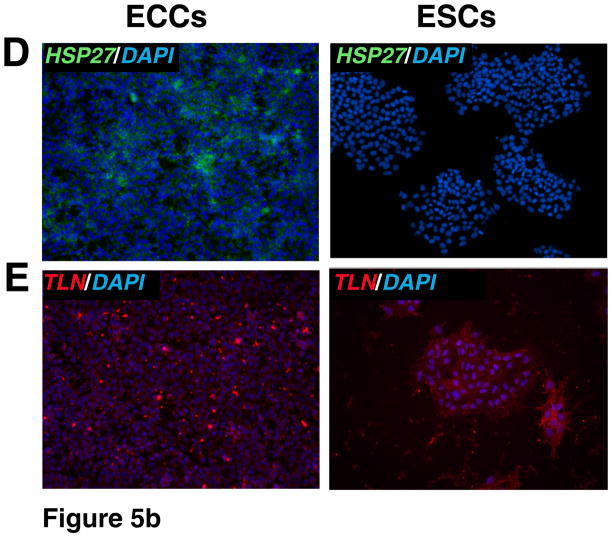
To corroborate proteomic analyses, the relative expression of selected proteins that were differentially expressed was also compared in cell lines by immunostaining. Relative levels of expression were consistent with proteomic analysis for the following proteins: UTF1, DNMT3B, CTNNB, GSN, BGN, and LGALS1 which showed a higher expression of these proteins in ESCs versus ECCs (Fig. 4). HELLS showed slight higher level of expression in ESCs. We also investigated proteins which were expressed higher in ECCs compared to ESCs. These included DPPA4, GDF3, MFGE8 and HSPB1 which demonstrated similar results in immunostaining while TLN1 showed no significant difference in expression between populations in the iTRAQ or immunostaining (Fig. 5 and Supplementary Fig. 2).
Fig. 4. Immunocytochemical analysis of proteins expressed at high levels in ESCs.
Indirect immunofluorescence labeling of different cell types was carried out using Alexa Fluor 594 or Alexa Fluor 488 conjugated secondary antibodies. DAPI (blue) was used to stain nuclei. Panels A to H show proteins found to be expressed at higher levels in ESCs. Panel A to G includes immunocytochemical staining for proteins encoded by DNMT3B (blue-green nuclei), DNMT3A (blue-green nuclei), GSN (red cytoplasm), UTF1 (blue-green nuclei), BGN (red secretory), LGALS1 (green cell surface), CTNNB1 (green cell surface) and HELLS (blue-green nuclei), respectively.
Fig. 5. Immunocytochemical analysis of proteins expressed at high levels in ECCs.
Indirect immunofluorescence labeling of different cell types was carried out using Alexa Fluor 594 or Alexa Fluor 488 conjugated secondary antibodies. DAPI (blue) was used to stain nuclei. Panels A to D show proteins found to be expressed at high levels in ECCs. Panel includes immunocytochemical staining for proteins encoded by DPPA4 (blue-green nuclei), MFGE8 (red cytoplasm), GDF3 (blue-green nuclei) and HSPB1 (also known as HSP27, green cytoplasm), respectively. Panel E shows similar expression level of TLN1 (red cytoplasm) in ESCs and ECCs.
4 Discussion
Factors expressed in early development and those associated with pluripotency
Comparisons between the expression profiles of ESCs and ECCs have been recently highlighted by the growing interest in identifying factors which distinguish pluripotency and oncogenesis. Although, ESC and ECCs provide a model to study these attributes, only a handful of comparisons have been performed on their transcriptomes and even less comparing their protein expression. What is known is that both cell types express markers associated with pluripotency including the three, well-established transcription factors which regulate this process - Oct4, Nanog and Sox2. Although these factors are expressed at such low abundance as to be detected by current proteomic technologies, other, more abundant members were found by this study to be expressed in both ESCs and ECCs. Several of these are known markers of undifferentiated ESC but whose relative protein levels compared to human ECCs have not been reported until now. These include, lin-28 homolog, THY1 cell surface antigen, UTF1, and GDF3, which showed 0.8, 1.1, 14 and 0.3 fold changes respectively in ESCs compared ECCs.
