Abstract
The mitochondrial protein apoptosis-inducing factor (AIF) plays a pivotal role in poly(ADP-ribose) polymerase-1 (PARP-1)-mediated cell death (parthanatos), during which it is released from the mitochondria and translocates to the nucleus. Here, we show that AIF is a high affinity poly(ADP-ribose) (PAR)–binding protein and that PAR binding to AIF is required for parthanatos both in vitro and in vivo. AIF bound PAR at a site distinct from AIF’s DNA binding site and this interaction triggered AIF release from the cytosolic side of the mitochondrial outer membrane. Mutation of the PAR binding site in AIF did not affect its NADH oxidase activity, its ability to bind FAD or DNA, or its ability to induce nuclear condensation. However, this AIF mutant was not released from mitochondria and did not translocate to the nucleus or mediate cell death following PARP-1 activation. These results suggest a mechanism for PARP-1 to initiate AIF-mediated cell death and indicate that AIF’s bioenergetic cell survival-promoting functions are separate from its effects as a mitochondrially-derived death effector. Interference with the PAR-AIF interaction or PAR signaling may provide unique opportunities for preventing cell death following activation of PARP-1.
INTRODUCTION
Poly(ADP-ribose) (PAR) polymerase-1 (PARP-1) is an important nuclear enzyme that responds to DNA damage and is required for DNA repair (1, 2). Upon activation, PARP-1 catalyzes the transfer of ADP-ribose from nicotinamide adenine dinucleotide (NAD+) and conjugates PAR onto a variety of nuclear proteins such as histones, DNA polymerases, topoisomerases, and transcription factors as well as automodification of PARP-1 itself, thus regulating a variety of physiologic processes. Excessive activation of PARP-1 leads to an intrinsic cell death program, which has been designated parthanatos (PARP-1-dependent cell death) to distinguish it from necrosis and apoptosis (1, 2). PARP inhibition or PARP-1 gene deletion is markedly protective in models of many cell injury paradigms, including stroke, trauma, ischemia-reperfusion injury, diabetes, and neurodegenerative diseases, indicating that parthanatos plays a prominent role in these disorders (3).
Apoptosis-inducing factor (AIF) is a mitochondrial oxidoreductase. It, like cytochrome C, has two independent functions: One within mitochondria, involving cell survival probably by assembly or stabilization of respiratory complex I (4), and another as a promoter of cell death, as AIF is released into the cytoplasm following PARP-1 activation, ultimately entering the nucleus to induce cell death (1, 5, 6). It has been difficult to separate AIF’s dual functions, because complete loss of AIF disrupts mitochondrial function and energy metabolism, preventing an assessment of its role as a death effector (7). Blocking mitochondrial AIF release or reducing AIF abundance protects cells against parthanatos, indicating that AIF plays a crucial role in cell death induced by PARP-1 activation (2, 8–10). Calpain, a calcium-dependent intracellular cysteine protease, has been suggested to cleave AIF at the N-terminus and cause AIF release from mitochondria after transient focal ischemia (11). However, calpain is not involved in mitochondrial AIF release during parthanatos (12, 13).
PAR acts as a pro-death signaling molecule in parthanatos. Following excessive PARP-1 activation, PAR, which is mainly produced in the nucleus, translocates to the cytosol and interacts with the mitochondrial outer surface where it induces AIF release (8, 14). Decreasing PAR abundance by PAR glycohydrolase (PARG), which degrades the PAR polymer, prevents PARP-1-dependent AIF release and reduces parthanatos in N-methyl-D-aspartate (NMDA) receptor-mediated glutamate excitotoxicity and N-methyl-N-nitro-N-nitrosoguanidine (MNNG) toxicity, and markedly reduces infarct volume in mice after two hours of transient middle cerebral artery occlusion (MCAO) (2, 8). How PAR stimulates the release of AIF is not known.
We recently performed an unbiased proteomic screen for PAR-binding proteins and identified AIF as a candidate PAR-binding protein (15). Here, we show that AIF contains a PAR-binding motif (PBM), and that PAR binding to AIF is required for its release from the mitochondria and its ability to induce cell death in parthanatos. Moreover, mutating the PAR-binding site in AIF allows the separation of AIF’s role in energy metabolism from its cell death function.
RESULTS
PAR binds to AIF
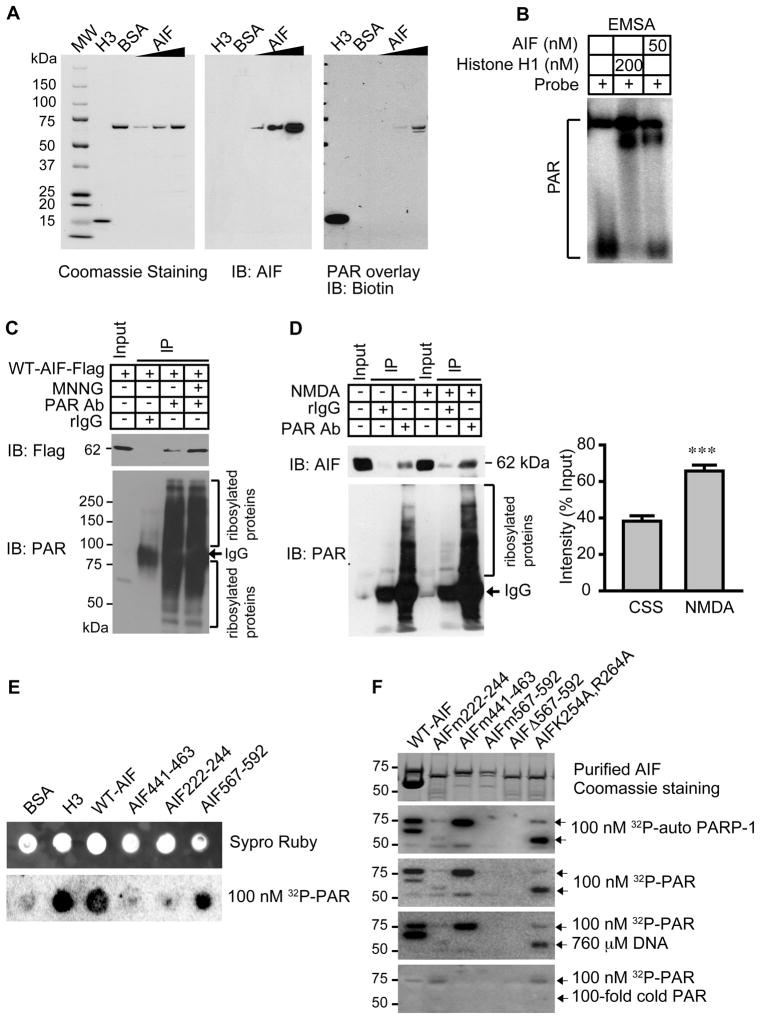
To ascertain whether AIF binds to PAR, we performed an overlay assay on recombinant AIF with affinity-purified biotin-labeled PAR (Fig. 1A). Histone H3, which binds to PAR with high affinity, was included as a positive control and bovine serum albumin (BSA) was included as a negative control (16). AIF bound to biotin-labeled PAR in a concentration-dependent manner (Fig. 1A). Furthermore, an electrophoretic mobility shift assay (EMSA) revealed that AIF caused a shift in PAR mobility (Fig. 1B). To ascertain whether PAR binds AIF in intact cells, we exposed HeLa cells stably transduced with lentivirus C-terminal Flag-tagged mouse wild type (WT) AIF (WT-AIF-Flag) to MNNG, a DNA alkylating agent that activates PARP-1 and kills cells primarily through parthanatos (3) (Fig. 1C). PAR immunoprecipitation was performed from postnuclear fractions, which is the fraction prepared from whole cell lysates after removing nuclear proteins. WT-AIF-Flag co-immunoprecipitated with PAR in resting cells. Following MNNG treatment, the interaction between AIF and PAR was significantly increased (Fig 1C, fig. S1, A and B, p < 0.001). The interaction between endogenous PAR and AIF was explored in primary cortical neurons under both resting conditions and after NMDA glutamate receptor stimulation, which activates PARP-1 and kills neurons through parthanatos (17). Endogenous AIF interacted with PAR in non-stimulated cortical neurons, an interaction that was significantly increased following NMDA treatment (Fig. 1D). Together, these data suggest that AIF is a PAR-binding protein and that PARP-1 activation increases the AIF-PAR interaction.
Fig. 1. PAR binds to AIF.
(A) Overlay assay of mouse AIF and biotin-labeled PAR. H3 and BSA were used as positive and negative controls, respectively. n = 3. (B) EMSA of AIF using 32P-labeled PAR. Histone H1 was used as a positive control, n = 3. (C) Co-immunoprecipitation of WT-AIF-Flag with PAR in post-nuclear fractions isolated from HeLa cells 2 h after MNNG treatment (50 μM for 15 min). rIgG, rabbit IgG. n = 3. (D) Co-immunoprecipitation of endogenous AIF with PAR in post-nuclear fractions isolated from cortical neurons 2 h after NMDA treatment (500 μM for 5 min). The intensity of AIF signal was quantified and normalized to input (right panel). *** p < 0.001 by Student’s t-test, n = 6. (E) Dot-blot analysis of full length WT-AIF, three AIF peptides, histone H3, and BSA in presence of 100 nM purified 32P-labeled PAR, n = 4. (F) Overlay assay of His-tagged WT-AIF, AIFm222-244, AIFm441-463, AIFm567-592, AIFΔ567-592, and AIF (K254A, R264A) mutants with 32P-labeled automodified PARP-1 (equivalent to 100 nM of PAR), purified 32P-labeled PAR (100 nM), or purified 32P-labeled PAR with either a 100-fold excess of DNA or cold PAR.
