Abstract
Cadherins mediate cell-cell adhesion and catenin (ctn)-related signaling pathways. Liver fibrosis is accompanied by the loss of E-cadherin (ECAD), which promotes the process of epithelial-mesenchymal transition. Currently, no information is available about the inhibitory role of ECAD in hepatic stellate cell activation. Because of ECAD’s potential for inhibiting the induction of transforming growth factor β1 (TGFβ1), we investigated whether ECAD overexpression prevents TGFβ1 gene induction; we also examined what the molecular basis could be. Forced expression of ECAD decreased α-smooth muscle actin and vimentin levels and caused decreases in the constitutive and inducible expression of the TGFβ1 gene and its downstream genes. ECAD overexpression decreased Smad3 phosphorylation, weakly decreased Smad2 phosphorylation, and thus inhibited Smad reporter activity induced by either treatment with TGFβ1 or Smad3 overexpression. Overexpression of a dominant negative mutant of ras homolog gene family A (RhoA) diminished the ability of TGFβ1 to elicit its own gene induction. Consistently, transfection with a constitutively active mutant of RhoA reversed the inhibition of TGFβ1-inducible or Smad3-inducible reporter activity by ECAD. Studies using the mutant constructs of ECAD revealed that the p120-ctn binding domain of ECAD was responsible for TGFβ1 repression. Consistently, ECAD was capable of binding p120-ctn, which recruited RhoA; this prevented TGFβ1 from increasing RhoA-mediated Smad3 phosphorylation. In the liver samples of patients with mild or severe fibrosis, ECAD expression reciprocally correlated with the severity of fibrosis.
Conclusion
Our results demonstrate that ECAD inhibits Smad3/2 phosphorylation by recruiting RhoA to p120-ctn at the p120-ctn binding domain, whereas the loss of ECAD due to cadherin switching promotes the up-regulation of TGFβ1 and its target genes, and facilitates liver fibrosis.
E-cadherin (ECAD), a transmembrane glycoprotein that mediates adherens junctions, is developmentally restricted to polarized epithelial cells.1,2 Repeated extracellular domains of ECAD are responsible for binding cells to neighboring ones and maintaining the structural integrity and polarization of epithelia. ECAD also regulates signaling pathways through the intracellular catenin (ctn) binding domains.1,2 Cadherin switching is a characteristic behavior of the process of epithelial-mesenchymal transition (EMT). An important phenotypic change in cadherin switching is the loss of ECAD expression. The loss of ECAD causes cells to dissociate from their neighbors and results in a loss of cell polarity. This, in turn, leads to the activation of cell signaling pathways that regulate the mesenchymal transition. On the contrary, an increase in ECAD expression inhibits cell transformation and tumor cell invasion in an adhesion-independent manner.3,4
Myofibroblasts play a key role in wound healing and pathological organ remodeling.5 The most accepted myofibroblast progenitors in the liver are hepatic stellate cells (HSCs),5,6 although various other resident cells are recognized as sources of liver myofibroblasts.5 As HSCs activate, the level of ECAD expression decreases.7 Activated HSCs then promote the synthesis and deposition of the extracellular matrix (ECM) component and the induction of α-smooth muscle actin (αSMA). In addition, multiple signaling cascades accelerate the growth of activated HSCs6 and contribute to the development of liver fibrosis. Although the link between cadherin switching and the EMT process in HSCs has been studied,7,8 it is yet unclear whether ECAD affects the activation of HSCs. Moreover, the potential signaling and molecular regulatory mechanism by which ECAD antagonizes profibrogenic gene expression in quiescent HSCs has not been explored.
Several lines of evidence indicate that transforming growth factor β1 (TGFβ1) from autocrine or paracrine sources plays a role in activating HSCs and increasing the synthesis of ECM proteins and cellular receptors for various matrix proteins.6 TGFβ1 is regulated transcriptionally by transcription factors and posttranslationally by the maturation of the precursors.6 In response to TGFβ1, type I and II TGFβ1 receptors form a complex and induce receptor autophosphorylation. TGFβ1 is also known as a cytokine that induces EMT, which inhibits ECAD expression by up-regulating transcriptional repressors such as Snail, Zeb, and Twist.9 Activated TGFβ1 receptors transmit the signal by which regulatory Smad molecules (Smad3/2) are phosphorylated and form an active complex with co-Smad (Smad4). The transcription factor complex then moves to the nucleus, in which it promotes the transcription of target genes through interactions with specific Smad binding elements (SBEs; also called the CAGA box).10 It has been reported that single or multiple copies of SBEs are located in the upstream regions of TGFβ1’s target genes, such as plasminogen activator inhibitor 1 (PAI-1), matrix metalloproteinases (MMPs), and collagen type I.11,12
Despite the finding that TGFβ1 leads to HSC activation with a phenotypic change of ECAD loss and causes hepatic ECM accumulation, it has not yet been determined whether ECAD overexpression inhibits the expression of TGFβ1 and its downstream target genes. We investigated whether ECAD negatively regulates TGFβ1 expression; we also examined what the molecular basis could be. Our findings demonstrate transcriptional repression of the TGFβ1 gene by ECAD. Thus, the loss of ECAD initiates TGFβ1 induction and consequently promotes the expression of genes for ECM accumulation. Moreover, the results of this study led to the identification of the p120-ctn binding domain of ECAD as the site required for complex formation with p120-ctn, which recruits ras homolog gene family A (RhoA); this results in the inhibition of RhoA activity. The data presented here support the ability of ECAD to hinder RhoA activity, which is critical for the Smad signaling pathway.
Materials and Methods
Materials
Information on the materials used in this study is given in the Materials and Methods section of the supporting information.
Animal Treatment
Animal experiments were conducted under the guidelines of the institutional animal use and care committee at Seoul National University. Male Sprague-Dawley rats at 6 weeks of age (140–160 g) were used for liver fibrosis induction as described previously.13
Patient Samples
Human liver tissues with fibrosis were obtained from 81 patients who had been diagnosed with liver fibrosis or liver cirrhosis by histological examination and ultrasonography in seven different hospitals in South Korea.14,15 This human investigation was performed after approval by the institutional review board.
Immunohistochemistry
Liver specimens were fixed in 10% formalin, embedded in paraffin, cut into 4-μm-thick sections, and mounted onto slides. Tissue sections were immunostained with antibodies directed against ECAD, glial fibrillar acidic protein (GFAP), and αSMA.
Cell Culture
Murine embryonic fibroblasts (MEFs), LX-2 cells (immortalized human activated HSCs), and HepG2 cells were supplied by Dr. M. Simon (Caltech Institute, Pasadena, CA), Dr. S. L. Friedmann (Mount Sinai School of Medicine, New York, NY), and the American Type Culture Collection (Manassas, VA), respectively. The isolation of primary HSCs and hepatocytes from rat livers and the establishment of a stable cell line expressing ECAD are described in the supporting information.