In addition to these factors, we were also able to detect differences in protein levels in three well-established factors of pluripotency which have been previously reported in the only other study to date comparing these lines using proteomic analysis [17]. Dormeyer et al reported that tissue non-specific alkaline phosphatase precursor (ALPL), CD9, and beta-catenin (CTNNB) were similarly expressed in HUES-7 ESCs (Doug Melton’s Harvard line) and NT2/D1 ECCs. However, our results revealed that although ALPL levels were similar in both lines, H1 ESCs expressed higher levels of CD9 and CTNNB than the NTera2 ECCs. However, it remains to be determined whether these inconsistencies are the result of differences in the sensitivity in the proteomic analysis or the result of subtle differences in expression between cell lines. Furthermore, our study was able to detect differences in expression for three other pluripotent associated factors. These included UTF1, which demonstrated higher levels of expression in ESCs compared to ECCs while both lines expressed similar levels of LIN28 and THY1.
In addition to these established regulators of pluripotency, this report demonstrates, for the first time, relative abundance of proteins associated with early development that also have implications in stem cell regulation. These include DPPA4, DNMT3A, DNMT3B, MAGEA4, IFITM1, left-right determination factor-B (LEFTB), CD9, helicase lymphoid-specific protein (HELLS), LIN28, insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), podocalyxin-like 1 (PODXL), cellular retinoic acid binding protein 2 (CRABP2) and DPPA4. Many of these factors were recently recognized by The International Stem Cell Initiative based on gene expression across 59 human embryonic stem cell lines. In fact, our study was able to compare the protein expression of 8 of the 20 transcripts described by this report as positively correlated with NANOG expression in undifferentiated ESCs. These include DNMT3B, GDF3, LEFTB, IFITMI1, UTF1, LIN28, PODXL and CD9.
Of significance is the ability of our analyses to detect quantifiable differences in the expression of these factors. While this has been performed for a number of these markers at the transcriptional level, this report signifies the importance of investigating directly differences in protein expression. Specifically many of the early developmental markers we investigated were high in ESCs compared to ECCs consistent with the premise that ECCs is derived from more mature germ line precursors. For instance, members of a family of proteins which play an important role in DNA methylation and genomic imprinting such as DMNT3B was high in ESCs compared to ECCs (5.6 fold) [26, 27]. This is also consistent with results we previously reported that demonstrated decreased levels of DNMT3B level during ESC differentiation into motor neurons [28]. Hence DNMT levels may be useful to delineate undifferentiated ESCs from differentiated cells as well ECCs. Interestingly, HELLS, also known as LSH, which supports transcription repression by interacting with DNMTs [29] was found to be same in both cell types.
DPPA4, another early developmental marker, was also found to be low in ESCs when compared to ECCs consistent with previous reports demonstrating the expression of this protein in ECCs and germline cells [30, 31]. Furthermore, DPPA4 has also been implemented in the inhibition of ESC differentiation into the ectoderm lineage in mouse [32] which is consistent with our earlier study showing decreased expression during human ESC differentiation into motor neurons [28]. Other early developmental factors that were also highly expressed in ESCs included IFITM1 and LEFTY1, while PODXL and IGF2BP2 levels were low. Similarly, a suspected markers of pluripotency CRABP2 demonstrated higher expression in ECCs than ESCs as well as key factors of chromatin remodeling. Immunocytochemical staining also showed increased ESC expression of gelsolin (GSN), which is an early developmental protein involved in actin restructuring [33, 34].
