Abstract
With the aim of exploring the anticancer properties of organometallic compounds with bioactive ligands, Ru(arene) compounds of the antibacterial quinolones nalidixic acid (2) and cinoxacin (3) were synthesized, and their physicochemical properties were compared to those of chlorido(η6-p-cymene)(ofloxacinato-κ2O,O)ruthenium(II) (1). All compounds undergo a rapid ligand exchange reaction from chlorido to aqua species. 2 and 3 are significantly more stable than 1 and undergo minor conversion to an unreactive [(cym)Ru(μ-OH)3Ru(cym)]+ species (cym = η6-p-cymene). In the presence of human serum albumin 1−3 form adducts with this transport protein within 20 min of incubation. With guanosine 5′-monophosphate (5′-GMP; as a simple model for reactions with DNA) very rapid reactions yielding adducts via its N7 atom were observed, illustrating that DNA is a possible target for this compound class. A moderate capacity of inhibiting tumor cell proliferation in vitro was observed for 1 in CH1 ovarian cancer cells, whereas 2 and 3 turned out to be inactive.
Introduction
In the past two decades ruthenium coordination compounds (Figure 1) have attracted considerable interest as potential anticancer agents because of their low toxicity and their efficacy against platinum-drug-resistant tumors, reflected in promising results in various stages of preclinical to early clinical studies.1−6 Organometallic ruthenium complexes bearing a π-bonded arene ligand and other simple mono- or bidentate ligands are considered promising candidates for cancer treatment.(7) Compounds containing phosphatriazaadamantane (pta) or its derivatives were developed,8,9 which show antimetastatic activity but low cytotoxicity in vitro.9−11 On the other hand, complexes bearing N,N-chelating ligands (Figure 1) have shown cytotoxicity comparable to that of cisplatin in a number of cell lines.12−14 The first organometallic ruthenium compound with chelating O,O-ligand systems were reported to undergo relatively fast decomposition due to hydrolysis. The biological activity of O,O- and S,O-chelates coordinated to the Ru(II) metal center was investigated recently,15−22 and some of the compounds were shown to be potent protein kinase inhibitors.(20)
Figure 1.
Structures of anticancer ruthenium complexes: RAPTA-C and [Ru(η6-biphenyl)Cl(en)]PF6 (en = ethylene-1,2-diamine).
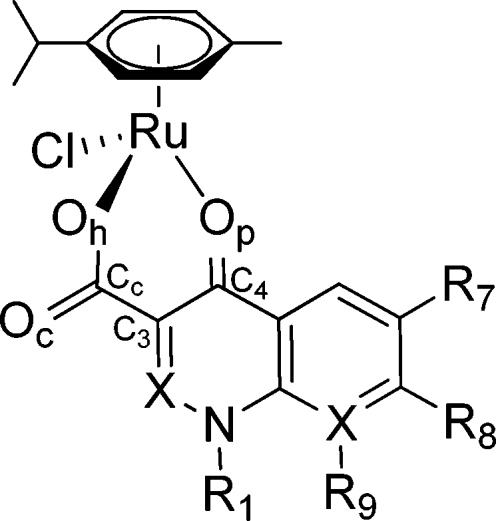
Quinolones are synthetic antibacterial agents, which are widely used in clinical practice. They are also suitable as ligands, featuring an O,O-chelate motif (Figure 2). Since the introduction of nalidixic acid into clinical use in 1962 more than 10 000 related compounds were synthesized and tested as potential antibacterial agents, and more than 30 were or still are in clinical or veterinary use.23−25 In addition to their antibacterial activity, they were also shown to exhibit tumor-inhibiting properties.26,27
Figure 2.
Chemical structures of the clinically applied quinolone antibacterials ofloxacin (oflo-H), nalidixic acid (nal-H), and cinoxacin (cin-H) used in this study.