Generation of the Recombinant Adenovirus
For the generation of an adenoviral ECAD construct, the ECAD gene was subcloned into the attL-containing shuttle plasmid pENTR-BHRNX (Newgex, Seoul, Korea). The recombinant adenovirus was constructed and generated with the pAd/CMV/V5-DEST gateway plasmid (Invitrogen, Carlsbad, CA). In Supporting Fig. 1, green fluorescence protein (GFP)–expressed images are shown to confirm the transduction efficiency.
Immunoblot Analysis
The preparation of whole cell lysates and immunoblot analyses were performed according to the established procedures.13
Real-Time Polymerase Chain Reaction (PCR) Assay
Total RNA was extracted with TRIzol (Invitrogen) and was reverse-transcribed. The resulting complementary DNA was amplified by PCR.
Luciferase Reporter Assay
The sources of the vectors and the procedures used in this study for transient transfection and reporter gene assays are described in the supporting information.
Immunoprecipitation and Immunoblot Assay
For the assessment of protein interactions, cell lysates were incubated with an anti-ECAD or anti-RhoA antibody overnight at 4°C. The antigen-antibody complex was immunoprecipitated and then solubilized in a 2× Laemmli buffer for immunoblotting.
Rho Activity Assay
Rho activity was measured with a Rho assay kit (Upstate Biotechnology, Lake Placid, NY).
Small Interfering RNA (siRNA) Knockdown
Scrambled siRNA (control) and siRNAs of human or rat p120-ctn were supplied by Santa Cruz Biotechnology (Santa Cruz, CA). Cells were cotransfected with an ECAD construct and siRNA (100 pmol) with Lipofectamine 2000 according to the manufacturer’s instructions, and then they were treated with 5 ng/mL TGFβ1 for the indicated time periods.
Results
Repression of N-Cadherin (NCAD), αSMA, and Vimentin Levels by ECAD
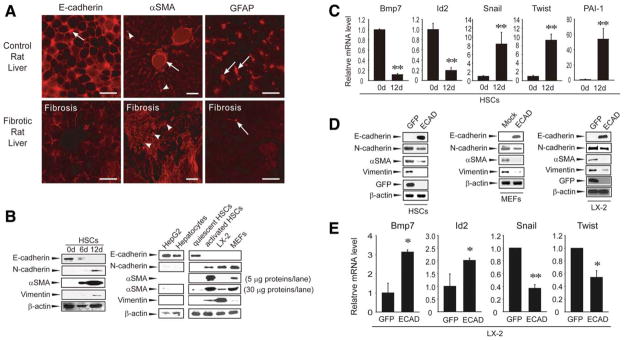
Liver injury induced by repeated dimethylnitrosamine (DMN) treatments activates HSCs and leads to liver fibrosis.13 For confirmation of the association of ECAD loss with HSC activation in vivo, immunohistochemistry was performed in the livers of rats that had been exposed to multiple doses of DMN. After DMN treatments, body weight gain in the animals decreased by 10% in comparison with a healthy control. Necrosis and fiber accumulation were observed in the rat livers with increases in the necrotic and fibrotic scores (data not shown). Liver sections of control rats exhibited simultaneous staining of GFAP (a maker of quiescent HSCs) and ECAD (a marker of the epithelial phenotype; Fig. 1A, top). In contrast, DMN treatments caused decreases in GFAP and ECAD expression in a liver section with a reciprocal increase in αSMA, and this indicated HSC activation (Fig. 1A, bottom). Immunohistochemical analysis confirmed ECAD loss in the fibrotic liver. These results raise the possibility that ECAD loss facilitates HSC activation in vivo.
Fig. 1.
Effects of ECAD on the expression of αSMA and vimentin. (A) ECAD loss in the fibrotic liver. Replicate liver sections prepared from rats that had been treated with vehicle (top) or DMN (10 μL/kg of body weight intraperitoneally 3 times per week for 4 weeks; bottom) were immunochemically stained for ECAD, αSMA, or GFAP (red). Arrowheads and arrows indicate the strong intensities of ECAD, αSMA, and GFAP. An arrow in the middle column shows αSMA expression around the vascular smooth muscle. The pictures are representative images from at least six independent experiments. The scale bar represents 20 (left and right) or 50 μm (middle). (B) Cadherin switching in HSCs. Primary HSCs were cultured in a growth medium for 6 or 12 days, and the cell lysates (30 μg each) were subjected to immunoblotting (left). The expression levels of ECAD and NCAD were also determined in the lysates of HepG2 cells, primary hepatocytes, or quiescent HSCs (epithelial type) and in the lysates of activated HSCs, LX-2 cells, or MEF cells (mesenchymal type; right). (C) Real-time PCR assays. The data are the means and standard errors of at least six separate experiments (significantly different versus day 0: **P < 0.01). (D) Effects of forced ECAD overexpression on αSMA and vimentin levels. HSCs and LX-2 cells were infected with adenoviral recombinant ECAD (multiplicity of infection = 50). Adenoviral GFP was used as a control. Results were confirmed by three separate experiments. (E) Real-time PCR assays in LX-2 cells infected with adenoviral recombinant ECAD (significantly different versus an adenoviral GFP infection: **P < 0.01 and *P < 0.05).
To understand the relationship between cadherin switching and αSMA expression, time-dependent changes in ECAD expression were monitored in primary cultured HSCs. GFAP and ECAD were colocalized in a normal rat liver (Supporting Fig. 2A,B), and this verified the expression of ECAD by HSCs.7 Quiescent HSCs on day 0 exhibited the expression of ECAD but not αSMA or vimentin, as did hepatocytes (Fig. 1B, left); this indicates that HSCs isolated from a healthy liver have an epithelial phenotype. After cultivation, the expression level of ECAD decreased, whereas the levels of the transdifferentiation markers (NCAD, αSMA, and vimentin) increased. Moreover, NCAD expression was prominent in LX-2 cells (an activated HSC cell line), and there were increases in the levels of αSMA and vimentin (Fig. 1B, right). As expected, MEF cells (a fibroblast cell line) also showed increased expression of NCAD, αSMA, and vimentin. We assessed the expression of TGFβ target genes regulating EMT in HSCs on days 0 and 12 (ECAD expression was distinctly different at these times). The messenger RNA (mRNA) levels of bone morphogenetic protein 7 (Bmp7) and inhibitor of DNA binding 2 (Id2) markedly decreased on day 12, but the levels of Snail, Twist, and PAI-1 increased (Fig. 1C); this suggests that ECAD loss in activated HSCs may promote EMT by inducing TGFβ target genes.