A meta-analysis across 38 studies of hESC transcriptomes between undifferentiated versus differentiated cells showed a subset of nearly thousand genes from ESCs common to at least three studies [35]. This list of genes was considered as potential differentiation genes based on their high expression levels in ESCs compared to differentiated cells. We have compared our entire protein list with this subset of genes to study the status of differentiation genes among ESCs and ECCs. Among these genes, our proteomic analyses found 265 proteins of which 193 proteins showed no change (using 2 fold change as cutoff) in protein abundance levels in ESCs and ECCs (Supplementary Table 4). Further, our proteomic analysis confirmed that 19 proteins were highly expressed in ESCs compared to ECCs such as CASP3, DNMT3B, ETV1, FABP5, GMFB, HMGA1, KPNA2, LGALS1, LIG1, MTHFD2, PAK1, PSIP1, RUVBL1, SERPINB9, SLC3A2, UCHL1, UGP2, UTF1 and WDR3. Notably, BCAT1, CSE1L, GDF3, DPPA4, MFGE8 and CRABP2 were highly expressed in ECCs compared to ESCs. We carried out correlation analysis among iTRAQ ratios from whole cell, cytosolic and non-cytosolic fractions, which showed significant correlations (r value around 0.4 to 0.5). This data is shown in Supplementary Table 5.
Novel candidate markers of pluripotency found in this study
Several overexpressed proteins identified in this study have not been reported earlier in the context of pluripotency or differentiation. Interestingly, three ECM glycoproteins were highly expressed in ESCs compared to ECCs, including biglycan (BGN), tectorin alpha (TECTA), and galectin 1 (LGALS1) (3.5, 34 and 4.8 fold, respectively). Although the relationship between cell-matrix interactions and pluripotency is a well recognized phenomenon in culture, to date nothing is known regarding the molecules or mechanisms involved in stem cell survival or maintenance. BTB (POZ) domain containing 5 (KLHL28) belongs to a BTB/POZ zinc finger domain family known to play important roles in transcriptional regulation [36]. KLHL28 is 13 fold more abundantly expressed in ESCs when compared to ECCs.
Comparison of known factors associated with oncogenesis in ESCs and ECCs
The protein dataset from his study also included ~480 proteins associated with cancer. Many cancer specific genes (15%) were also found to be highly expressed in ESCs when compared to ECCs. P21-activated kinase 1 (PAK1) overexpression (1.8 fold) has been reported in breast cancer [37] and anti-PAK1 drugs have been used for pancreatic cancer therapy [38]. Similarly cancer/testis antigen melanoma antigen family A, 4 (MAGEA4) is overexpressed in many cancers including oral squamous cell carcinoma [39] and non-small cell lung cancer [40]. Both PAK1 and MAGEA4 levels were high (6.0 and 3.2 fold respectively) in ESCs when compared to ECCs. Some proteins involved in limiting proliferation were also overexpressed in ESCs. For example, the HECT-domain ubiquitin ligase, Huwe1, expressed >2.8 fold higher in ESCs. This protein has been described in controlling differentiation and proliferation through nMyc ubiquitin mediated degradation [41] and as a key player in multiple cancers by degrading tumor suppressor genes [42]. Likewise, A-kinase anchor protein 12 isoform 2 (AKAP12), a tumor suppressor gene whose inactivation has been implicated in gastric cancer [43] and myeloid malignancies [44], was also detected 2.5 fold higher in ESCs than ECC. Another marker associated with certain types of cancer, MCAM, an adhesion molecule was also more highly expressed in ESCs.