Arg588, Lys589 and Arg592 in AIF are critical for PAR-binding
The PBM comprises approximately 20 amino acids and is characterized by hydrophobic amino acids separated by basic amino acids, and by a cluster of basic amino acid residues at the N-terminal side of the motif (18). A comparison of the AIF amino acid sequence with the sequences of previously-identified PBMs revealed three putative PAR-binding sites—amino acid residues 222–244, 441–463, and 567–592 in mouse AIF—that are evolutionally conserved in mouse, human, rat, and chicken AIF (fig. S2). To determine whether these PBMs bind to PAR, we performed a dot-blot assay with peptides corresponding to these motifs in the presence of 100 nM purified 32P-labeled PAR. A peptide composed of amino acids 567–592 showed similar binding to PAR as full length WT-AIF and histone H3 (Fig. 1E). A peptide composed of AIF amino acids 222–244 bound less well to 32P-labeled PAR, and a peptide composed of AIF amino acids 441–463 failed to bind PAR appreciably (Fig. 1E). Nitrocellulose saturation binding experiments indicated that PAR bound AIF 567–592 with a Kd of 1.24 × 10−6Mand Bmax of 4.55 pmol. To further study these potential AIF PBMs, we determined the effect of mutating AIF amino acids 222–244, 441–463, or 567–592 to poly-alanine on the ability of AIF to bind PAR (Fig. 1F). Binding of a His-tagged form of AIF in which only amino acids 441 to 463 were mutated (AIFm441–463) to 32P-labeled automodified PARP-1 or purified 32P-labeled PAR was comparable to that of unmutated AIF (WT-AIF) (Fig. 1F). The AIFm222-244 mutant also bound PAR, but at reduced abundance (Fig. 1F). Unlike AIFm441-463 or AIFm222-244, however, AIFm567-592 failed to bind to 32P-labeled automodified PARP-1 or purified 32P-labeled PAR. A His-tagged AIF deletion mutant lacking amino acids 567–592 also failed to bind PAR, confirming that amino acids 567–592 contain the primary PAR-binding site in the intact mouse AIF protein (Fig. 1F). Together, these results indicate that amino acids 567–592 contain the major PBM; however we cannot exclude the possibility that amino acids 222–244 contribute to PAR binding.
Human AIF binds to DNA through a groove between the FAD-binding domain (D1) and C-terminal domain (D3) (19). A mutant form of human AIF in which lysine 255 is substituted with alanine and arginine 265 is substituted with alanine (AIFK255A;R265A) completely lacks DNA binding (19). To investigate the possible contribution of the AIF DNA-binding domain to PAR binding, we assessed PAR binding to mouse AIF mutated in the same way (AIFK254A;R264A). AIFK254A; R264A bound to 32P-labeled automodified PARP-1 and purified 32P-labeled PAR (Fig. 1F). Moreover, salmon sperm DNA at 760 μM failed to disrupt PAR binding to WT-AIF, AIFm441-463, AIFm222-244, or AIFK254A;R264A, whereas cold PAR blocked 32P-labeled PAR binding, indicating that the AIF PAR-binding site is separable from its DNA-binding site (Fig. 1F).
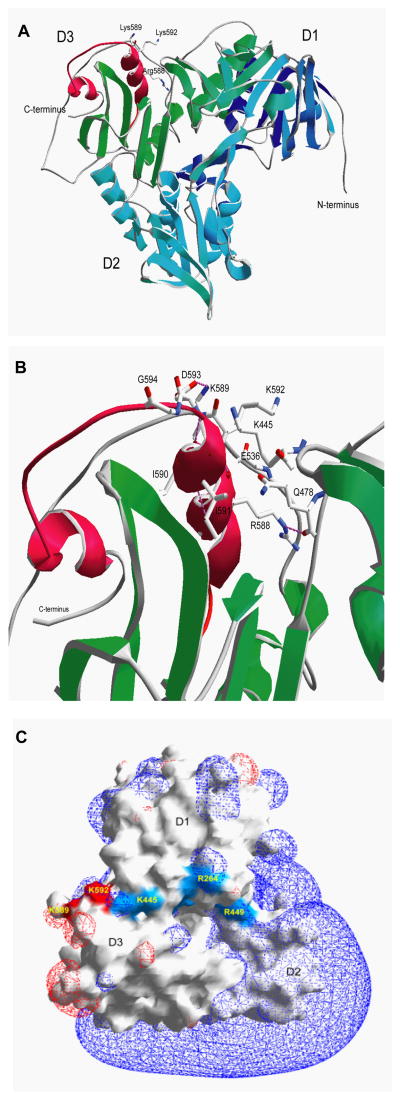
Analysis of the crystal structure of AIF (20) revealed that amino acids 567–592 (the PAR binding site we identified) reside within an alpha helix and an external loop that is well suited to mediate the interaction between AIF and PAR (Fig. 2, A to C). We identified that amino acids 567–592 are located in a predominantly hydrophobic cleft maintained in position through four hydrogen bonds. Several hydrogen bonds are involved in the interaction between side chains and backbone amine bonds inside the alpha helix located at the C-terminal domain of the AIF (the D3 domain). A cluster of positively-charged basic amino acids (Arg583, Arg588, Lys589 and Lys592) potentially essential for PAR binding are located in the AIF D3 domain in close proximity to the DNA-binding domain, but distinct from it. Thus, the PAR-binding site does not overlap with the DNA binding site (Fig. 2, A to C), consistent with the failure of the DNA binding mutant AIFK254A;R264A or excess DNA to affect PAR binding (see Fig. 1F).
Fig. 2. Mouse AIF structure.
(A) Ribbon diagram of the AIF structure showing the three domains (D1 in blue, D2 in cyan and D3 in green) along with the basic helix-loop-helix domain constituting the PAR-binding domain (in red). Side chains of essential basic amino acids are indicated. Modified from Mate et al. (20) (B) Structural details of the PAR-binding domain of AIF. Stabilizing hydrogen bonds are shown in pink. D, aspartic acid; E, glutamic acid; G, glycine; I, isoleucine; Q, glutamine. (C) The surface and electrostatic potential of AIF. DNA binding sites are shown in blue and the PAR binding sites that do not overlap with the DNA binding sites are shown in red.
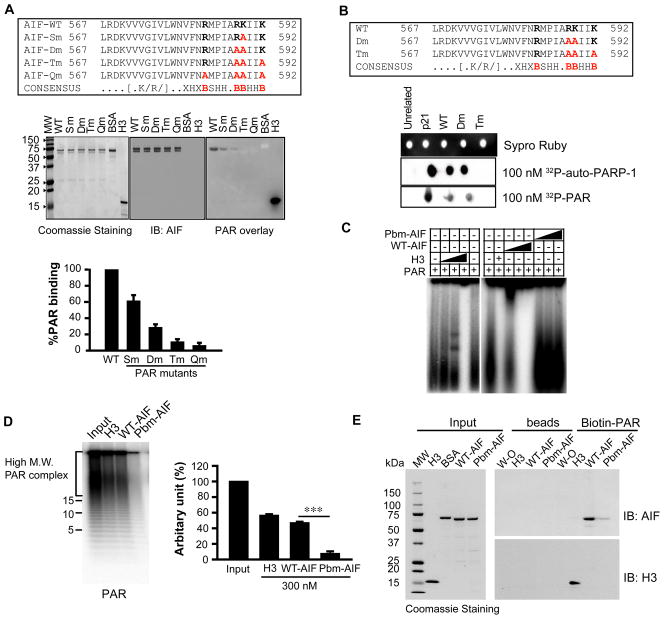
Based on the 3D structure, and the PAR-binding consensus sequence, we analyzed the effects of mutating basic amino acids in the AIF PBM likely to be critical for PAR binding (Fig. 3A). Substituting arginine 588 with alanine reduced AIF binding to PAR by 40%; substituting both arginine 588 and lysine 589 with alanines reduced PAR binding by 80%; and substituting arginine 588, lysine 589, and lysine 592 with alanines reduced PAR binding by 90%, as did mutating arginine 588, lysine 589, lysine 592, and arginine 583 (Fig. 3A). We further explored PAR binding to AIF by examining double amino acid mutants obtained by substituting combinations of arginine 588, lysine 589, and lysine 592 with leucine, which is more similar to the structure of arginine and lysine than is alanine. All double mutants (AIFR588L;K589L, AIFR588L;K592L, and AIFK589L;K592L) significantly reduced PAR binding by about 85% (fig. S3). Similar to the triple alanine mutant (R588A; K589A; K592A), the triple leucine mutant (R588L;K589L;K592L) nearly abolished PAR-binding (fig. S3).
Fig. 3. Determination of amino acids in AIF responsible for PAR binding.