The mechanism of action of quinolones is not yet fully understood. It is supposed that the quinolones bind to DNA, inhibiting bacterial topoisomerase and thus preventing the bacteria from replicating.23−25 The DNA interactions of quinolones28,29 and their metal complexes30−32 as well as their affinity to serum proteins(33) have been studied by applying different techniques. However, their mode of binding to DNA is uncertain. The quinolone molecule either forms hydrogen bonds to the nucleobases through the ketocarboxylate moiety or may bind to the phosphate backbone with the aid of magnesium ions that act as a bridge between the ketocarboxylate moiety and the DNA phosphates.(34) Recently, the crystal structure of a topoisomerase−DNA−quinolone complex was reported which shows that the magnesium ion is bidentately coordinated by the quinolone and four additional aqua ligands, which in turn form hydrogen bonds with DNA nucleobases.(35)
Since the approach to attach a bioactive ligand to a Ru(arene) moiety has been previously successfully used,36−38 and keeping in mind the various biological properties of quinolones, we have recently prepared the first organometallic ruthenium complex with ofloxacin (oflo-H; Figure 2) and studied its interactions with DNA.(39) Herein, we describe an extended study comprising the synthesis and characterization of Ru(arene) complexes of the first-generation quinolone agents nalidixic acid (nal-H) and cinoxacin (cin-H) and comparisons of these complexes to the analogous ofloxacin derivative with regard to stability in aqueous solution and reactivity toward the DNA model 5′-guanosine monophosphate (5′-GMP) and the serum transport protein human serum albumin (HSA) as well as anticancer activity in human tumor cell lines.
Experimental Section
Materials and Methods
The starting materials were purchased from Sigma-Aldrich and were used as received. All the solvents were of reagent grade and were purchased from Fluka. The synthesis of [(cym)RuCl(oflo)] (1) was performed as reported recently.(39)1H NMR spectra were recorded with Bruker Avance DPX 300 (at 29 °C and 300.13 MHz) or Avance III 500 spectrometers (at 25 °C and 500.10 MHz). 31P{1H} NMR spectra were recorded at 25 °C on the latter instrument at 161.98 MHz. Elemental analyses (C, H, N) were performed with a Perkin-Elmer 2400 Series II CHNS/O analyzer. Infrared spectra were recorded with a Perkin-Elmer Spectrum 100 FTIR spectrometer, equipped with a Specac Golden Gate Diamond ATR as a solid sample support. X-ray diffraction data (Supporting Information) for 2 and 3 were collected on a Nonius Kappa CCD difractometer at 293(2) K equipped with a Mo anode (Kα radiation, λ = 0.710 73 Å) and a graphite monochromator. The structures were solved by direct methods implemented in SIR92(40) and refined by a full-matrix least-squares procedure based on F2 using SHELXL-97.(41) All non-hydrogen atoms were refined anisotropically. The hydrogen atoms were either placed at calculated positions and treated using appropriate riding models or determined from the difference Fourier map. The programs Mercury(42) and ORTEP(43) were used for data analysis and figure preparation.
Synthesis
Chlorido(η6-p-cymene)(nalidixicato-κ2O,O)ruthenium(II) (2)
[(Cym)RuCl(μ-Cl)]2 (40.0 mg, 0.065 mmol) and nalidixic acid sodium salt hydrate (33.2 mg, 0.130 mmol) were dissolved in chloroform/methanol (1/1; 15 mL), and the reaction mixture was refluxed for 6 h. The obtained NaCl was removed by filtration through Celite, toluene (10 mL) was slowly added, and the solution was left in an open flask. Orange-brown crystals were obtained after 3 days at room temperature. The crystals were collected and washed with hexane. Yield: 50 mg, 65%.