Lower expression of some of the known cancer genes were also detected in ESCs compared to ECCs. These included well known cell-cycle regulators such as S100A4 and p53 induced protein (in non-cytosolic) (TPS3I11) as well as the signal transduction molecules such as heat shock 27kDa protein 1 (HSPB1), MAP3K1 (MEK1) and nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NFKBIL2). Invasive factors such as non-metastatic cells 4 protein (NME4) and milk fat globule-EGF factor 8 proteins were also down regulated in ESCs compared to ECCs. These results suggest that the regulation of these factors in ESC may prevent the tumorigenicity found in ECCs. Other cancer markers such as the adhesion molecules pinin, desmosome-associated protein (PNN) and integrin, beta 1 (ITGB1) were similarly expressed between ESC and ECCs as well as factors identified in metastatic tumors such as MTA2, MTA3 and non-metastatic cells 1 protein (NME1) suggesting that these are not contributing factors in oncogenesis. Interestingly GDF3 was also found to be high in ECC versus normal testis consistent with our data showing increase in ECCs compared to ESCs. Another study also compared the gene expression of human ECCs to normal testis [31]. Compared to normal testis, ECCs expressed more branched chain aminotransferase 1, cytosolic (BCAT1), DNMT3B, N-acylaminoacyl-peptide hydrolase (APEH), visinin-like 1 (VSNL1), metallothionein 2A (MT2A), CD9 and GDF3. In our study, comparing ECCs to ESCs, ECCs expressed less DNMT3B (5.6 and 3.5 fold), VSNL1 (1.8 fold) and more CD9 (1.6 fold) and, GDF3 (3.0 fold) and BCAT1 (4.0 fold, cytosolic) while APEH showed similar expression.
Currently, there are only a few markers that can distinguish human ECCs from human ESCs. For instance, various proteins encoded on chromosome 12p, duplicated in testicular cancer, were uniquely high in human ECCs [17]. Of the 8 proteins originally reported by Dormeyer et al. [17] that were unique to ECCs, we found five proteins to be consistently high in ECCs although still expressed in ESCs. These included GAPDH, lactate dehydrogenase B (LDHB), tyrosyl-tRNA synthetase 2, mitochondrial (YARS2), moesin (MSN) and nucleolar protein 1 (NOP2) which are also known to be high in testicular cancer. Furthermore, the germ cell marker recently used to derive adult male germ-line stem cells, ITGA6 (CD49f), showed no change in expression level in ECCs versus ESCs consistent with the theory of germ cell origin for ESCs. There are two other markers that have been used previously to distinguish ECCs and ESCs. One is the well-established germ cell-specific marker VASA or DDX4 (DEAD (Asp-Glu-Ala-Asp) box polypeptide 4). Another is a protein marker recently discovered for its role in identifying ECCs in patient semen, known as AP2-gamma or transcription factor activating protein 2-gamma (TFAP2C) [45]. However neither transcription factor was detected in our analyses. Like Oct4, Nanog and Sox2 expression, this is consistent with the inability to detect low abundance transcription factors by these analyses emphasizing the current need for various approaches trying to identify factors contributing to cell identity.
5 Concluding Remarks
Isotope labeling based quantitative proteomics using mass spectrometry is a powerful approach for global characterization of proteins highlighting cell specific key molecules. This is the largest report using this approach to compare ESCs versus ECCs, which provided a number of candidate factors to study for roles in oncogenesis versus pluripotency. Interestingly, cells from different genetic backgrounds are expected to show extensive quantitative differences, while the ESC and ECC proteomes identified in this study shared several common factors. Thus this technology had greater sensitivity to detect proteins, we are able to report on a number of pluripotent-associated factors and their relative expression levels between two fundamentally similar pluripotent cell lines which differ in their oncogenic tendencies. Significant changes were observed for many targets in this study which could not be detected using label-free methods of quantitation. For instance, there is one other study that reports on the proteomic comparisons between human ECCs and ESCs but it was limited to membrane proteins and used methods with reduced sensitivity for detecting differences in levels of protein concentration compared to the analyses performed here. Nonetheless, it provides comparisons to study which are relatively consistent with the results shown here. This provides a powerful model to distinguish those factors associated with developmental potency from those regulating tumorigenicity. Large numbers of proteins reported in this study have not been studied in the context of ESCs and ECCs. All the peptides and corresponding protein data found in this study has been deposited in Human Proteinpedia [46] (www.humanproteinpedia.org, identification number HuPA 00641) to facilitate the dissemination of this data set.