(A) PAR overlay assay of full length WT and mutant AIFs (amino acid 567–592 sequences shown in upper panel). The radioactive signal was quantified and normalized to WT-AIF (lower panel). n = 3. V, valine; W, tryptophan; N, asparagine; F, phenylalanine; M, methionine; P, proline; X, any amino acid; H, hydrophobic amino acid; B, basic amino acid. (B) Dot-blot analysis of WT and mutant AIF peptides (sequences shown in upper panel), and p21, a peptide that binds strongly to PAR, (43) in the presence of 100 nM purified 32P-labeled PAR, n = 3. (C) EMSA of WT-AIF and Pbm-AIF using 32P-labeled PAR, n = 3. (D) [32P]-PAR bound to WT-AIF or Pbm-AIF was analyzed in 20% TBE-PAGE. Values represent ADP-ribose units. The radioactive signal was quantified and normalized to Input.*** p < 0.001 by one-way ANOVA, n = 5. (E) Pulldown assay of WT-AIF and Pbm-AIF using biotin-labeled PAR-conjugated NeutrAvidin beads. W/O, buffer. Beads, NeutrAvidin beads without biotin-labeled PAR polymer, n = 4.
Binding assays with wild type and mutant AIF peptides spanning amino acids 567–592 were consistent with those using full-length AIF. Unlike the WT peptide, the triple amino acid mutant (Tm) AIF peptide (R588A;K589A;K592A) failed to bind 32P-labeled automodified PARP-1 or 32P-labeled purified PAR (Fig. 3B). The double amino acid mutant (Dm) AIF peptide (R588A; K589A) bound PAR (Fig. 3B). Nitrocellulose saturation binding experiments revealed that the WT-AIF (Kd, 1.24 × 10−6M; Bmax, 4.57 pmol) and Dm-AIF peptides (Kd, 3.3 × 10−6M; Bmax, 3.97 pmol) bound PAR in a saturable manner, whereas the Tm-AIF peptide (Tm) (Kd, 1.07 × 10−4M; Bmax, 1.77 pmol) had clearly reduced PAR binding (fig. S4A).
To ascertain whether full length recombinant Pbm-AIF (R588A;K589A;K592A) binds PAR, we performed an EMSA assay for PAR binding with recombinant WT-AIF and Pbm-AIF and determined that WT-AIF caused a PAR mobility shift in a concentration-dependent manner, whereas Pbm-AIF had no effect on PAR mobility (Fig. 3C). Twenty percent Trisborate-EDTA(TBE) PAGE analysis indicated that WT-AIF bound to 32P-PAR of different length and Pbm-AIF failed to bind to 32P-PAR (Fig. 3D). To confirm that Pbm-AIF fails to bind PAR, we also performed biotin-labeled PAR pull-down assays on WT-AIF, Pbm-AIF, or histone H3. WT-AIF and histone H3 were pulled down with biotin-labeled PAR, whereas only a barely detectable amount of Pbm-AIF was pulled down (Fig. 3E). Recombinant AIFs and histone H3 did not bind directly to NeutrAvidin beads in the absence of biotin-labeled PAR under the conditions used, confirming the specificity of the assay (Fig. 3E). Nitrocellulose saturation binding experiments indicated that full-length recombinant His-WT-AIF bound to PAR with high affinity in a saturable manner (Kd, 1.0 × 10−7M; Bmax, 200.83 pmol), whereas full-length His-Pbm-AIF showed much less (and virtually linear) binding (fig. S4B). Because this residual PAR binding was likely non-specific, we subtracted the PAR-binding values obtained with His-Pbm-AIF from those for His-WT-AIF and subjected the values thus determined for specific binding to Scatchard analysis (fig. S4C). We found that the affinity of purified mouse His-WT-AIF for PAR was high, with a calculated Kd of 6.63 ×10−8 M and a Bmax of 116.208 pmol, a concentration consistent with those found in intact HeLa cells after MNNG treatment or in cortical neurons after exposure to NMDA (8).
WT-AIF and Pbm-AIF have similar PAR-independent activities
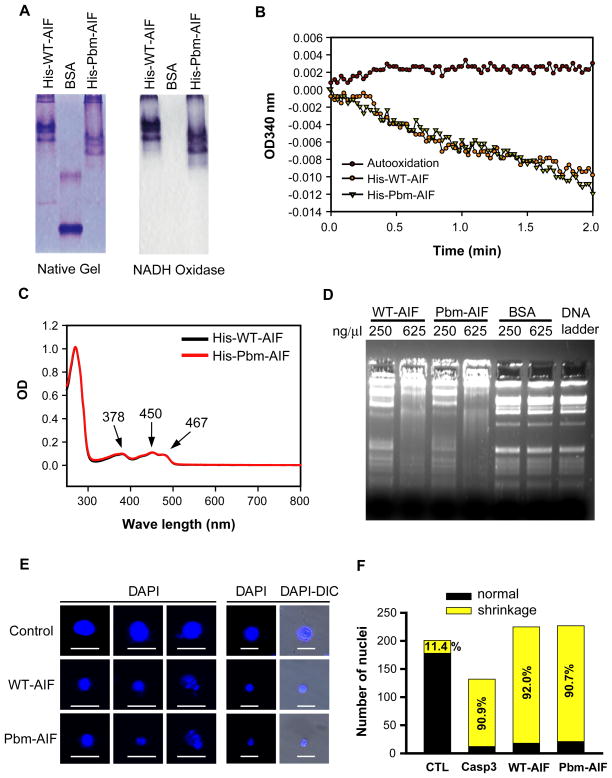
Independent of its role in parthanatos, AIF has NADH (the reduced form of nicotinamide adenine dinucleotide) oxidase activity, binds to FAD (flavin adenine dinucleotide) and DNA, and causes nuclear condensation (19, 21). To determine whether the triple amino acid mutation (R588A;K589A;K592A) of the PBM of AIF interferes with these properties, we assessed the NADH oxidase activity, FAD-binding capacity, and DNA-binding capacity of full-length His-WT-AIF and His-Pbm-AIF. His-Pbm-AIF showed NADH oxidase activity (Fig. 4, A and B), FAD binding (Fig. 4C), and DNA-binding properties (Fig. 4D) comparable to those of His-WT-AIF. Moreover, non-tagged forms of WT-AIF or Pbm-AIF caused chromatin condensation and nuclear shrinkage when incubated with isolated HeLa nuclei (Fig. 4, E and F). Together, these results indicate that AIF is a high affinity PAR binding protein and that the triple amino acid (R588A;K589A;K592A) mutation in AIF substantially eliminates PAR binding without affecting its PAR-independent functions.
Fig. 4. Characterization of PAR-independent properties of WT-AIF and Pbm-AIF.
(A) NADH oxidase activity of His-WT-AIF and His-Pbm-AIF was visualized on native gel by NBT reduction. BSA was used as a negative control. (B) NADH oxidase activity of His-WT-AIF and His-Pbm-AIF was determined by monitoring the changes in absorbance at OD 340 nm. (C) FAD binding of His-WT-AIF and His-Pbm-AIF was determined by spectrophotometric wave length scanning of purified proteins. (D) DNA retardation assay of His-WT-AIF and His-Pbm-AIF by incubating with DNA (200 ng) for 30 min. BSA was used as a negative control. (E) Non-tagged WT-AIF and Pbm-AIF cause chromatin condensation and nuclear shrinkage in isolated HeLa cell nuclei. Scale bar, 20 μm. (F) Quantification of the number of nuclei treated by WT-AIF, Pbm-AIF, and control (CTL) in E. Caspase 3 (Casp3) was applied as a positive control, n = 5.
To determine whether Pbm-AIF fails to bind to PAR in cultured cortical neurons, neuronal cultures were transduced by lentivirus with WT-AIF-Flag or Flag-tagged Pbm-AIF (Pbm-AIF-Flag). We used neurons from Harlequin (Hq) mice, which have an 80% reduction in WT-AIF due to a proviral insertion (22), for these analyses to reduce interference from endogenous AIF (fig. S5A). WT-AIF-Flag and Pbm-AIF-Flag were equally abundant in Hq cortical neurons (Fig. 5, A to C) and both localized to the mitochondria, as determined by their co-localization with the mitochondrial enzyme manganese superoxide dismutase (MnSOD) (fig. S5B). Treatment of Hq cortical cultures with 500 μM NMDA for 5 minutes activated PARP-1, leading to the time-dependent formation of PAR in both the nuclear fraction and the postnuclear fraction (fig. S5C). Two hours after a 5 minute treatment with 500 μM NMDA, we measured PAR binding to AIF from postnuclear fractions and compared it to that in control (non-NMDA-treated) cultures (Fig. 5, A and B). Immunoprecipitation with PAR pulled down WT-AIF-Flag from both non-NMDA- and NMDA-treated cultures, whereas Pbm-AIF-Flag failed to appreciably co-immunoprecipitate with PAR (Fig. 5, A and B). WT-AIF-Flag interacted with PAR in unstimulated Hq cortical neurons, and this interaction significantly increased following NMDA treatment (Fig. 5, A and B). The PAR-AIF interaction was not reduced by benzonase, DNAse I, RNAse A, or ethidium bromide (fig. S6), indicating that it was not mediated by DNA- or RNA-binding.