1H NMR (CDCl3, 300.13 MHz): δ 9.02 (s, 1H, H2 nal), 8.67 (d, 3J(H,H) = 8 Hz, 1H, H6 nal), 7.34−7.13 (m, 1H, H-2 nal), 5.64−5.57 (m, 2H, Ar-H cym), 5.35−5.32 (m, 2H, Ar-H cym), 4.50 (q, 3J(H,H) = 7 Hz, 2H, NCH2CH3), 3.12−3.03 (m, 1H, Ar-CH(CH3)2 cym), 2.69 (s, 3H, Ar-CH3 cym), 2.35 (s, 3H, Ar-CH3 nal), 1.42 (t, 3J(H,H) = 7 Hz, 3H, NCH2CH3 nal), 1.39 (d, 3J(H,H) = 7 Hz, 6H, Ar-CH(CH3)2 cym). IR (cm−1, ATR): 3045, 2963, 1624, 1607, 1560, 1517, 1490, 1444, 1362, 1342, 1315, 1286, 1250, 1231, 1160, 1124, 1091, 1031, 895, 858, 805, 771, 736, 700, 670, 636. Anal. Calcd for C22H27ClN2O3Ru·C7H8: C, 58.43; H, 5.92; N, 4.70. Found: C, 58.63; H, 5.60; N, 4.72.
Chlorido(η6-p-cymene)(cinoxacinato-κ2O,O)ruthenium(II) (3)
[(Cym)RuCl(μ-Cl)]2 (40.0 mg, 0.065 mmol) and cinoxacin (34.2 mg, 0.130 mmol) were dissolved in chloroform/methanol (1/1; 15 mL). NaOMe (5.2 mg, 0.130 mmol) was added, and the reaction mixture was refluxed for 6 h. The obtained NaCl was removed by filtration through Celite, toluene (10 mL) was slowly added, and the solution was left in an open flask. Orange-brown crystals were obtained after 3 days at room temperature. The crystals were collected and washed with hexane. Yield: 42 mg, 55%.
1H NMR (CDCl3, 300.13 MHz): δ 7.69 (s, 1H, H5 cin), 6.94 (s, 1H, H8 cin), 6.20 (s, 2H, O−CH2O cin), 5.65−5.58 (m, 2H, Ar-H cym), 5.36−5.33 (m, 2H, Ar-H cym), 4.58 (q, 3J(H,H) = 7 Hz, 2H, NCH2CH3), 3.11−3.06 (m, 1H, Ar-CH(CH3)2 cym), 2.35 (s, 3H, Ar-CH3 cym), 1.48 (t, 3J(H,H) = 7 Hz, 3H, NCH2CH3 cin), 1.40 (d, 6H, 3J(H,H) = 7 Hz, Ar-CH(CH3)2 cym). IR (cm−1, ATR): 3538, 3436, 2970, 1620, 1517, 1494, 1471, 1461, 1277, 1241, 1159, 1124, 1086, 1034, 938, 899, 882, 868, 850, 812, 787, 747, 700, 666, 648, 610. Anal. Calcd for C22H23ClN2O5Ru·H2O·0.5C7H8: C, 51.39; H, 4.90; N, 4.70. Found: C, 51.40; H, 4.75; N, 4.74.
Aquation Experiments by Means of NMR Spectroscopy
For aquation studies, 1−3 (1−2 mg/mL) were dissolved in D2O and the samples were analyzed by 1H NMR spectroscopy immediately after dissolution and after 18 h.
pKa Determination
pKa values were determined by dissolving 1−3 in MeOD-d4/D2O (5/95). The pH values were measured directly in the NMR tubes with an Eco Scan pH6 pH meter equipped with a glass micro combination pH electrode (Orion 9826BN) and calibrated with standard buffer solutions of pH 4.00, 7.00, and 10.00. The pH titration was performed with NaOD (0.4−0.0004% in D2O) and DNO3 (0.4−0.0004% in D2O).
5′-GMP Interaction Study
5′-GMP binding experiments were carried out by titrating solutions of 1−3 (1−2 mg/mL) in D2O with a 5′-GMP solution (10 mg/mL D2O) in 50 μL increments. The reaction was monitored by 1H and 31P{1H} NMR spectroscopy until unreacted 5′-GMP was detected.