Supplementary Material
Immunocytochemical staining of ECCs and ESCs. A. Oct4, B. SSEA4, and C, Tra-1-81. DAPI (blue) was used to stain nuclei.
MS/MS spectra and the iTRAQ reporter ion spectra of 12 peptides. A. BGN, B. GSN, C. LGAL1, D., TLN1, E., NOP1, F. CRABP2, G. DPPA4 and H. HELLS.
Methods describing details of karyotyping, mass spectrometry analysis and immunocytochemical labeling experiments.
Complete list of proteins quantitated from ESCs and ECCs using iTRAQ.
Subcellular localization of proteins identified in ESCs and ECCs.
The list of proteins quantitated from ESCs and ECCs, which are found to be associated in different types of cancer.
Proteins identified in this study that have previously been reported in a meta analysis of 36 studies pertaining to ESC differentiation (Assou et al. 2006[47])
Correlation analysis among iTRAQ ratios from whole cell, cytosolic and non-cytosolic fractions,
Acknowledgments
This work was supported by a grant from the Maryland Stem Cell Research Fund, State of Maryland (2007-MSCRFE-0137-01) to C.L.K., J.D.G. and A.P. and an NIH Roadmap grant “Technology Center for Networks and Pathways” (U54 RR 020839) to A.P and the Maryland Stem Cell Research Fund, State of Maryland (2007-MSCRFE-0210-01) to C.L.K. We thank Marjan Gucek and Robert Cole for assistance with mass spectrometry and Ms. Fei Fei (Cyndi) Liu for help with immunocytochemistry.
Abbreviations
- ESCs
embryonic stem cells
- ECCs
embryonal carcinoma cells
References
- 1.Gearhart J. New potential for human embryonic stem cells. Science. 1998;282:1061–1062. doi: 10.1126/science.282.5391.1061. [DOI] [PubMed] [Google Scholar]
- 2.Rossant J. Stem cells: the magic brew. Nature. 2007;448:260–262. doi: 10.1038/448260a. [DOI] [PubMed] [Google Scholar]
- 3.Andrews PW, Damjanov I, Berends J, Kumpf S, et al. Inhibition of proliferation and induction of differentiation of pluripotent human embryonal carcinoma cells by osteogenic protein-1 (or bone morphogenetic protein-7) Lab Invest. 1994;71:243–251. [PubMed] [Google Scholar]
- 4.Blelloch RH, Hochedlinger K, Yamada Y, Brennan C, et al. Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci U S A. 2004;101:13985–13990. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondziolka D, Wechsler L, Goldstein S, Meltzer C, et al. Transplantation of cultured human neuronal cells for patients with stroke. Neurology. 2000;55:565–569. doi: 10.1212/wnl.55.4.565. [DOI] [PubMed] [Google Scholar]
- 6.Watson DJ, Longhi L, Lee EB, Fulp CT, et al. Genetically modified NT2N human neuronal cells mediate long-term gene expression as CNS grafts in vivo and improve functional cognitive outcome following experimental traumatic brain injury. J Neuropathol Exp Neurol. 2003;62:368–380. doi: 10.1093/jnen/62.4.368. [DOI] [PubMed] [Google Scholar]
- 7.Hara K, Yasuhara T, Maki M, Matsukawa N, et al. Neural progenitor NT2N cell lines from teratocarcinoma for transplantation therapy in stroke. Prog Neurobiol. 2008;85:318–334. doi: 10.1016/j.pneurobio.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Baker M. Stem cells by any other name. Nature. 2007;449:389. doi: 10.1038/449389a. [DOI] [PubMed] [Google Scholar]
- 9.Couzin J. Biotechnology. Celebration and concern over U.S. trial of embryonic stem cells. Science. 2009;323:568. doi: 10.1126/science.323.5914.568. [DOI] [PubMed] [Google Scholar]
- 10.Maherali N, Sridharan R, Xie W, Utikal J, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 12.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 13.Wernig M, Meissner A, Foreman R, Brambrink T, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, Ku M, Jaffe DB, Issac B, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano K, Taoka M, Yamauchi Y, Itagaki C, et al. Large-scale identification of proteins expressed in mouse embryonic stem cells. Proteomics. 2005;5:1346–1361. doi: 10.1002/pmic.200400990. [DOI] [PubMed] [Google Scholar]