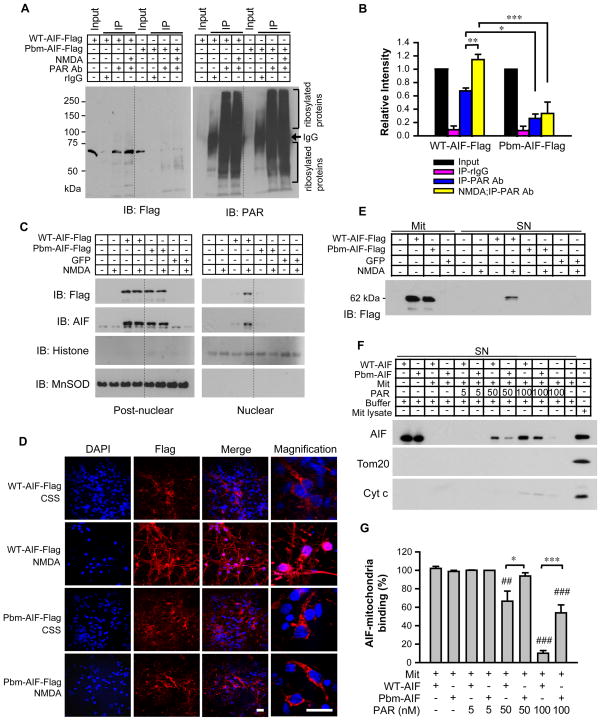
Figure 5. Pbm-AIF is resistant to release from mitochondria and subsequent nuclear translocation.
(A) Co-immunoprecipitation of PAR with WT-AIF-Flag and Pbm-AIF-Flag in post-nuclear fractions isolated from Hq cortical neurons 2 h after NMDA treatment (500 μM for 5 min), rabbit IgG (rIgG). (B) The signal was quantified and normalized to input. n = 5. (C) Subcellular localization of WT-AIF-Flag and Pbm-AIF-Flag in Hq cortical neurons 2 h after NMDA treatment, n = 4. (D) NMDA-induced AIF nuclear translocation in cortical neurons. DAPI, nuclei staining. Scale, 20 μm, n = 3. (E) Mitochondria isolated from cortical neurons were incubated with PAR for 30 min. The levels of AIF were determined in the mitochondria (left) and in the supernatant (right), n = 6. (F & G) WT-AIF-Flag or Pbm-AIF-Flag (500 ng/ml) were incubated with isolated mitochondria (1 mg/ml) in the absence or presence of PAR for 30 min. The supernatant was collected for analyses of AIF, Tom20, and Cyt C. AIF binding to mitochondria in the absence of PAR was regarded as 100% binding. ##p < 0.01, ###p < 0.001, compared to control (absence of PAR); * p < 0.05, **p < 0.01, ***p <0.001 by one-way ANOVA, n = 5.
We observed similar results following treatment of HeLa cells transduced with WT-AIF-Flag or Pbm-AIF-Flag with MNNG. PAR co-immunoprecipitated with WT-AIF-Flag but not Pbm-AIF-Flag in postnuclear fractions of both non-MNNG- and MNNG-treated cultures, and quantification revealed that this interaction was significantly increased by MNNG treatment (fig. S7, A and B). Rabbit IgG, used as a negative control in both Hq cortical cultures and HeLa cells, failed to co-immunoprecipitate with WT-AIF-Flag or Pbm-AIF-Flag (Fig. 5, A and B; fig. S7, A and B). Together, these results indicate that Pbm-AIF fails to bind to PAR either in cells at rest or following PARP-1 activation.
PAR disrupts AIF binding to mitochondria and causes AIF release from mitochondria
We next determined whether Pbm-AIF was released from the mitochondria and translocated to the nucleus after PARP-1 activation in Hq mouse cortical neuronal cultures transduced with WT-AIF-Flag or Pbm-AIF-Flag lentiviruses. GFP lentivirus-transduced and non-transduced neurons served as negative controls. AIF translocation to the nucleus was monitored two hours after NMDA (500 μM, 5 minutes) treatment. Immunoblot analysis of nuclear and postnuclear fractions using antibodies to Flag or AIF (Fig. 5C) revealed that WT-AIF-Flag translocated to the nucleus following NMDA administration, whereas Pbm-AIF-Flag failed to do so (Fig. 5C). Moreover, confocal image analysis confirmed that, two hours after NMDA stimulation (500 μM, 5 minutes), WT-AIF-Flag was observed in the nucleus whereas Pbm-AIF-Flag remained in the cytosol (Fig. 5D). Both WT-AIF and Pbm-AIF-Flag translocated from the mitochondria to the nucleus 24 h after 100 nM staurosporine treatment (fig. S7C), indicating that both forms of AIF are able to translocate, but in parthanatos PAR binding to AIF is required for the translocation.
Endogenous AIF, WT-AIF-Flag, and Pbm-AIF-Flag were monitored for nuclear translocation by confocal image analysis, following MNNG treatment of HeLa cells (fig. S7, D and E) under conditions in which MNNG induces parthanatos (8). In cells transfected with WT-AIF-Flag, both endogenous AIF and WT-AIF-Flag translocated to the nucleus following MNNG treatment; the PARP-1 inhibitor, 3,4-Dihydro-5[4-(1-piperindinyl)butoxy]-1(2H)-isoquinoline, (DPQ), prevented the nuclear translocation of both endogenous AIF and WT-AIF-Flag (fig. S7D), confirming the dependence of AIF translocation on PARP-1 activation. In contrast, MNNG treatment failed to induce the nuclear translocation of Pbm-AIF-Flag, despite inducing the translocation of endogenous AIF (fig. S7E).
Next, we used Hq cortical neurons transduced with WT-AIF-Flag or Pbm-AIF-Flag lentiviruses to determine whether PAR plays a direct role in mitochondrial AIF release (Fig. 5E). GFP lentivirus transduced and non-transduced neurons served as negative controls (Fig. 5E). Three days after lentivirus infection, mitochondria were isolated and exposed to purified PAR to elicit in vitro AIF release (14). WT-AIF-Flag and Pbm-AIF-Flag were equally abundant in mitochondria (Fig. 5E, left). PAR induced the release of WT-AIF-Flag from mitochondria, but failed to release Pbm-AIF-Flag (Fig. 5E, right). Similar results were observed in HeLa cells transiently transfected with WT-AIF-Flag or Pbm-AIF-Flag. Two days after transfection, mitochondria were isolated and exposed to purified PAR, which induced the release of WT-AIF-Flag, but failed to release Pbm-AIF-Flag (fig. S8A). Together, these results indicate that PAR binding to AIF is required for AIF release following PARP-1 activation.
Approximately 20%–30% of AIF resides on the cytosolic side of the outer membrane of mitochondria, where it is poised to be rapidly released by PAR following PARP-1 activation (23). An in vitro assay of AIF binding to mitochondria showed that both purified WT-AIF and Pbm-AIF bound saturably to mitochondria in a time- and concentration-dependent manner (fig. S8, B and C). To investigate the mechanism underlying mitochondrial AIF release following PARP-1 activation, we incubated isolated mitochondria with purified non-tagged forms of AIF in the presence or absence of PAR and followed this by centrifugation to remove the mitochondria and detection of AIF remaining in the supernatant through immunoblot analysis (Fig. 5, F and G). Untreated mitochondria effectively depleted both WT-AIF and Pbm-AIF from the supernatant, confirming that both WT-AIF and Pbm-AIF bind to mitochondria. 5 nM PAR failed to affect association of WT-AIF or Pbm-AIF with mitochondria, whereas 50 nM PAR increased the concentration of WT-AIF in the supernatant, indicating that it disrupted its binding to mitochondria, with barely detectable effects on the concentration of Pbm-AIF (Fig. 5, F and G). 100 nM PAR almost completely abolished the ability of WT-AIF to bind to mitochondria and partially reduced mitochondrial binding of Pbm-AIF (Fig. 5, F and G). 5 nM and 50 nM PAR failed to induce the release of endogenous cytochrome c from mitochondria under the same experimental conditions, although a weak cytochrome c signal could be detected with exposure to 100 nM PAR (Fig. 5F). Mitochondrial outer membrane protein Tom20 was also used to monitor mitochondrial integrity and it could not be detected in the supernatant at any concentration of PAR used (Fig. 5F), indicating that mitochondrial integrity was maintained throughout the experiment. These data suggest that PAR can disrupt binding of WT-AIF to intact mitochondria, thereby causing AIF release, whereas Pbm-AIF is relatively resistant to the releasing effects of PAR.