Aqueous Stability and Interactions with Human Serum Albumin
Instrumentation
Capillary zone electrophoresis (CZE) separations were carried out on an HP3D CE system (Agilent, Waldbronn, Germany) equipped with an on-column diode array detector. Detection was carried out either by UV (200 nm) or with an Agilent 7500ce inductively coupled plasma mass spectrometer (ICP-MS) interfaced to the CE system utilizing a CETAC CEI-100 microconcentric nebulizer. For all experiments with UV detection, capillaries of 48.5 cm total length (40 cm effective length; 50 μm i.d.) were used (Polymicro Technologies, Phoenix, AZ); in the case of ICP-MS detection (Supporting Information), the capillary length was extended to 60 cm. For the hydrolysis studies in water the capillary and sample tray were thermostated at 25 °C, whereas the HSA binding experiments were done at 37 °C. Injections were performed by applying a pressure of 25 mbar for 4 s and a constant voltage of 25 kV. Prior to the first use, the capillary was flushed at 1 bar with 0.1 M HCl, water, 0.1 M NaOH, and again with water (10 min each). Before each injection, the capillary was purged for 2 min with both water and the background electrolyte (BGE). The nebulizer was employed in self-aspiration mode with the sheath liquid closing the electrical circuit and spraying a fine aerosol. The working conditions were daily optimized using a 1 μg L−1 tuning solution containing 7Li, 89Y, and 205Tl in 2% HNO3. Doubly charged ions and oxide levels were minimized by using 140Ce and were typically <2.5%. To improve precision and to ensure interday reproducibility, the peak area responses of the two monitored Ru isotopes as well as of the 34S trace were normalized with the total ion current of the internal standard (72Ge). Analyses were only started if a sufficiently stable signal (RSD 72Ge <5%) was attained. The kinetics of the hydrolysis and the binding of the three compounds toward HSA were determined by monitoring the time-dependent changes in the peak area.
Reagents
Sodium hydroxide, sodium chloride, sodium dihydrogenphosphate, and sodium bicarbonate were obtained from Fluka (Buchs, Switzerland). Disodium hydrogenphosphate was purchased from Riedel-de Haen (Seelze, Germany), and 1,2-dibromoethane and human serum albumin (ca. 99%) were obtained from Sigma-Aldrich (Vienna, Austria). The ICP-MS tuning solution was from Agilent Technologies (Vienna, Austria) and the 72Ge standard from CPI international (Santa Rosa, CA). High-purity water used throughout this study was obtained from a Millipore Synergy 185 UV Ultrapure water system (Molsheim, France).
Sample Preparation
The hydrolysis studies were done in water at 25 °C, and solutions of the Ru complexes 1−3 were analyzed immediately and after 1 h and 1 and 2 days. In order to work at concentrations similar to those used in NMR investigations, 1 mM solutions of the complexes were prepared in water and diluted 1/10 with water before analysis by CZE-ICP-MS. Due to the poor aqueous solubility of the complexes, the dissolution was supported by ultrasonification for 15, 20, and 5 min for 1−3, respectively. For the in vitro protein binding studies, solutions containing 0.1 mM of the ruthenium compound and 0.05 mM of HSA in physiological buffer (25 mM NaHCO3, 4 mM NaHPO4, 100 mM NaCl) were prepared and incubated at 37 °C in order to simulate physiological conditions. The samples were analyzed by CZE-ICP-MS immediately after mixing and after 0.5, 1, and 1.5 h.