- 16.Van Hoof D, Passier R, Ward-Van Oostwaard D, Pinkse MW, et al. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Dormeyer W, van Hoof D, Braam SR, Heck AJ, et al. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J Proteome Res. 2008;7:2936–2951. doi: 10.1021/pr800056j. [DOI] [PubMed] [Google Scholar]
- 18.Sperger JM, Chen X, Draper JS, Antosiewicz JE, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100:13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watkins J, Basu S, Bogenhagen DF. A quantitative proteomic analysis of mitochondrial participation in p19 cell neuronal differentiation. J Proteome Res. 2008;7:328–338. doi: 10.1021/pr070300g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Shin S, Zeng X, Zhan M, et al. Genome wide profiling of human embryonic stem cells (hESCs), their derivatives and embryonal carcinoma cells to develop base profiles of U.S. Federal government approved hESC lines. BMC Dev Biol. 2006;6:20. doi: 10.1186/1471-213X-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graumann J, Hubner NC, Kim JB, Ko K, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Prokhorova TA, Rigbolt KT, Johansen PT, Henningsen J, et al. SILAC-labeling and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800287-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andrews PW, Damjanov I, Simon D, Banting GS, et al. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50:147–162. [PubMed] [Google Scholar]
- 24.Tan SM, Wang ST, Hentze H, Droge P. A UTF1-based selection system for stable homogeneously pluripotent human embryonic stem cell cultures. Nucleic Acids Res. 2007;35:e118. doi: 10.1093/nar/gkm704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaneda M, Okano M, Hata K, Sado T, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 27.Kato Y, Kaneda M, Hata K, Kumaki K, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- 28.Chaerkady R, Kerr CL, Marimuthu A, Kelkar DS, et al. Temporal Analysis of Neural Differentiation Using Quantitative Proteomics (dagger) J Proteome Res. 2009 doi: 10.1021/pr8006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldonado-Saldivia J, van den Bergen J, Krouskos M, Gilchrist M, et al. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- 31.Skotheim RI, Lind GE, Monni O, Nesland JM, et al. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res. 2005;65:5588–5598. doi: 10.1158/0008-5472.CAN-05-0153. [DOI] [PubMed] [Google Scholar]
- 32.Masaki H, Nishida T, Kitajima S, Asahina K, Teraoka H. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J Biol Chem. 2007;282:33034–33042. doi: 10.1074/jbc.M703245200. [DOI] [PubMed] [Google Scholar]
- 33.Dieffenbach CW, SenGupta DN, Krause D, Sawzak D, Silverman RH. Cloning of murine gelsolin and its regulation during differentiation of embryonal carcinoma cells. J Biol Chem. 1989;264:13281–13288. [PubMed] [Google Scholar]
- 34.Campbell HD, Fountain S, McLennan IS, Berven LA, et al. Fliih, a gelsolin-related cytoskeletal regulator essential for early mammalian embryonic development. Mol Cell Biol. 2002;22:3518–3526. doi: 10.1128/MCB.22.10.3518-3526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assou S, Le Carrour T, Tondeur S, Strom S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007;25:961–973. doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi J, Zhang X, Zhang HK, Yang HM, et al. ZBTB34, a novel human BTB/POZ zinc finger protein, is a potential transcriptional repressor. Mol Cell Biochem. 2006;290:159–167. doi: 10.1007/s11010-006-9183-x. [DOI] [PubMed] [Google Scholar]