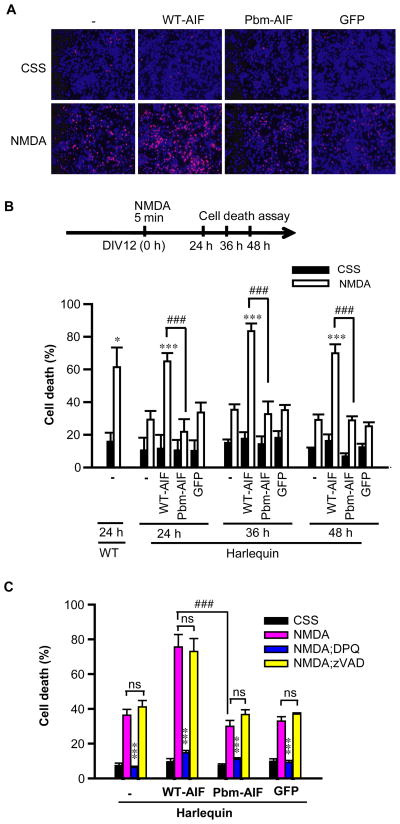
WT-AIF, but not Pbm-AIF, sensitizes cells to parthanatos in vitro and in vivo
To ascertain whether the failure of the Pbm-AIF to translocate to the nucleus following PARP-1 activation has implications for PARP-1-dependent cell death, we monitored the susceptibility of cells expressing WT-AIF-Flag or Pbm-AIF-Flag to parthanatos (Fig. 6 and fig. S9). In WT mouse embryonic fibroblasts (MEFs), WT-AIF-Flag and Pbm-AIF-Flag were equally abundant and failed to affect susceptibility to MNNG-induced cell death (fig. S9, A and B). Hq MEFs showed significantly reduced cell death in response to MNNG compared to WT MEFs, and WT-AIF-Flag restored their susceptibility to MNNG toxicity. In contrast, Pbm-AIF-Flag failed to restore the susceptibility of Hq MEFs to MNNG toxicity (fig. S9A). The broad-spectrum caspase inhibitor zVAD also failed to influence MNNG-induced cell death in MEFs consistent with a lack of involvement of caspases in parthanatos (fig. S9C).
Figure 6. WT-AIF, but not Pbm-AIF, sensitizes cells to parthanatos in vitro.
(A) The effect of WT-AIF-Flag or Pbm-AIF-Flag on NMDA-induced cytotoxicity in Hq cortical neurons 24 h after NMDA (500 μM for 5 min). Cells were transduced with lentivirus carrying WT-AIF-Flag or Pbm-AIF-Flag. Non-transduced neurons and GFP-lentivirus infected cells were used as negative controls. Blue indicates Hoechst 33342 staining and red indicates propidium iodide staining. (B) Cytotoxicity in Hq cortical neurons was determined at 24 h, 36 h, and 48 h after NMDA (500 μM for 5 min) treatment. (C) DPQ (30 μM) but not zVAD (100 μM) inhibited NMDA-induced cortical neuron death. *p < 0.05, ***p < 0.001 as compared to its control group treated with CSS. ###p < 0.001, n = 4. By one-way ANOVA analysis. Neurons prepared from wild type mice were used as a control.
We next assessed NMDA excitotoxicity in Hq cortical neurons transduced with WT-AIF-Flag or Pbm-AIF-Flag (Fig. 6, A to C). As previously reported (14), Hq cortical neurons were resistant to NMDA excitotoxicity. Experiments performed 24 h, 36 h, or 48 h after NMDA treatment indicated that WT-AIF-Flag restored Hq cortical neuron susceptibility to NMDA excitotoxicity, whereas Pbm-AIF-Flag failed to do so (Fig. 6, A and B). The PARP-1 inhibitor, DPQ, largely prevented NMDA-induced excitotoxicity (Fig. 6C), whereas zVAD failed to do so (Fig. 6C).
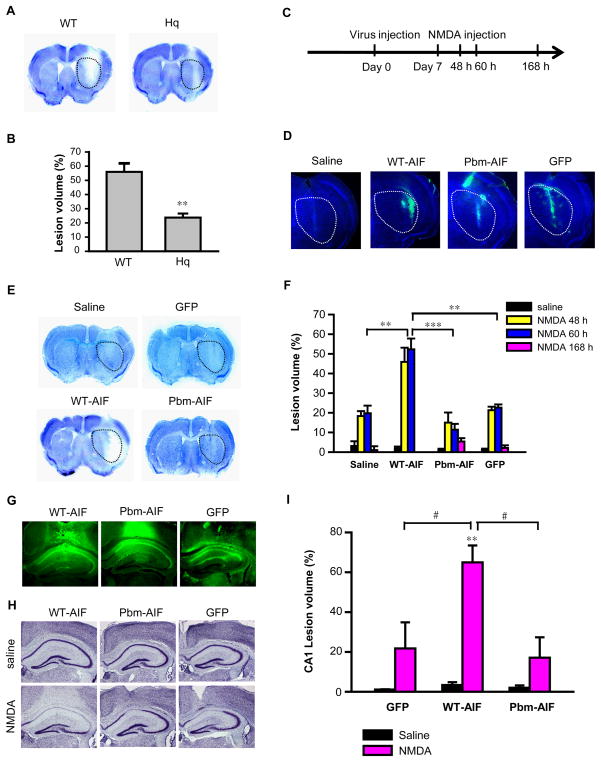
We used Hq mice to determine whether our in vitro findings are relevant in vivo. We examined NMDA receptor-mediated glutamate excitotoxicity, which is a major component of neuronal death in neurodegenerative diseases and stroke (24). We administered NMDA (20 nmol) stereotactically into the striata of Hq or WT mice and assessed lesion volume in the striatum by Nissl staining 60 h later (Fig. 7, A and B). As previously reported (14), NMDA administration caused a larger lesion in WT mice than in Hq mice (Fig. 7, A and B). We next assessed whether replacement of WT-AIF-Flag or Pbm-AIF-Flag through lentiviral transduction restored the sensitivity of Hq mice to NMDA excitotoxicity (Fig. 7, C to F). Lentiviruses carrying WT-AIF-Flag, Pbm-AIF-Flag, or GFP were injected in both the left and right side striata of Hq mice seven days before injection with NMDA in the right striatum and saline in the left striatum. WT-AIF, Pbm-AIF, and GFP were successfully expressed in striatum (Fig. 7D). WT-AIF-Flag restored sensitivity to NMDA excitotoxicity, so that Hq mice showed larger lesions volume 48 h or 60 h after the NMDA injection (Fig. 7, E and F). In contrast, Pbm-AIF-Flag failed to restore NMDA excitotoxicity, and lesions following NMDA injection were similar to those seen in mice transduced with GFP or injected with saline (Fig. 7, E and F). Mice injected with Pbm-AIF-Flag lentivirus survived for at least 168 h after the NMDA injection, whereas, with one exception, mice injected with WT-AIF-Flag lentivirus failed to survive for 168 h. The lesion volume of mice injected with Pbm-AIF-Flag lentivirus remains reduced at 168 h suggesting that the protection is long-lasting (Fig. 7F).
Figure 7. Pbm-AIF, but not WT-AIF, protects cells from parthanatos in vivo.
(A & B) Nissl staining and lesion volume of WT and Hq mice 60 h after 20 nmols NMDA injection in striatum. **p < 0.01 by Student’s t-test, n = 6 mice. (C) Time windows for virus injection. (D) Transduction of WT-AIF-Flag, Pbm-AIF-Flag, or GFP in striatum of Hq mouse brains. (E) Nissl staining of WT-AIF-, Pbm-AIF-, and GFP-lentivirus-, and saline-injected mice 60 h after NMDA injection in striatum. A dotted line demarcates the striatum for panels A, D and E. (F) Lesion volumes of WT-AIF-, Pbm-AIF-, GFP-, and saline-injected mice assessed at 48 h, 60 h, and 168 h after injection of NMDA or saline. n (WT-AIF) = 11, n (Pbm-AIF) = 10, n (saline) = 9, n (GFP) =9. (G) Transduction of WT-AIF-Flag, Pbm-AIF-Flag, and GFP in CA1 of Hq mouse brains. (H & I) Nissl staining and lesion volumes of WT-AIF, Pbm-AIF, and GFP AAV2 injected mice 60 h after NMDA injection in CA1. (**p < 0.01, as compared to saline injection. ***p < 0.001, #p < 0.05, by one-way ANOVA, n (WT-AIF) = 4, n (Pbm-AIF) = 3, n (GFP) =3.
In a second model of NMDA excitotoxicity, we injected adeno-associated virus (AAV2) carrying WT-AIF-Flag, Pbm-AIF-Flag, or GFP into the CA1 region of both the left and right hippocampus of Hq mice. WT-AIF-Flag, Pbm-AIF-Flag, and GFP were strongly transduced throughout the whole CA1 region (Fig. 7G). Seven days later, we injected NMDA into the right CA1 region and saline into the left CA1 region; we assessed CA1 lesion volume 60 h later. NMDA injection caused a large lesion volume in WT-AIF-Flag injected Hq mice, whereas Pbm-AIF-Flag- and GFP-injected Hq mice had significantly smaller and equivalent lesions (Fig. 7, H and I). Together, our results indicate that PAR binding to AIF is required for NMDA excitotoxicity both in vitro and in vivo.