In Vitro Anticancer Activity
Cell Lines and Cell Culture Conditions
The human nonsmall cell lung carcinoma cell line A549 and colon adenocarcinoma cell line SW480 were kindly provided by Brigitte Marian (Institute of Cancer Research, Department of Medicine I, Medical University of Vienna, Vienna, Austria). CH1 cells (ovarian cancer, human) were a gift from Lloyd R. Kelland (CRC Centre for Cancer Therapeutics, Institute of Cancer Research, Sutton, U.K.). Cells were grown in 75 cm2 culture flasks (Iwaki/Asahi Technoglass, Gyouda, Japan) in complete medium [Minimum Essential Medium supplemented with 10% heat-inactivated fetal bovine serum, 1 mM sodium pyruvate, 4 mM l-glutamine, and 1% nonessential amino acids (100×)] as adherent monolayer cultures. All media and supplements were purchased from Sigma-Aldrich, Vienna, Austria. Cultures were maintained at 37 °C under a humidified atmosphere containing 5% CO2 and 95% air.
MTT Assay
Cytotoxicity was determined by a colorimetric microculture assay (MTT assay, MTT = 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide, Fluka). For this purpose, cells were harvested from culture flasks by use of trypsin and seeded in 100 μL per well into 96-well plates (Iwaki/Asahi Technoglass, Gyouda, Japan) in cell densities of 4 × 103 (A549), 1.5 × 103 (CH1), and 2.5 × 103 (SW480) cells per well, respectively. These cell numbers ensure exponential growth of untreated controls throughout drug exposure of treated microcultures. Cells were allowed to adhere and resume proliferation in drug-free complete culture medium for 24 h. Drugs were dissolved in complete medium and appropriately diluted, and instantly 100 μL of the drug dilutions were added per well. After exposure for 96 h at 37 °C and 5% CO2, drug solutions were replaced by 100 μL/well RPMI 1640 culture medium (supplemented with 10% heat-inactivated fetal bovine serum and 4 mM l-glutamine) plus 20 μL/well MTT solution in phosphate-buffered saline (5 mg/mL) and incubated for 4 h. Subsequently, the medium/MTT mixture was removed and the formazan crystals that were formed in vital cells were dissolved in 150 μL of DMSO (dimethyl sulfoxide) per well. Optical densities were measured with a microplate reader (Tecan Spectra Classic) at 550 nm (and a reference wavelength of 690 nm) to yield relative quantities of viable cells as percentages of untreated controls, and 50% inhibitory concentrations (IC50) were calculated from concentration−effect curves by interpolation. Calculations are based on at least three independent experiments, each consisting of three replicates per concentration level.
Results and Discussion
The combination of biologically active precursors with metals is a promising strategy to develop new anticancer agents.7,44−46 In a recent study, the antibacterial quinolone ofloxacin was used as a bidentate chelating ligand to form a Ru(cym) complex.(39) In an attempt to extend the series of compounds the quinolones nalidixic acid and cinoxacin were included into this study. The synthesis of 2 and 3 differs slightly from that of 1, which is obtained by reaction of the precursor [(cym)RuCl(μ-Cl)]2 with ofloxacin and NaOH in MeOH. In contrast, 2 was prepared in chloroform/methanol (1/1) by reaction of the sodium salt of nalidixic acid (nal-Na), due to the commercial availability of the latter (Scheme 1). Compound 3 was synthesized in dry methanol/chloroform (1:1) by addition of sodium methoxide, in order to avoid aquation in the reaction mixture. In all cases, the solvent system was evaporated from the reaction mixture, the crude product was dissolved in dichloromethane, and insoluble NaCl was removed by filtration over Celite. Finally, toluene was added to aid crystallization, which yielded crystals suitable for X-ray diffraction analysis.
Scheme 1. Synthetic Pathways Applied to Prepare 1−3.