Ectopic expression of ECAD prevents cell transformation as well as tumor cell invasion in an adhesion-independent manner.3,4 Next, the effects of ECAD overexpression on the levels of NCAD, αSMA, and vimentin were examined in primary cultured HSCs that had been activated (Fig. 1D, left). Forced expression of ECAD repressed the expression levels of NCAD, αSMA, and vimentin in HSCs. Consistently, ECAD overexpression resulted in similar changes in both MEFs and LX-2 cells (Fig. 1D, middle and right). Moreover, forced expression of ECAD increased negative regulators of EMT, Bmp7, and Id2 but reciprocally decreased positive regulators of EMT, Snail, and Twist (Fig. 1E). These results indicate that the expression of ECAD alters the activation status of HSCs.
Inhibition of the Basal TGFβ1 and Target Gene Expression
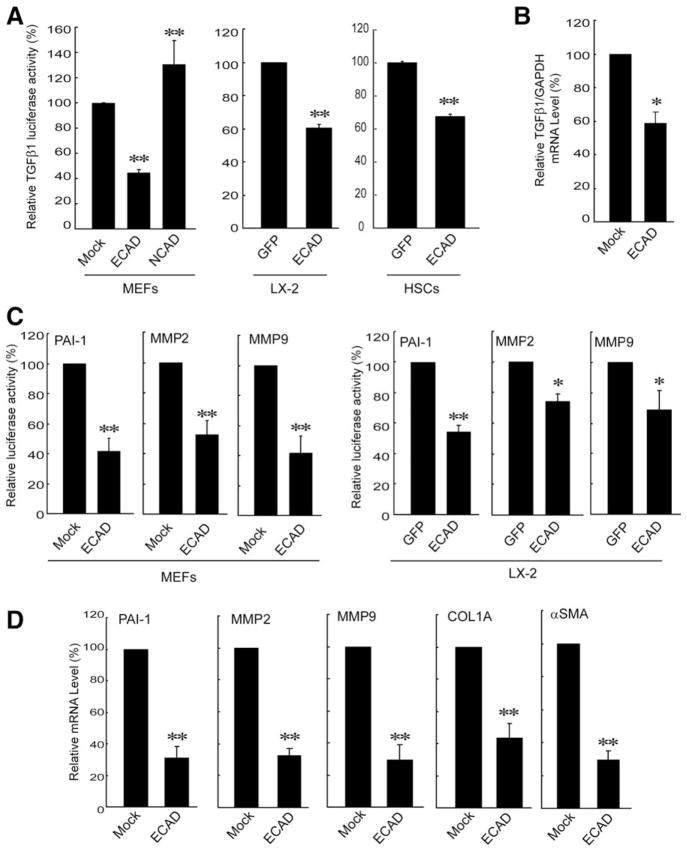
TGFβ1 stimulates HSC activation and liver fibrosis.6,13 Therefore, the effect of forced expression of ECAD on the TGFβ1 gene was monitored in subsequent experiments. Overexpression of ECAD elicited a significant decrease in luciferase activity from a TGFβ1 promoter construct in MEF cells, but this was not observed in cells transfected with NCAD (Fig. 2A, left). Instead, TGFβ1 luciferase activity was moderately increased by NCAD overexpression. A decrease in the basal TGFβ1 expression by ECAD was also confirmed in LX-2 cells and HSCs (Fig. 2A, middle and right). Real-time PCR analysis verified the ability of ECAD to repress the TGFβ1 gene (Fig. 2B). TGFβ1 promotes the remodeling and deposition of ECM through the activation of downstream target genes such as PAI-1 and MMPs.6,11,12 In agreement with the repression of TGFβ1, the basal luciferase activities of PAI-1, MMP2, and MMP9 were all inhibited by ECAD overexpression in both MEFs and LX-2 cells (Fig. 2C). As expected, ECAD expression decreased the transcript levels of PAI-1, MMP2, MMP9, collagen type IA (COL1A), and αSMA (Fig. 2D). Our results support the contention that ECAD has the ability to inhibit TGFβ1 gene expression and thereby repress its downstream target genes.
Fig. 2.
Effects of ECAD on TGFβ1 and its target gene transactivation. (A) Inhibition of the basal TGFβ1 gene expression by ECAD. MEFs were transfected with a TGFβ1 luciferase construct in combination with the construct encoding for ECAD or NCAD (pCEP4 for mock transfection). LX-2 cells or HSCs were infected with adenoviral GFP or ECAD. The activity of luciferase was normalized by the activity of β-galactosidase. (B) Real-time PCR assays. TGFβ1 mRNA levels were measured in MEFs that had been stably transfected with ECAD. (C) Reporter gene activities. (D) Realtime PCR assays in MEFs stably transfected with ECAD. The data are the means and standard errors of at least three separate experiments (significantly different in comparison with a mock transfection or adenoviral GFP infection: **P < 0.01 and *P < 0.05).
Inhibition of TGFβ1-Inducible SBE Activity and Smad3/2 Phosphorylation
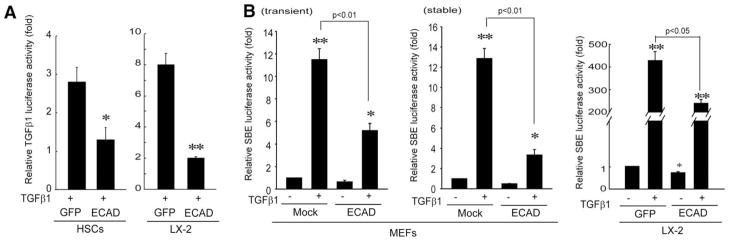
The signaling pathway activated by TGFβ1 involves TGFβ1 receptor–mediated cell signaling. In an effort to identify the basis for ECAD’s regulation of the TGFβ1 signaling pathway, we measured the effect of forced expression of ECAD on the TGFβ1-mediated induction of its own gene. The exposure of primary HSCs to TGFβ1 (5 ng/mL for 12 hours) caused a 2.8-fold increase in TGFβ1 reporter gene activity in comparison with a control, and this was abolished by ECAD (Fig. 3A, left). Similarly, TGFβ1-inducible TGFβ1 luciferase activity was also reduced by ECAD in LX-2 cells (Fig. 3A, right). To assess whether ECAD inhibits SBE-mediated gene induction in response to TGFβ1 treatment, we performed reporter gene assays in MEFs or LX-2 cells transfected with pGL3-(CAGA)9-MLP luciferase. As expected, ECAD overexpression significantly decreased TGFβ1-inducible SBE luciferase activity in these cells (Fig. 3B): the effects of transient and stable transfections were comparable. The SBE reporter activity in MEFs was less than 10% of that in LX-2 cells.
Fig. 3.