DISCUSSION
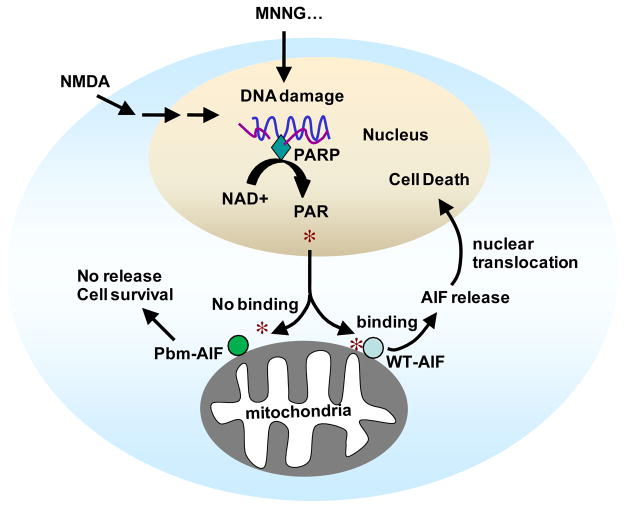
Our data show that AIF is a PAR-binding protein and that PAR binding to AIF is critical for AIF release from the mitochondria following PARP-1 activation (Fig. 8). Moreover, PAR binding was required for AIF to induce cell death in parthanatos. PAR bound to AIF saturably and with high affinity. Amino acids 567–592 in the D3 domain of AIF comprised the major PAR binding site in AIF, as determined by PAR binding assays to peptides corresponding to the AIF PAR-binding domain and mutational analysis of both these mimetic peptides and full length AIF. The basic amino acids arginine 588, lysine 589, and lysine 592 were critical for PAR binding to AIF. Their mutation to alanine or leucine markedly interfered with PAR binding to AIF. Moreover, mutation of these amino acids to alanine prevented the release of AIF from mitochondria following PAR treatment of purified mitochondria or PARP-1 activation with MNNG or NMDA. The failure of the Pbm-AIF to be released from the mitochondria following PARP-1 activation attenuated AIF-mediated cell death (Fig. 8). The PAR binding site is distinct from the AIF DNA binding site; thus, AIF appears to have evolved separate binding properties for DNA and PAR. Moreover, AIF’s cell death functions can be separated from its role in mitochondrial respiration and cell survival.
Figure 8. Model of PAR-dependent AIF release in parthanatos.
The scheme shows that DNA damage induced by MNNG administration (or other alkylating agents, as indicated by the “…”) or NMDA excitotoxicity activates PARP-1, which catalyzes PAR formation. PAR then translocates from the nucleus to the cytosol and mitochondria where it binds to a pool of AIF that is on the cytosolic side of the outer membrane of the mitochondria (25) inducing its release. AIF then translocates to nucleus and causes cell death. In contrast, PAR fails to bind Pbm-AIF and Pbm-AIF is not released during PARP-1 activation and cells survive the toxic stimuli.
Parthanatos is a key cell death mechanism involved in various diseases including ischemic injury and excitotoxicity. One pivotal step of parthanatos is that nuclear PAR signals to mitochondrial AIF and causes its release, thereby initiating a deadly conversation between these two organelles (Fig. 8). How PAR exits the nucleus is not known, although this may be carried out by a PARylated nuclear protein. Moreover, reducing PAR abundance with PARG expressed in the cytosol or interfering with PAR through neutralizing antibodies reduces mitochondrial AIF release and subsequent cell death (8). At the mitochondria there are two pools of AIF. Approximately 80% of AIF is localized to the inner membrane and inner membrane space where it would be protected from direct actions of PAR. However, 20–30% of mitochondrial AIF is localized to the cytosolic side of the outer mitochondrial membrane where it is available to bind PAR and be released during parthanatos (25). Our findings indicate that PAR binding to AIF’s PAR-binding motif acts as the transducer that mediates AIF release from the mitochondria allowing AIF to translocate to the nucleus. PAR binding to AIF likely induces a conformation change in AIF that lowers its affinity for the mitochondrial outer membrane leading to its release, although the exact mechanism whereby PAR binding induces AIF release remains unclear.
PAR binds to various proteins, thereby affecting their physiologic function (15, 26–31). PAR also appears to function as a scaffold, assembling PAR-binding proteins into a signaling complex that confers DNA damage-induced NF-κB activation (32). AIF is the first identified PAR-binding protein involved in cell death. How PAR-AIF binding modulates mitochondrial function remains unclear, as does the mechanism whereby AIF enters the nucleus and causes chromatinolysis and cell death.
It is likely that PAR has both inhibitory and stimulatory effects on its binding partners. Further elucidation of the parthanatos signaling pathway will provide new opportunities for discovery of heretofore unidentified biologic functions. In particular, identification of AIF as a PAR-binding protein opens up opportunities for the development of compounds that inhibit the interaction of PAR with AIF, thus potentially protecting against parthanatos. Alternatively, agents could be identified that enhance the release of AIF, thereby promoting parthanatos and serving as potential cancer chemotherapeutic agents.
MATERIALS AND METHODS
Protein expression and Purification
Mouse AIF cDNA was subcloned into 6 X His-tagged pET28b vector (Novagen) or GST-tagged pGex-6P-1 vector (GE Health Care) by EcoR I and Xho I restriction sites. The protein was expressed and purified from E. coli using metal-affinity chromatography and glutathione sepharose, respectively. Flag-tagged WT-AIF and Pbm-AIF were subcloned into pCI vector (Promega) by Xho I and Sal I restriction sites. The recombinant AIF either with His-tag or Flag-tag is used in this study as indicated. To exclude interference from the tag, we also used non-tagged recombinant AIF in the PAR-binding assay and other AIF biochemical determinations. Recombinant AIF was initially prepared from GST-tagged-AIF, purified by Glutathione Sepharose, with the GST-tag subsequently proteolytically removed. PBM mutants (AIFm222-244, AIFm441-463 and AIFm567-592), deletion mutant (AIFΔ567-592) and point mutants (K254A, R264A, R583A, R588A, K589A, K592A, R588L, K589L, K592L) were constructed by PCR and verified by sequencing. The primers used in sequential point mutation are summarized in Table S1.
Biotin- and 32P-labeled automodified PARP-1 synthesis and PARP-free PAR preparation
Biotin- and 32P-labeled automodified PARP-1 were synthesized as described previously (18, 33). Briefly, PARP-1 purified up to the DNA-cellulose step (600 U/mg) was incubated with biotin-labeled NAD+ and 32P-labeled NAD+ for 2 min at 30 °C, thereafter a nonlabeled and nonisotopic NAD+ was added to the reaction mixture, which was incubated for further 28 min at 30 °C. High specific activity biotin-labeled NAD+ and 32P-labeled automodified PARP-1 (80 cpm/nmol) were precipitated as described (34). Biotin-labeled, non-radioactive, and 32P-labeled free PAR were prepared and purified on a DHBB column as described (35). Polymer size was assessed by 20% TBE-PAGE (90 mM Tris-borate pH 8.0, 2 mM EDTA) and HPLC chromatography using a DEAE-NPR column (36). PAR has a mean length of 40 ADP-ribose residues as determined by HPLC methods and gel electrophoresis. The range of size of PAR in this mix is 6-mer through 100-mer ADP-ribose units (8, 14).
Structural Characterization
Ribbon models of the murine AIF monomer (Protein Data Bank ID no.: 1GV4) were created using Rastop molecular visualization software, and molecular surface representation and electrostatic potential were calculated using Swiss-PBD software.
Nitrocellulose PAR-Binding Assay and EMSA
Synthetic peptides or purified proteins were diluted in TBS-T buffer (0.5μg/μl) and loaded onto a nitrocellulose membrane (0.1 μm) using a dot blot manifold system (Life Technologies, Gaithersburg, MD). The membranes were washed once with TBS-T buffer and air dried followed by incubation with indicated concentrations of 32P-labeled automodified PARP-1, 32P-labeled PAR, or biotin-labeled PAR for 1 h at room temperature. After washing, the membranes were analyzed by autoradiography on Bio-Max MR (Kodak), or probed with anti-biotin antibody and visualized with X-ray films by the SuperSignal West Pico Chemiluminescent Substrate (Pierce). Fluorescent Sypro Ruby protein staining was performed on the nitrocellulose membrane to demonstrate equal loading. For EMSA analysis, 50 ng of purified proteins (100 ng/ml) were incubated with 32P-labeled PAR for 1 min at RT; thereafter samples were resolved in 5% PAGE-gel. The gel was heat dried and developed using a Typhoon 9400 Imager (GE Health Care). PAR binding affinity was determined by nitrocellulose saturation binding experiments using the ligand binding program of Sigma plot (9.0 software-SYSTAT).
In vitro PAR-binding assay
300 nM of purified wild type (WT)-AIF or PAR binding mutant (Pbm)-AIF were incubated with 32P-labeled PAR for 10 min at RT. Thereafter, samples were incubated with AIF antibody-linked protein G slurry for 1 h. Samples were washed two times with PBS and boiled with 1 X Laemmli sample buffer (BIO-RAD). Each soluble fraction was resolved in 20% TBE-PAGE. The gel was heat dried and developed using a Typhoon 9400 Imager (GE Health Care).
Determination of NADH Oxidase Activity and FAD Binding of AIF
AIF’s NADH oxidase activity was determined as described (21). In brief, the NADH oxidase activity was measured at RT in substrate solution containing 250 μM NADH in air-saturated 50 mM Tris-HCl, pH 8.0. The decrease in absorbance at OD340 nm was monitored right after the addition of recombinant His-tagged AIF into the substrate solution. In situ redox activity of recombinant His-tagged AIF was performed as previously described (21). The AIF protein was separated on a 10% native gel and then the gel was briefly washed in distilled water. The gel was equilibrated in 2,2′-Di-p-nitrophenyl-5-5′-diphenyl-3,3′ (3-3′-dimetoxy-4-4′ difenilen) tetrazolium chloride (NBT) solution for 20 min in the dark followed by the addition of 1 mM NADH to NBT solution to reduce NBT. FAD binding to AIF was monitored at 13 μM concentration of His-tagged AIF by wave length scanning ranging from 250–800 nm using Beckman Coulter DU800 UV/Vis spectrophotometer (Fullerton).