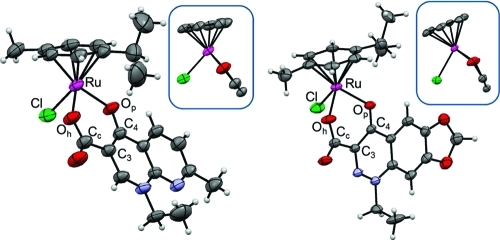
The molecular structures of the Ru(cym) complexes 2 and 3 adopt a pseudo-octahedral “piano-stool” geometry, which is typical for this compound class, with ruthenium(II) π-bonded to the p-cymene ring and σ-bonded to a chloride as well as the pyridone and carboxylato oxygen atoms of the chelating quinolone ligands (Figure 3, Table 1, and the Supporting Information). In case of 1 and 3, the Ru atom is located out of the plane of the six-membered chelate ring with Ru−centroidO,O−C3 angles of 151.72 and 151.39°, respectively, whereas this kinking is only marginal in the structure of 2 (175.72°). The unit cell of 2 contains a toluene solvate, while 3 cocrystallized with a water molecule, which forms a hydrogen bond with the carbonyl oxygen (d(Oc−Owater) = 2.917 Å). The Ru−O (Ru−Op, Ru−Oh) distances in both 2 and 3 as in the structure of 1 range from 2.070 to 2.099 Å, while the Ru−Cl bond is longer (2.415−2.418 Å).(39) In contrast to the case for 1, the molecules in the structures of 2 and 3 show distinct π-interactions between the quinolone ligands with a more stacked arrangement in the case of 2 as compared to 3 (Supporting Information). The difference Fourier map of 3 showed high residual density peaks which could not be refined due to high disorder and partial occupancy. In the final model, the scattering contributions were removed from all of these diffuse moieties using the SQUEEZE routine in PLATON.(47) A potential solvent-accessible volume of 192.5 Å3 was found, which corresponds to the volume of a small organic molecule such as toluene. 1H NMR spectroscopy of a crystal sample revealed peaks corresponding to toluene protons, which confirms the used model. The O−Ru−Cl and O−Ru−O angles are between 83.73 and 87.30°. This is in the same range as for other organometallic ruthenium compounds bearing maltolato ligands, although six-membered chelate ring systems are present in 2 and 3, whereas five-membered rings are formed with maltol.(20)
Figure 3.
X-ray structures of 2 (left) and 3 (right). The insets show the Ru atom and its coordinated ligands along the plane of the chelating ring system. The thermal ellipsoids are shown at the 50% probability level, and the solvent molecules are omitted for clarity.
Table 1. X-ray Structure Numbering Scheme and Selected Bond Lengths (Å) and Angles (deg) for 1−3.
| bond length/angle | 1·2.8H2O(39) | 2·C7H8 | 3·H2O |
|---|---|---|---|
| Ru−Op | 2.0713(18) | 2.0866(19) | 2.099(2) |
| Ru−Oh | 2.069(2) | 2.070(2) | 2.071(2) |
| Ru−Cl | 2.4183(7) | 2.4155(10) | 2.4153(8) |
| Ru−cymcentroid | 1.6345(14) | 1.6421(15) | 1.644(3) |
| Op−C4 | 1.275(3) | 1.273(3) | 1.272(3) |
| Cc−Oh | 1.293(3) | 1.276(4) | 1.280(3) |
| Cc−Oc | 1.232(4) | 1.226(4) | 1.226(4) |
| Oh−Ru−Op | 85.30(7) | 87.30(8) | 84.94(8) |
| Op−Ru−Cl | 86.92(6) | 84.73(7) | 84.95(6) |
| Oh−Ru−Cl | 83.73(6) | 85.63(8) | 86.89(7) |
Behavior and Stability in Aqueous Solution
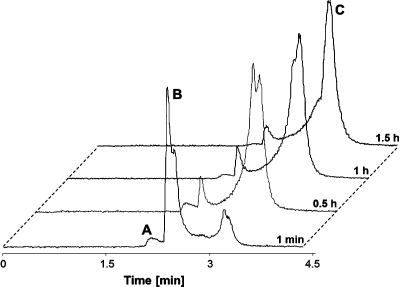
Aquation of Ru(arene) complexes is supposed to be an essential step for activation of these compounds.(48) Replacing the chlorido ligand by a water molecule leads to the more reactive aqua species, which can react with biological target molecules. The stability of these aqua species constitutes an essential requirement for pharmaceutical formulation and subsequently also intravenous administration of the substance. The aqueous stability of 1−3 was studied by means of 1H NMR spectroscopy (Supporting Information) and CZE with spectrophotometric and ICP-MS detection (Table 2). All of the complexes undergo a quick first hydrolytic step by releasing the chlorido ligand and filling the coordination sphere with a water molecule. The stability of the aqua species