Effects of ECAD on TGFβ1-inducible reporter gene activities. (A) TGFβ1 reporter gene activity. Cells were treated with TGFβ1 (5 ng/mL for 12 hours). (B) Smad reporter activity. MEFs or LX-2 cells that had been transiently transfected with ECAD in combination with an SBE-driven reporter construct were treated with TGFβ1 for 12 hours. The reporter activity was also measured in MEFs stably transfected with ECAD. The data are the means and standard errors of three separate experiments (significantly different in comparison with a mock transfection: *P < 0.05 and **P < 0.01).
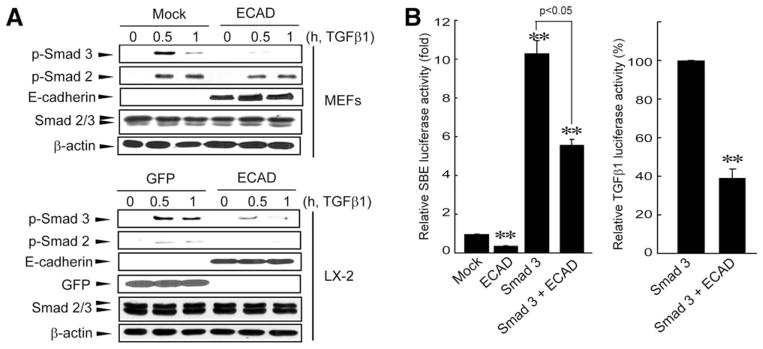
TGFβ1 receptor–mediated cell signaling depends on Smad3/2 phosphorylation; this allows phosphorylated Smad3/2 to form oligomers with Smad4. The resultant complex translocates into the nucleus and there acts as a transcriptional activator.10 To address the downstream link between ECAD and TGFβ1 repression, we assessed the inhibitory effect of ECAD on TGFβ1-dependent Smad3/2 phosphorylation. The treatment of mock-transfected MEFs or GFP-infected LX-2 cells with TGFβ1 enhanced Smad3/2 phosphorylation (Fig. 4A). Intriguingly, ECAD overexpression attenuated the phosphorylation of Smad3 and, to a minor extent, that of Smad2. A similar change was observed in LX-2 cells treated with TGFβ1 after the adenoviral infection of ECAD. As we anticipated, ECAD inhibited the ability of Smad3 to induce luciferase activity from an SBE-driven reporter or TGFβ1 reporter construct (Fig. 4B). Our results indicate that ECAD inhibits Smad3/2 phosphorylation and thus antagonizes Smad-dependent gene transcription.
Fig. 4.
Effect of ECAD overexpression on Smad activity. (A) Immunoblots of phosphorylated Smad3 and Smad2. MEFs stably transfected with ECAD or LX-2 cells infected with ECAD were treated with TGFβ1 (5 ng/mL). (B) Smad and TGFβ1 reporter gene activities. The data are the means and standard errors of three separate experiments [significantly different in comparison with a mock transfection (left) or a Smad3 transfection (right): **P < 0.01].
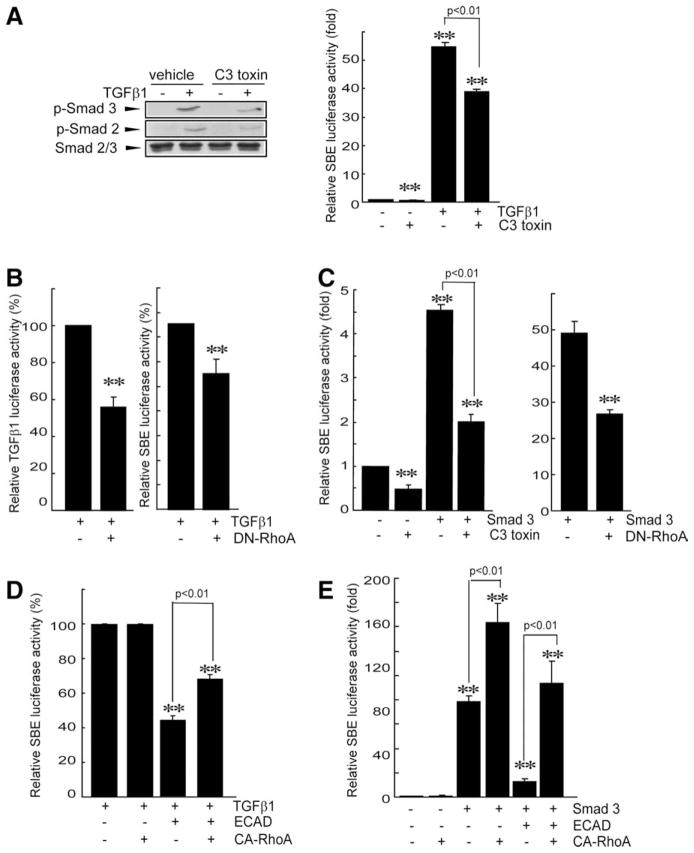
RhoA Regulation of Smad3/2 Phosphorylation and Gene Induction
In addition to the Smad pathway, TGFβ1 receptor signaling activates other pathways such as small guanosine triphosphatase (GTPase), mitogen-activated protein kinases, and phosphatidylinositol 3-kinase.10 These pathways may crosstalk with Smad signaling.10,16 In particular, RhoA regulates Smad phosphorylation and Smad-dependent gene induction in response to TGFβ1.16 To verify the regulatory role of RhoA in TGFβ1-dependent Smad activation, the phosphorylation status of Smad3/2 was monitored in cells treated with cell-permeable C3 toxin (a RhoA inhibitor) or cells transfected with a dominant negative mutant of ras homolog gene family A (DN-RhoA). The treatment of LX-2 cells with C3 toxin led to a reduction in Smad3/2 phosphorylation and, consequently, inhibited the ability of TGFβ1 to induce SBE luciferase activity (Fig. 5A). In addition, transfection with DN-RhoA repressed the TGFβ1-dependent induction of its own gene or the SBE reporter gene (Fig. 5B). Similarly, either C3 toxin treatment or DN-RhoA transfection significantly attenuated Smad3-inducible SBE reporter gene activity (Fig. 5C), and this confirmed that RhoA inhibition antagonizes Smad3-dependent gene transcription. Moreover, transfection with a construct encoding for the constitutively active mutant of ras homolog gene family A (CA-RhoA) reversed the ability of ECAD to inhibit TGFβ1-inducible or Smad3-inducible SBE luciferase activity (Fig. 5D,E). These results indicate that the inhibition of Smad activity by ECAD may be associated with RhoA inhibition.
Fig. 5.