- 37.Li Q, Mullins SR, Sloane BF, Mattingly RR. p21-Activated kinase 1 coordinates aberrant cell survival and pericellular proteolysis in a three-dimensional culture model for premalignant progression of human breast cancer. Neoplasia. 2008;10:314–329. doi: 10.1593/neo.07970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirokawa Y, Levitzki A, Lessene G, Baell J, et al. Signal therapy of human pancreatic cancer and NF1-deficient breast cancer xenograft in mice by a combination of PP1 and GL-2003; anti-PAK1 drugs (Tyr-kinase inhibitors) Cancer Lett. 2007;245:242–251. doi: 10.1016/j.canlet.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 39.Ries J, Vairaktaris E, Mollaoglu N, Wiltfang J, et al. Expression of melanoma-associated antigens in oral squamous cell carcinoma. J Oral Pathol Med. 2008;37:88–93. doi: 10.1111/j.1600-0714.2007.00600.x. [DOI] [PubMed] [Google Scholar]
- 40.Peikert T, Specks U, Farver C, Erzurum SC, Comhair SA. Melanoma antigen A4 is expressed in non-small cell lung cancers and promotes apoptosis. Cancer Res. 2006;66:4693–4700. doi: 10.1158/0008-5472.CAN-05-3327. [DOI] [PubMed] [Google Scholar]
- 41.Zhao X, Heng JI, Guardavaccaro D, Jiang R, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell. 2008;14:10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 43.Choi MC, Lee YU, Kim SH, Park JH, et al. A-kinase anchoring protein 12 regulates the completion of cytokinesis. Biochem Biophys Res Commun. 2008;373:85–89. doi: 10.1016/j.bbrc.2008.05.184. [DOI] [PubMed] [Google Scholar]
- 44.Flotho C, Paulun A, Batz C, Niemeyer CM. AKAP12, a gene with tumour suppressor properties, is a target of promoter DNA methylation in childhood myeloid malignancies. Br J Haematol. 2007;138:644–650. doi: 10.1111/j.1365-2141.2007.06709.x. [DOI] [PubMed] [Google Scholar]
- 45.Hoei-Hansen CE, Nielsen JE, Almstrup K, Sonne SB, et al. Transcription factor AP-2gamma is a developmentally regulated marker of testicular carcinoma in situ and germ cell tumors. Clin Cancer Res. 2004;10:8521–8530. doi: 10.1158/1078-0432.CCR-04-1285. [DOI] [PubMed] [Google Scholar]
- 46.Kandasamy K, Keerthikumar S, Goel R, Mathivanan S, et al. Human Proteinpedia: a unified discovery resource for proteomics research. Nucleic Acids Res. 2009;37:D773–781. doi: 10.1093/nar/gkn701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assou S, Lecarrour T, Tondeur S, Strom S, et al. A meta-analysis of human embryonic stem cells transcriptome integrated into a web-based expression atlas. Stem Cells. 2007 doi: 10.1634/stemcells.2006-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunocytochemical staining of ECCs and ESCs. A. Oct4, B. SSEA4, and C, Tra-1-81. DAPI (blue) was used to stain nuclei.
MS/MS spectra and the iTRAQ reporter ion spectra of 12 peptides. A. BGN, B. GSN, C. LGAL1, D., TLN1, E., NOP1, F. CRABP2, G. DPPA4 and H. HELLS.
Methods describing details of karyotyping, mass spectrometry analysis and immunocytochemical labeling experiments.
Complete list of proteins quantitated from ESCs and ECCs using iTRAQ.
Subcellular localization of proteins identified in ESCs and ECCs.
The list of proteins quantitated from ESCs and ECCs, which are found to be associated in different types of cancer.
Proteins identified in this study that have previously been reported in a meta analysis of 36 studies pertaining to ESC differentiation (Assou et al. 2006[47])
Correlation analysis among iTRAQ ratios from whole cell, cytosolic and non-cytosolic fractions,