differs significantly among the three compounds. The aqua species of 1 decomposes slowly over 24 h, resulting in more than 40% of free ligand and the previously reported hydrolytic product [(cym)Ru(μ-OH)3Ru(cym)]+,16,17,39,49 whereas complexes 2 and 3 were found to possess a higher stability, with only a small fraction of the ligand dissociating. The hydrolyzed fraction was determined by 1H NMR spectroscopy, comparing the peak area of the aromatic hydrogen atoms of cymene (δ 5.6 and 6.0 ppm) and of the hydroxido-bridged dimer (δ 5.2 and 5.5 ppm), and by a complementary CZE-ICP-MS study. The migration behavior of the two Ru species in the CZE mode is an indication of the charge of the analyte. The electroosmotic flow (EOF), and therewith neutral species in the sample, was marked by adding 1,2-dibromoethane (Figure 4). The peak migrating with higher velocity is a positively charged species, most probably the highly stable dinuclear complex [Ru2(cym)2(OH)3]+. The second peak is a negatively charged complex, possibly a carbonate adduct formed by replacement of the chlorido/aqua ligand due to the use of carbonate buffer as BGE. The behavior in aqueous solution was monitored for 2 days, but no additional peaks were detected.
Table 2. Stability (%) of Compounds 1−3 as Determined by 1H NMR and CZE-ICP-MSa.
The values represent the amount of aqua complex in the solution.
Determined after 18 or 24 h incubation.
Figure 4.
CZE-ICP-MS electropherogram of 1 in water at 25 °C after 1 h. Shown are the traces of 102Ru and 79Br (1,2-dibromoethane as EOF marker). Peak identifications: (A) hydrolysis product [Ru2(cym)2(OH)3]+; (B) [Ru(cym)(CO3)(oflo)]−.
The pKa values of the aquated Ru(arene) complexes can be determined by stepwise titration of the primary hydrolysis products and analysis by 1H NMR spectroscopy. In this case, fast decomposition of the compounds was observed under alkaline conditions (pD >9). The major products were identified as the released ligand and dimeric [Ru2(cym)2(OD)3]+. Similar observations were made by CZE with spectrophotometric detection. Accordingly, the pKa values of 1−3 can only be estimated from the amount of hydroxido species formed at pD <9, which indicates pKa values greater than 8.5. This fact confirms that the complexes are present as reactive aqua species under physiological conditions.
Interactions with Human Serum Albumin (HSA) and the DNA Model 5′-GMP
As the most abundant protein in the circulatory system, HSA plays an important role in the binding and delivery of many pharmaceuticals to sites of disease.(50) Ruthenium complexes such as KP1019 and NAMI-A have a high affinity for HSA and other serum proteins,51,52 which may also contribute to the selective accumulation of ruthenium complexes within tumor cells.(5)
The binding kinetics for the reactions of the three ruthenium compounds with HSA were characterized by CZE-ICP-MS. Electropherograms illustrating the interaction with HSA were recorded immediately after mixing HSA and the complex and after 0.5, 1, and 1.5 h of incubation. Quantification of non-metal-containing proteins by ICP-MS is only feasible via determination of the sulfur content (the amino acids methionine and cysteine are present in many proteins).(53) The results demonstrate that the protein binding occurs rapidly, as indicated by a fast disappearance of the peaks of the unbound complex (Figure 5). The binding kinetics for the ruthenium complexes are rather similar, and 90% of the total ruthenium content is bound to HSA within 20 min (Supporting Information). Only two minor additional peaks which do not correspond to HSA-bound ruthenium but might be attributable to hydrolysis products were observed in the 102Ru trace of the electropherogram (Figure 5).