Association of RhoA with ECAD for the repression of Smad activity. (A) Effects of C3 toxin on Smad3/2 phosphorylation and Smad-driven reporter activity. The levels of phosphorylated Smad3/2 were measured in LX-2 cells that had been treated with TGFβ1 for 1 hour after a cell-permeable C3 toxin treatment for 4 hours. (B) Inhibition by DN-RhoA of TGFβ1-inducible TGFβ1 reporter or Smad reporter activities. Luciferase activities were measured in LX-2 cells that had been treated with TGFβ1 for 12 hours after DN-RhoA transfection. (C) Repression of Smad3-inducible SBE reporter activity by RhoA inhibition. SBE luciferase activities were measured on the lysates of cells that had been treated with C3 toxin or transfected with DN-RhoA after Smad3 transfection. (D) Reversal by CA-RhoA of the ability of ECAD to inhibit TGFβ1-inducible SBE luciferase activity. (E) Reversal by CA-RhoA of the ability of ECAD to inhibit Smad3-inducible SBE luciferase activity. The data are the means and standard errors of three separate experiments (significantly different in comparison with a mock transfection: **P < 0.01).
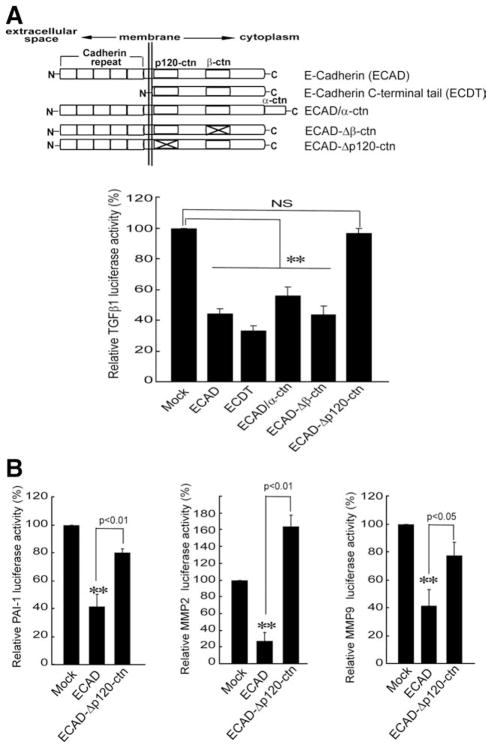
Role of the p120-ctn Binding Domain of ECAD in TGFβ1 Repression
ECAD has an impact on adherens junctions through extracellular repeated domains of cadherin, whereas intracellular domains of ECAD regulate signaling pathways.1,2 ECAD contains intracellular binding domains that directly interact with p120-ctn or β-ctn.1 In order to understand in more depth the mechanism underlying ECAD and RhoA, we measured the abilities of several mutant constructs of ECAD to inhibit TGFβ1 reporter gene activity (Fig. 6A, upper). Transfection (transient) with a construct encoding the C-terminal intracellular domain of E-cadherin (ECDT) resulted in a decrease in TGFβ1 luciferase activity comparable to that obtained with full-length ECAD (Fig. 6A, bottom), and this supports the concept that the intracellular domain is responsible for TGFβ1 gene repression. Either ECAD/α-ctn, which encodes for ECAD fused with α-ctn, or ECAD–Δβ-ctn, which encodes for an ECAD mutant deficient in β-ctn binding domain, also inhibited TGFβ1 reporter activity to a similar extent. In contrast, the transfection of ECAD–Δp120-ctn, which expresses a mutant ECAD in the p120-ctn binding domain, failed to repress TGFβ1 gene transcription. Therefore, repression of the TGFβ1 gene by ECAD may rely on the p120-ctn binding domain. In addition, the lack of inhibitory effects of ECAD–Δp120-ctn on the PAI-1, MMP2, or MMP9 luciferase activities verified the important role of the p120-ctn binding domain in the repression of the genes (Fig. 6B). In light of these results, we conclude that the p120-ctn binding domain of ECAD may be involved in repressing TGFβ1 or its downstream gene induction.
Fig. 6.
Effects of ECAD mutant constructs on the expression of the TGFβ1 gene or its target genes. (A) TGFβ1 luciferase activity. The schematic representation shows the major protein binding domains. The basal TGFβ1 reporter activities were measured in MEF cells that had been transiently transfected with each mutant construct of ECAD. (B) Reporter gene activities. The data are the means and standard errors of at least three separate experiments (significantly different in comparison with a mock transfection: **P < 0.01). Abbreviation: NS, not significant.
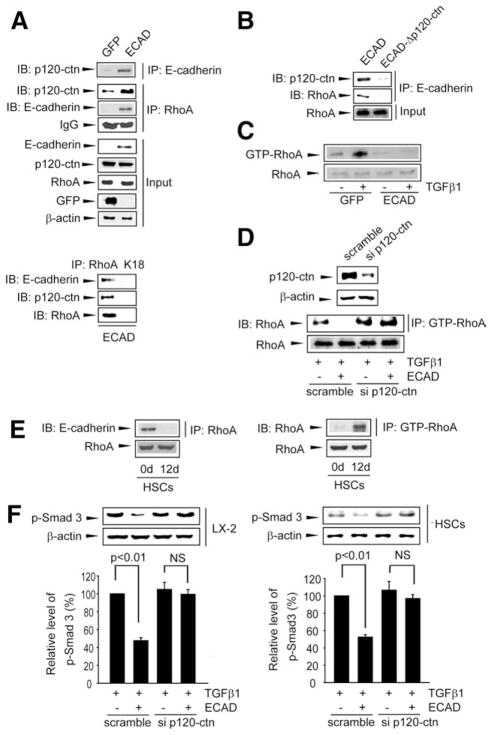
Recruitment of RhoA to p120-ctn Bound to ECAD
ECAD regulates the activity of small GTPase via p120-ctn.1,17 In another effort to understand the association between ECAD and RhoA, we explored the role of p120-ctn in the interaction of these molecules (Fig. 7A). As expected, the forced expression of ECAD notably increased its association with p120-ctn in LX-2 cells according to immunoprecipitation and immunoblot assays. Also, enforced ECAD expression increased the interaction between p120-ctn and RhoA: ECAD promoted RhoA recruitment to its complex with p120-ctn, although it did not alter the basal expression levels of p120-ctn and RhoA. The hypothesis that RhoA is recruited to ECAD through p120-ctn binding was verified by the lack of ECAD binding to RhoA in cells transfected with ECAD–Δp120-ctn (Fig. 7B).
Fig. 7.