DNA Gel Retardation Assay
AIF binding-mediated DNA mobility retardation was performed as described (19). Recombinant His-tagged AIF (10 μg or 25 μg) was incubated with 1 kb DNA molecular weight marker (Fermentas) for 30 min at RT and loaded on 2.5 % agarose gel prestained with ethidium bromide (1 μg/ml). Mobility shift was visualized under the Alpha Innotech UV illuminator (San Leandro).
Lentivirus Construction and Virus Production
WT-AIF-Flag or Pbm-AIF-Flag was subcloned into a lentiviral cFugw vector by Age I and EcoR I restriction sites, and its expression was driven by human ubiquitin C (hUBC) promoter. The lentivirus was produced by transient transfection of the recombinant cFugw vector into 293T cells with three packaging vectors: pLP1, pLP2 and pVSV-G (1.3: 1.5:1:1.5). The viral supernatants were collected at 48 h and 72 h after transfection and concentrated by ultracentrifuge for 3 h at 50,000 g.
Cell Culture, transfection, lentiviral transduction and Cytotoxicity
MEF cells from wild type (WT) or Hq mice and HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen) supplemented with 10% Fetal Bovine Serum (HyClone), 100 μg/ml streptomycin, and 100 U/ml penicillin (Invitrogen). Flag-tagged AIF (WT and Pbm) was transfected using lipofectamine Plus (Invitrogen). Primary neuronal cultures from cortex were prepared as previously described (37). Briefly, the cortex was dissected and the cells dissociated by trituration in modified Eagle’s medium (MEM), 20% horse serum, 30 mM glucose, and 2 mM L-glutamine following a 10 min digestion in 0.027% trypsin/saline solution (Gibco BRL, Gaithersburg, Maryland). The neurons were plated on 15 mm multiwell plates coated with polyornithine or on coverslips coated with polyornithine. Neurons were maintained in MEM, 10% horse serum, 30 mM glucose, and 2 mM L-glutamine in a 7% CO2 humidified 37°C incubator. The growth medium was replaced twice per week. In mature cultures, neurons represent 70–90% of the total number of cells. In day in vitro (DIV) 7–9, neurons were infected by lentivirus carrying WT-AIF-Flag, Pbm-AIF-Flag, or GFP (1×109 T.U./ml) for 72 h. PARP-1 dependent cell death was induced by either MNNG (Sigma) in MEFs or HeLa cells or NMDA (Sigma) in neurons. HeLa cells or MEFs were exposed to MNNG (50 μM and 500 μM, respectively) for 15 min and neurons (DIV10-14) were exposed to NMDA as described previously (6, 38). Neurons were washed with control salt solution [CSS, containing (in mM): 120 NaCl, 5.4 KCl, 1.8 CaCl2, 25 Tris-Cl, 20 glucose, pH 7.4], exposed to 500 μM NMDA plus 10 μM glycine in CSS for 5 min, and then postexposed in MEM, containing 10% horse serum, 30 mM glucose, and 2 mM L-glutamine for various times before fixation, immunocytochemical staining, and confocal laser scanning microscopy. Cell viability was determined the following day by unbiased objective computer-assisted cell counting after staining of all nuclei with 7 μM Hoechst 33342 (Invitrogen) and dead cell nuclei with 2 μM propidium iodide (Invitrogen). The numbers of total and dead cells were counted with the Axiovision 4.3 software (Carl Zeiss). At least three separate experiments using at least six separate wells were performed with a minimum of 15,000–20,000 neurons or cells counted per data point. For neuronal toxicity assessments, glial nuclei fluoresce at a different intensity than neuronal nuclei and were gated out. The percentage of cell death was determined as the ratio of live to dead cells as compared with the percentage of cell death in control wells to account for cell death attributed to mechanical stimulation of the cultures.
Pulldown, co-immunoprecipitation, and immunoblotting
For the pulldown assay, NeutrAvidin beads or biotin-labeled PAR -immobilized NeutrAvidin beads were incubated with 500 ng recombinant WT-AIF or Pbm-AIF, washed in lysis buffer, and eluted by boiling in sample buffer. For Co-immunoprecipitation, the post-nuclear cell extracts, which is the fraction prepared from whole cell lysates after removing nuclear proteins, were isolated in hypotonic buffer (6) and incubated overnight with an antibody against PAR in the presence of protein A/G Sepharose (Santa Cruz), followed by immunoblot analysis with mouse anti-Flag antibody (Sigma). The proteins were separated on denaturing polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. The membrane was blocked and incubated overnight with the 1 mg/ml primary antibody (mouse anti-Flag; rabbit anti-AIF; or rabbit anti-PAR 96–10) at 4°C, followed by donkey anti-mouse or goat anti-rabbit IgG conjugated to HRP for 1 h at room temperature. After washing, the immune complexes were detected by the SuperSignal West Pico Chemiluminescent Substrate (Pierce). The integrity of the nuclear and postnuclear subcellular fractions was determined by monitoring histone and MnSOD immunoreactivity, respectively.
Mitochondria isolation and in vitro AIF release assay
Mitochondria were prepared from HeLa cells and cortical neurons as follows. The cells were harvested and resuspended in 2 ml MIB buffer (10 mM HEPES, pH 7.4, 300 mM sucrose, 1 mM EGTA) and homogenized by 60 strokes with B type Dounce homogenizer. Nuclei were pelleted at 750 g for 5 min and mitochondria were spun down at 3,000 g for 10 min. Isolated mitochondria were dissolved in a minimum volume of sucrose buffer (10 mM HEPES, pH 7.4, 300 mM sucrose) and protein concentration was adjusted to 2 mg/ml. The functional integrity of isolated mitochondria was verified by measuring the acceptor control ratio using a Clark type oxygen electrode (Hansatech). The ratio was determined by dividing the ADP (0.8 mM)-stimulated, state 3 respiration rate with the state 4 respiration rate in the presence of oligomycin (2.5 μg/ml) (39). Mitochondria preparations giving a ratio higher than 5 were used for the experiments. In vitro AIF release was carried out at RT for 30 min in assay buffer (20 mM HEPES, pH 7.4, 125 mM KCl, 2 mM K2HPO4, 4 mM MgCl2, 5 mM succinate, 2 mM rotenone, 3 μM ATP and 1 μM ADP) in the presence of 100 nM PAR and 1 mg/ml isolated mitochondria. Proteins released into the assay buffer were saved by centrifugation at 12,000 g for 5 min.
Nuclear shrinkage assay
Nuclei were purified from HeLa cells as described previously (40). For the nuclear shrinkage assay, nuclei were incubated with recombinant WT-AIF or Pbm-AIF (20 ng/ml) in the presence of ATP (2 mM), phosphocreatine (20 mM) and creatine kinase (50 μg/ml) for 90 min at 37 °C. Nuclei were stained by DAPI and examined by fluorescence microscopy.
Immunocytochemistry, immunohistochemistry and Confocal Microscopy
For immunocytochemistry, cells were fixed 2 h after treatment with 4% paraformaldehyde, permeabilized with 0.05% Triton X-100, and blocked with 3% bovine serum albumin in PBS. AIF was visualized by 2 μg/ml mouse anti-Flag/Cy2 AffiniPure Donkey Anti-Mouse IgG or rabbit anti-AIF/Cy3 AffiniPure Donkey Anti-Rabbit IgG. Immunohistochemistry was performed using the antibody against Flag. Immunofluorescence analysis was carried out by using a LSM510 confocal laser scanning microscope (Carl Zeiss) as described earlier (41).
Injection of lentivirus and NMDA in Harlequin mouse Striatum
Striatal injections of NMDA were performed as reported previously (42). Briefly, 3-month-old Hq mice were anesthetized with pentobarbital (45 mg/kg, i.p.). 1 μl Lentivirus (5 × 1010 T.U./ml) carrying WT-AIF-Flag, Pbm-AIF-Flag, or GFP was injected into striatum on both sides of the brain (rostral, 0.4 mm; lateral +/− 1.7 mm; ventral 3.5 mm from bregma), and 1 μl AAV2 (1 × 1013 T.U./ml) carrying WT-AIF-Flag, Pbm-AIF-Flag or GFP was injected into CA1 in hippocampi on both sides of the brain (posterior, 2.0 mm; lateral +/− 1.5 mm; ventral 1.5 mm from bregma) using a stereotactic frame (Kopf) (0.02 ml/min). The needle was left in place for an additional 8 min after injection. One week after the first injection, 20 nmol NMDA or normal saline in 0.8 μl was injected into the right and left striatum (0.02 μl/min), respectively. The needle was left in place for an additional 8 min after injection. Lesions volumes were assessed 48 h, 60 h, or 168 h after NMDA administration. At the indicated time point after the second injection, the mice were deeply anesthetized, and then perfused with saline and 4% paraformaldehyde. After postfixation and freezing in 20% glycerol-PBS, brain sections (30 μm) were obtained for Nissl staining and immunohistochemistry.