Interaction of ECAD with RhoA through p120-ctn binding. (A) IP-IB assays. ECAD immunoprecipitates were immunoblotted for p120-ctn. For the assessment of protein interactions, RhoA immunoprecipitates were immunoblotted for p120-ctn or ECAD; 10% of the total lysates were used as an input control. K18 was used as a negative control. (B) Association of ECAD with RhoA through p120-ctn. LX-2 cells were transfected with ECAD or ECAD–Δp120-ctn. (C) RhoA activity assay. RhoA activity was measured in LX-2 cells treated with TGFβ1 5 minutes after ECAD infection. (D) Effect of p120-ctn knockdown on the ability of ECAD to inhibit RhoA. LX-2 cells were infected with an adenoviral ECAD construct, were transfected with p120-ctn siRNA, and then were exposed to TGFβ1. (E) RhoA binding to ECAD and RhoA activity. The interactions between RhoA and ECAD (left) and RhoA activity (right) were measured in HSCs on days 0 and 12. (F) Effect of p120-ctn knockdown on TGFβ1-inducible Smad3 phosphorylation. The data are the means and standard errors of at least three separate experiments (significantly different in comparison with GFP-infected cells treated with TGFβ1). Abbreviations: IB, immunoblotting; IgG, immunoglobulin G; IP, immunoprecipitation; K18, cytokeratin 18; NS, not significant.
Because of the regulatory role of RhoA in the Smad signaling pathway, the effect of ECAD on RhoA activity was determined in cells treated with TGFβ1. As expected, TGFβ1 treatment increased RhoA activity in comparison with a control, which was completely antagonized by ECAD overexpression (Fig. 7C). The ECAD-mediated RhoA inhibition was reversed by siRNA targeting p120-ctn (Fig. 7D). In addition, we examined the physical interaction between RhoA and ECAD in HSCs on days 0 and 12. As expected, ECAD interacted with RhoA on day 0, but this was abrogated by a deficiency in ECAD on day 12 (Fig. 7E, left). Consistently, RhoA activity increased in the activated HSCs (Fig. 7E, right). Likewise, the ability of ECAD to inhibit Smad3 phosphorylation was attenuated by p120-ctn knockdown in either LX-2 cells or primary HSCs (Fig. 7F).
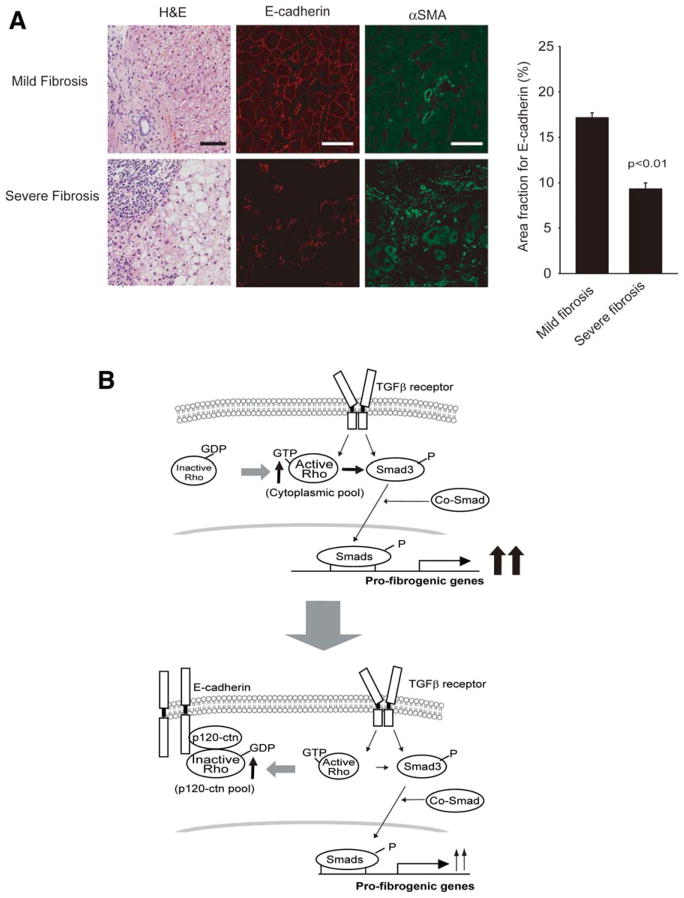
In an effort to show the biological relevance of ECAD function in clinical situations, we compared ECAD expression levels in groups of patients with mild or severe fibrosis. The levels of ECAD were clearly higher in patients with mild fibrosis versus patients with severe fibrosis (Fig. 8A, left). In contrast, αSMA expression levels increased as the disease progressed. Multiple analyses of the human liver samples indicated that ECAD expression reciprocally correlated with the severity of fibrosis (Fig. 8A, right) and verified the biological function and relevance of ECAD in human liver fibrosis.
Fig. 8.
ECAD expression and fibrosis. (A) ECAD expression in patients with mild fibrosis (Ishak fibrosis score ≤ 2) or severe fibrosis (Ishak fibrosis = 6). Liver sections (nine patients in each group) were stained for H&E and for ECAD (red) and αSMA (green) immunohistochemically. The pictures are representative images. The scale bars represent 50 μm. The area fraction for ECAD (%) was determined on the liver sections with image analysis software. The data are means and standard errors (n = 9; significantly different in comparison with mild fibrosis). (B) Proposed signaling pathway by which ECAD prevents Smad3-dependent TGFβ1 gene induction. Abbreviation: H&E, hematoxylin and eosin.
Collectively, all these results provide compelling evidence that ECAD inhibits RhoA activity by recruiting RhoA to p120-ctn bound to the p120-ctn binding domain, and this prevents RhoA-dependent Smad signaling pathway in HSCs (Fig. 8B).
Discussion
In the healthy liver, quiescent HSCs show no fibrogenic phenotype and have a low proliferative capacity. These HSCs are the major vitamin A storage sites. Repeated injury of any etiology triggers various inflammatory processes such as cytokine production, inflammatory cell recruitment, and a phenotypic transition of HSCs to more contractile and fibrogenic myofibroblasts. 6 Activated HSCs with a myofibroblast-like phenotype lose their lipid droplets, proliferate, migrate to zone 3 of the acinus, and produce collagen types I, III, and IV and laminin. Thus, activated HSCs are responsible for the development and establishment of fibrosis, a prepathological state of cirrhosis. Liver cirrhosis results in hepatic parenchymal cell destruction, the formation of septa and nodules, and alteration of the blood flow.6
ECAD is expressed as a major form in quiescent HSCs7 and most normal cells within epithelial tissues. When HSCs are activated, the level of ECAD expression decreases through the process of cadherin switching (i.e., a switch from ECAD expression to NCAD expression). Therefore, this is a conversion to NCAD expression followed by a loss of ECAD. Activated HSCs then alter the gene expression profile and acquire a migratory phenotype.1,6,7 It has been shown that during the culture-related transition of quiescent HSCs to myofibroblastic HSCs, ECAD and other epithelial markers are down-regulated, whereas mesenchymal markers are enhanced.7 Hedgehog and ras-related C3 botulinum toxin substrate (Rac) signaling may regulate the EMT of HSCs.7,8 However, no information was available about the significance of ECAD with respect to the inhibition of HSC activation. Our results demonstrate that ectopic ECAD expression prevents HSC activation. Thus, a deficiency of ECAD may facilitate the activation or motility of HSCs. Sometimes, increased expression of NCAD without a change in ECAD expression is called cadherin switching. In a study,18 increased motility of epithelial cells was claimed to be associated with NCAD up-regulation. Thus, HSC activation may result in part from the increased expression of NCAD as well as the loss of ECAD.
In addition to the fibrotic process in the liver, cadherin switching is involved in other physiological and pathological conditions such as the normal physiology of embryonic development, chronic inflammation, and the invasion and metastasis of cancer cells.1,2 In clinical studies, the loss of ECAD in many epithelium-derived cancer cells promotes the conversion of the epithelial phenotype into a more motile and less polarized mesenchymal phenotype.1,19 Consistently, decreased ECAD expression has been observed in approximately 40% of hepatocellular carcinoma samples. 20 Activated HSCs serve as liver-specific pericytes in hepatic carcinogenesis and may contribute to the remodeling and deposition of tumor-associated ECM.13 Because of the link between ECAD loss and the pathological process of EMT, information on the molecular basis of ECAD signaling may be helpful in understanding the development and progression of hepatocellular carcinoma.
TGFβ1 represses the expression of ECAD and promotes the following temporal sequence: disassembly of cell junctions, loss of epithelial polarity, cytoskeletal reorganization, and cell-matrix adhesion remodeling.9 Transcription factors such as Snail, Twist, Slug, and Zeb negatively regulate the expression of ECAD by binding to specific sequences within the ECAD gene, and these sequences are called E-boxes. These proteins are involved in the pathological process of EMT and thereby enhance the accumulation of ECM. Although ECAD deficiency or cadherin switching had been recognized during HSC activation in liver disease,7 the inhibitory role of ECAD in fibrogenesis had not been studied. Moreover, despite the well-known process of the disintegration and disassembly of cell-cell junctions by TGFβ1, information about whether ECAD has an inhibitory effect on TGFβ1 gene expression was not available. Our results demonstrate that ECAD prevents the induction of the TGFβ1 gene and its downstream genes, whereas the loss of ECAD initiates it and facilitates hepatic fibrosis.
Sustained injury to hepatocytes activates fibrogenic mechanisms in patients with chronic liver diseases induced by any means. Fibrogenic cells (i.e., myofibroblasts) may be derived from hepatocytes, biliary epithelial cells, portal fibroblasts, or local mesenchymal cells from bone marrow as well as HSCs.5,6 All these cells exhibit a reduction in ECAD expression with the increased expression of NCAD. Even though it was recognized that the expression of different cadherin forms allows a select population of cells to separate from other cell types, whether ECAD itself directly affects profibrogenic signaling was unclear. The intracellular region of ECAD contains ctn binding domains and regulates ctn-mediated signaling.1 The important finding of our study is the identification of p120-ctn as a docking molecule of RhoA in HSCs. This is supported by the following observations: ECAD–Δp120-ctn failed to inhibit the expression of TGFβ1 and its target genes, and siRNA knockdown of p120-ctn reversed ECAD’s inhibition of RhoA activity and Smad3 phosphorylation. Therefore, the signaling pathway mediated by p120-ctn bound to ECAD appeared to be responsible for TGFβ1 repression in cells of the epithelial type. It has also been shown that forced expression of NCAD in epithelial cells causes down-regulation of ECAD through increased degradation,18 and this may also be linked to the function of p120-ctn.
p120-ctn stabilizes cadherins and affects cell migration, morphogenesis, and proliferation.21 Therefore, altered localization and decreased expression of p120-ctn are associated with the malignancy of certain cancers. 21 Because the cadherin/p120-ctn complex regulates the activities of small GTPase (e.g., Rho),1,17 p120-ctn may inhibit RhoA activity in certain types of cells. In the present study, the inhibition of Rho activity prevented Smad3/2 phosphorylation and gene transactivation, and this is in line with the finding that Rho/Rho-associated protein kinase (ROCK) inhibitors ameliorate liver fibrosis and TGFβ1 expression.22 In addition, our data illustrate that ECAD’s inhibition of Smad activity was reversed by CA-RhoA, and this supports the physiological importance of RhoA recruitment to ECAD. Another important finding of this study is that ECAD-mediated stalling of RhoA depends on p120-ctn binding. In other studies, activated HSCs showed sustained activation of Rac1, another Rho family member, and perturbation of Rac1 activity blocked the phenotypic transition.8,23
Signals downstream from the TGFβ1 receptor activation merge on the major transcription factors (including Smads). Notably, Smad3 and Smad2 are differentially activated by TGFβ1 in HSCs; in quiescent HSCs, TGFβ1 receptor activation promotes Smad2 phosphorylation, whereas in transdifferentiated HSCs, it promotes Smad3 phosphorylation.24 Consistently, our findings indicate that the loss of ECAD activated Smad3 to a greater extent than Smad2 in both LX-2 cells (activated HSCs) and MEFs. This is consistent with the observation that a Smad3 deficiency ameliorates epithelial degeneration and fibrosis.25 Collectively, our results demonstrate that ECAD has the ability to inhibit Smad3/2 phosphorylation by recruiting RhoA to p120-ctn at the p120-ctn binding domain (i.e., an increase in inactive RhoA at the p120-ctn pool), and they provide important information about how ECAD antagonizes liver fibrosis. Consequently, the loss of ECAD due to cadherin switching up-regulates TGFβ1 and its target genes.
ECAD also interacts with the endothelial growth factor (EGF) receptor and, by restricting the mobility of the receptor, inhibits EGF-dependent signaling.26 Activating protein 1 is another transcription factor complex activated by TGFβ1,13 and it is required for EGF-mediated biological effects. However, the inhibition of activating protein 1 by a c-Jun N-terminal kinase deficiency does not affect Smad3/2 phosphorylation 27; no crosstalk is shown between the activation of these two transcription complexes. Thus, ECAD is likely to prevent the clustering of a set of cell surface receptors and inhibit receptor-mediated cell signaling and gene induction. Because the interaction of VE-cadherin with the cell surface receptor may also contribute to TGFβ1 signaling,28 ECAD overexpression and the resultant repression of other cadherins may work together to switch cell signaling and prevent the EMT process.
In conclusion, ECAD inhibits Smad3/2 phosphorylation by recruiting RhoA to p120-ctn at the p120-ctn binding domain, whereas the loss of ECAD due to cadherin switching promotes the expression of TGFβ1 and its target genes and facilitates liver fibrosis. Our results, showing a reciprocal correlation between ECAD expression and fibrosis severity in human liver samples, strengthens this concept.