Statistical Analysis
Statistical evaluation was carried out by Student’s t-test between two groups and by one-way analysis of variance (ANOVA) followed by post hoc comparisons with the Bonferroni test using GraphPrism software within multiple groups. Data are shown as mean ± S.E.M., p < 0.05 is considered significant.
Supplementary Material
Table S1. Primers used in sequential point mutation to construct single-, di-, triple-, and quadruple point mutations.
Fig. S1. PAR binds to AIF.
Fig. S2. Alignment of the three putative PAR binding motifs in AIF.
Fig. S3. Determination of amino acids in AIF responsible for PAR binding.
Fig. S4. AIF and PAR binding affinity.
Fig. S5. Subcellular localization of AIF and PAR in cortical neurons.
Fig. S6. DNA and RNA are not involved in the PAR-AIF interaction.
Fig. S7. Pbm-AIF fails to bind to PAR and is resistant to its release from mitochondria in HeLa cells.
Fig. S8. Kinetics of AIF-mitochondria binding.
Fig. S9. PAR-binding mutant AIF is resistant to PARP-1-dependent cell death.
Acknowledgments
Funding: This work was supported by the National Institutes of Health grants NINDS NS039148, NS067525 the American Heart Association (AHA, 0825413E), and Canadian Institutes Health Research grant (GGP). GGP holds a Research Chair in Proteomics. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
Author Contributions: Experiments were coordinated by G.G.P, V.L.D. and T.M.D. Experiments were conducted by Y.W., N.S.K., J.-F.H., H.K., K.K.D., S.A. The study was conceived and scientifically directed by V.L.D and T.M.D. The paper written by Y.W., N.S.K, V.L.D. and T.M.D. All authors contributed to the final editing of the manuscript.
Competing interests: We are planning to submit a report of invention and to patent the PAR binding to AIF.
REFERENCES AND NOTES
- 1.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pacher P, Szabo C. Role of the peroxynitrite-poly(ADP-ribose) polymerase pathway in human disease. Am J Pathol. 2008;173:2–13. doi: 10.2353/ajpath.2008.080019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, MacLaurin JG, Rippstein P, Park DS, Shore GC, McBride HM, Penninger JM, Slack RS. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. Embo J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang YH, Yi MJ, Kim MJ, Park MT, Bae S, Kang CM, Cho CK, Park IC, Park MJ, Rhee CH, Hong SI, Chung HY, Lee YS, Lee SJ. Caspase-independent cell death by arsenic trioxide in human cervical cancer cells: reactive oxygen species-mediated poly(ADP-ribose) polymerase-1 activation signals apoptosis-inducing factor release from mitochondria. Cancer Res. 2004;64:8960–8967. doi: 10.1158/0008-5472.CAN-04-1830. [DOI] [PubMed] [Google Scholar]
- 6.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 7.Brown D, Yu BD, Joza N, Benit P, Meneses J, Firpo M, Rustin P, Penninger JM, Martin GR. Loss of Aif function causes cell death in the mouse embryo, but the temporal progression of patterning is normal. Proc Natl Acad Sci U S A. 2006;103:9918–9923. doi: 10.1073/pnas.0603950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung EC, Melanson-Drapeau L, Cregan SP, Vanderluit JL, Ferguson KL, McIntosh WC, Park DS, Bennett SA, Slack RS. Apoptosis-inducing factor is a key factor in neuronal cell death propagated by BAX-dependent and BAX-independent mechanisms. J Neurosci. 2005;25:1324–1334. doi: 10.1523/JNEUROSCI.4261-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Culmsee C, Zhu C, Landshamer S, Becattini B, Wagner E, Pellecchia M, Blomgren K, Plesnila N. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and Bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25:10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao G, Xing J, Xiao X, Liou AK, Gao Y, Yin XM, Clark RS, Graham SH, Chen J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J Neurosci. 2007;27:9278–9293. doi: 10.1523/JNEUROSCI.2826-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi A, Bondada V, Geddes JW. Mitochondrial micro-calpain is not involved in the processing of apoptosis-inducing factor. Exp Neurol. 2009;218:221–227. doi: 10.1016/j.expneurol.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Kim NS, Li X, Greer PA, Koehler RC, Dawson VL, Dawson TM. Calpain activation is not required for AIF translocation in PARP-1-dependent cell death (parthanatos) J Neurochem. 2009;110:687–696. doi: 10.1111/j.1471-4159.2009.06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A. 2006;103:18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panzeter PL, Zweifel B, Malanga M, Waser SH, Richard M, Althaus FR. Targeting of histone tails by poly(ADP-ribose) J Biol Chem. 1993;268:17662–17664. [PubMed] [Google Scholar]
- 17.Eliasson MJ, Sampei K, Mandir AS, Hurn PD, Traystman RJ, Bao J, Pieper A, Wang ZQ, Dawson TM, Snyder SH, Dawson VL. Poly(ADP-ribose) polymerase gene disruption renders mice resistant to cerebral ischemia. Nat Med. 1997;3:1089–1095. doi: 10.1038/nm1097-1089. [DOI] [PubMed] [Google Scholar]
- 18.Gagne JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye H, Cande C, Stephanou NC, Jiang S, Gurbuxani S, Larochette N, Daugas E, Garrido C, Kroemer G, Wu H. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat Struct Biol. 2002;9:680–684. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- 20.Mate MJ, Ortiz-Lombardia M, Boitel B, Haouz A, Tello D, Susin SA, Penninger J, Kroemer G, Alzari PM. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struct Biol. 2002;9:442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 21.Miramar MD, Costantini P, Ravagnan L, Saraiva LM, Haouzi D, Brothers G, Penninger JM, Peleato ML, Kroemer G, Susin SA. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 22.Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman SL. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 23.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1:pii, e00021. doi: 10.1042/AN20090046. doi: 00010.01042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arundine M, Tymianski M. Molecular mechanisms of glutamate-dependent neurodegeneration in ischemia and traumatic brain injury. Cell Mol Life Sci. 2004;61:657–668. doi: 10.1007/s00018-003-3319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1 doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 28.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 29.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci U S A. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 31.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 32.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A nuclear poly(ADP-ribose)-dependent signalosome confers DNA damage-induced IkappaB kinase activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Shah GM, Kaufmann SH, Poirier GG. Detection of poly(ADP-ribose) polymerase and its apoptosis-specific fragment by a nonisotopic activity-western blot technique. Anal Biochem. 1995;232:251–254. doi: 10.1006/abio.1995.0016. [DOI] [PubMed] [Google Scholar]
- 34.Brochu G, Shah GM, Poirier GG. Purification of poly(ADP-ribose) glycohydrolase and detection of its isoforms by a zymogram following one- or two-dimensional electrophoresis. Anal Biochem. 1994;218:265–272. doi: 10.1006/abio.1994.1177. [DOI] [PubMed] [Google Scholar]
- 35.Affar EB, Duriez PJ, Shah RG, Winstall E, Germain M, Boucher C, Bourassa S, Kirkland JB, Poirier GG. Immunological determination and size characterization of poly(ADP-ribose) synthesized in vitro and in vivo. Biochim Biophys Acta. 1999;1428:137–146. doi: 10.1016/s0304-4165(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 36.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 37.Dawson VL, Dawson TM, Bartley DA, Uhl GR, Snyder SH. Mechanisms of nitric oxide-mediated neurotoxicity in primary brain cultures. J Neurosci. 1993;13:2651–2661. doi: 10.1523/JNEUROSCI.13-06-02651.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Zulueta M, Ensz LM, Mukhina G, Lebovitz RM, Zwacka RM, Engelhardt JF, Oberley LW, Dawson VL, Dawson TM. Manganese superoxide dismutase protects nNOS neurons from NMDA and nitric oxide-mediated neurotoxicity. J Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polster BM, Kinnally KW, Fiskum G. BH3 death domain peptide induces cell type-selective mitochondrial outer membrane permeability. J Biol Chem. 2001;276:37887–37894. doi: 10.1074/jbc.M104552200. [DOI] [PubMed] [Google Scholar]
- 40.Lazebnik YA, Cole S, Cooke CA, Nelson WG, Earnshaw WC. Nuclear events of apoptosis in vitro in cell-free mitotic extracts: a model system for analysis of the active phase of apoptosis. J Cell Biol. 1993;123:7–22. doi: 10.1083/jcb.123.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandir AS, Poitras MF, Berliner AR, Herring WJ, Guastella DB, Feldman A, Poirier GG, Wang ZQ, Dawson TM, Dawson VL. NMDA but not non-NMDA excitotoxicity is mediated by Poly(ADP-ribose) polymerase. J Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in sequential point mutation to construct single-, di-, triple-, and quadruple point mutations.
Fig. S1. PAR binds to AIF.
Fig. S2. Alignment of the three putative PAR binding motifs in AIF.
Fig. S3. Determination of amino acids in AIF responsible for PAR binding.
Fig. S4. AIF and PAR binding affinity.
Fig. S5. Subcellular localization of AIF and PAR in cortical neurons.
Fig. S6. DNA and RNA are not involved in the PAR-AIF interaction.
Fig. S7. Pbm-AIF fails to bind to PAR and is resistant to its release from mitochondria in HeLa cells.
Fig. S8. Kinetics of AIF-mitochondria binding.
Fig. S9. PAR-binding mutant AIF is resistant to PARP-1-dependent cell death.