Supplementary Material
Acknowledgments
The kind donation of pMLP-(SBE)-luciferase and pCDNA-flagSmad3 from Dr. H. S. Choi is gratefully acknowledged.
This work was supported by the World Class University project (which is funded by the Korean Ministry of Education, Science, and Technology) through grant R32-2008-000-10098-0 and by the National Research Foundation of Korea (which is also funded by the Korean Ministry of Education, Science, and Technology) through grant R11-2007-107-00000-0. Il Je Cho was supported by the Korea Research Foundation (which is funded by the Korean Ministry of Education and Human Resources Development) through grant KRF-2007-355-E00007.
Abbreviations
- αSMA
α-smooth muscle actin
- BMP
bone morphogenetic protein
- CA-RhoA
constitutively active mutant of ras homolog gene family A
- COL1A
collagen type IA
- ctn
catenin
- DMN
dimethylnitrosamine
- DN-RhoA
dominant negative mutant of ras homolog gene family A
- ECAD
E-cadherin
- ECDT
C-terminal intracellular domain of E-cadherin
- ECM
extracellular matrix
- EGF
endothelial growth factor
- EMT
epithelial-mesenchymal transition
- GFAP
glial fibrillar acidic protein
- GFP
green fluorescence protein
- GTPase
guanosine triphosphatase
- H&E
hematoxylin and eosin
- HSC
hepatic stellate cell
- IB
immunoblotting
- Id
inhibitor of DNA binding
- IgG
immunoglobulin G
- IP
immunoprecipitation
- K18
cytokeratin 18
- MEF
murine embryonic fibroblast
- MMP
matrix metalloproteinase
- mRNA
messenger RNA
- NCAD
N-cadherin
- NS
not significant
- PAI-1
plasminogen activator inhibitor 1
- PCR
polymerase chain reaction
- RAC
ras-related C3 botulinum toxin substrate
- RhoA
ras homolog gene family
- SBE
Smad binding element
- siRNA
small interfering RNA
- TGFβ
transforming growth factor β
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121:727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 2.Gumbiner BM. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- 3.Wong AS, Gumbiner BM. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol. 2003;161:1191–1203. doi: 10.1083/jcb.200212033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol. 2001;153:1049–1060. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170:1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi SS, Omenetti A, Witek RP, Moylan CA, Syn WK, Jung Y, et al. Hedgehog pathway activation and epithelial-to-mesenchymal transitions during myofibroblastic transformation of rat hepatic cells in culture and cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G1093–G1106. doi: 10.1152/ajpgi.00292.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi SS, Witek RP, Yang L, Omenetti A, Syn WK, Moylan CA, et al. Activation of Rac1 promotes hedgehog-mediated acquisition of the myofibroblastic phenotype in rat and human hepatic stellate cells. Hepatology. 2010;52:278–290. doi: 10.1002/hep.23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zavadil J, Böttinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 10.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto T, Takahashi S, Nakamura E, Nagaya K, Hayashi T, Fujieda K. Transforming growth factor-beta1 induces matrix metalloproteinase-9 expression in human meningeal cells via ERK and Smad pathways. Biochem Biophys Res Commun. 2009;383:475–479. doi: 10.1016/j.bbrc.2009.04.038. [DOI] [PubMed] [Google Scholar]
- 12.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SJ, Yang JW, Cho IJ, Kim WD, Cho MK, Lee CH, et al. The gep oncogenes, Gα12 and Gα13, upregulate the transforming growth factor-β1 gene. Oncogene. 2009;28:1230–1240. doi: 10.1038/onc.2008.488. [DOI] [PubMed] [Google Scholar]
- 14.Kim SG, Kim YM, Choi YH, Lee MG, Choi JY, Han JY, et al. Pharmacokinetics of oltipraz and its major metabolite in patients with liver fibrosis or cirrhosis: relationship with suppression of circulating TGF-β1. Clin Pharmacol Ther. 2010;88:360–368. doi: 10.1038/clpt.2010.89. [DOI] [PubMed] [Google Scholar]
- 15.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Crawford M, Day RM, Briones VR, Leader JE, Jose PA, et al. RhoA modulates Smad signaling during transforming growth factor-beta-induced smooth muscle differentiation. J Biol Chem. 2006;281:1765–1770. doi: 10.1074/jbc.M507771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, et al. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- 18.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147:631–644. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul R, Ewing CM, Jarrard DF, Isaacs WB. The cadherin cell-cell adhesion pathway in prostate cancer progression. Br J Urol. 1997;79:37–43. doi: 10.1111/j.1464-410x.1997.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 20.Yang MH, Chen CL, Chau GY, Chiou SH, Su CW, Chou TY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50:1464–1474. doi: 10.1002/hep.23221. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds AB. p120-catenin: past and present. Biochim Biophys Acta. 2007;1773:2–7. doi: 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura K, Tada S, Nakamoto N, Toda K, Horikawa H, Kurita S, et al. Rho/Rho kinase is a key enzyme system involved in the angiotensin II signaling pathway of liver fibrosis and steatosis. J Gastroenterol Hepatol. 2007;22:2022–2033. doi: 10.1111/j.1440-1746.2006.04735.x. [DOI] [PubMed] [Google Scholar]
- 23.Choi SS, Sicklick JK, Ma Q, Yang L, Huang J, Qi Y, et al. Sustained activation of Rac1 in hepatic stellate cells promotes liver injury and fibrosis in mice. Hepatology. 2006;44:1267–1277. doi: 10.1002/hep.21375. [DOI] [PubMed] [Google Scholar]
- 24.Liu C, Gaca MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem. 2003;278:11721–11728. doi: 10.1074/jbc.M207728200. [DOI] [PubMed] [Google Scholar]
- 25.Saika S, Kono-Saika S, Ohnishi Y, Sato M, Muragaki Y, Ooshima A, et al. Smad3 signaling is required for epithelial-mesenchymal transition of lens epithelium after injury. Am J Pathol. 2004;164:651–663. doi: 10.1016/S0002-9440(10)63153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian X, Karpova T, Sheppard AM, McNally J, Lowy DR. E-cadherin-mediated adhesion inhibits ligand-dependent activation of diverse receptor tyrosine kinases. EMBO J. 2004;23:1739–1748. doi: 10.1038/sj.emboj.7600136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcorn JF, Guala AS, van der Velden J, McElhinney B, Irvin CG, Davis RJ, et al. Jun N-terminal kinase 1 regulates epithelial-to-mesenchymal transition induced by TGF-beta1. J Cell Sci. 2008;121:1036–1045. doi: 10.1242/jcs.019455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudini N, Felici A, Giampietro C, Lampugnani M, Corada M, Swirsding K, et al. VE-cadherin is a critical endothelial regulator of TGF-beta signalling. EMBO J. 2008;27:993–1004. doi: 10.1038/emboj.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.