Figure 5.
Electropherograms illustrating the kinetics of the interaction of 3 with HSA by monitoring the 102Ru signal at different incubation times, normalized for the migration of 3 and HSA: (A) hydrolysis product; (B) 3; (C) HSA adduct.
DNA is one of the potential biological targets for metal-based anticancer drugs. Sadler et al. proposed that Ru complexes, such as [Ru(η6-biphenyl)Cl(en)]+ (see Figure 1), initially bind to the phosphate backbone of DNA, followed by rearrangement to adducts with softer nucleobase donor atoms.(14) Similarly, the interactions between 1 and DNA appear to be of an electrostatic kind initially, but the exact binding mode was not determined.(39) Furthermore, competitive DNA binding experiments revealed that 1 prevents the binding of cisplatin and vice versa, indicating either competition for the same binding sites or major conformational changes of the macromolecule.
In order to determine a possible binding site on DNA, 1−3 were reacted with 5′-GMP (used as a model for DNA binding), and these interactions were characterized by means of 1H and 31P{1H} NMR spectroscopy. Aqua species of 1−3 are formed immediately after dissolution (see above), and they react quickly and selectively with N7 of 5′-GMP, as indicated by a shift of the H8 proton signal.(54) However, it cannot be excluded that the first interaction occurs via the phosphate backbone, followed by the formation of a covalent bond to N7. Furthermore, the reaction of metal ions with isolated nucleotides is not necessarily comparable to that with the macromolecule DNA.(55)
In Vitro Anticancer Activity
Preliminary cytotoxicity experiments with rat skeletal myoblasts in vitro were previously reported for 1, but no significant activity was observed.(39) In this study, the biological activities of 1 and of the related quinolone complexes 2 and 3 were studied more in detail. The in vitro anticancer activities of compounds 1−3 were determined in human A549 (nonsmall cell lung carcinoma), CH1 (ovarian carcinoma), and SW480 (colon carcinoma) cells by means of the colorimetric MTT assay and compared to the tumor-inhibiting properties of the respective ligands (Table 3, Figure 6). CH1 cells were found to be about 10 times more sensitive to 1 (IC50 18 μM) than the intrinsically resistant SW480 cells (IC50 225 μM), whereas the complex does not show marked activity in A549 cells, which is reflected in an IC50 value higher than 320 μM. Moreover, 2 and 3 and all the ligands are inactive in the three cell lines, resulting in IC50 values higher than 320 μM. Even though the compounds were shown to be mostly noncytotoxic to the various cell lines, this is not necessarily a negative property for an anticancer drug candidate. The mechanisms of anticancer activity of ruthenium compounds are still not fully understood, and the example of NAMI-A, which is noncytotoxic in vitro but exhibits a high activity against metastases in vivo, has shown that mere IC50 values are not a sufficient reason to discard a compound as a potential drug candidate.2,56,57
Table 3. Cytotoxicity of 1−3 and the Respective Ligands in Human A549, CH1, and SW480 Cancer Cell Linesa.
| IC50 [μM] |
|||
|---|---|---|---|
| compd | A549 | CH1 | SW480 |
| oflo-H | >320 | >320 | >320 |
| 1 | >320 | 18 ± 7 | 225 ± 39 |
| nal-H | >320 | >320 | >320 |
| 2 | >320 | >320 | >320 |
| cin-H | >320 | >320 | >320 |
| 3 | >320 | >320 | >320 |
Presented are the 50% inhibitory concentrations obtained by the MTT assay. Values are the means ± standard deviations obtained from at least three independent experiments using exposure times of 96 h.
Figure 6.
Concentration−effect curves of 1 in human CH1 and SW480 cancer cells. Values were obtained by the MTT assay and are means ± standard deviations from at least three independent experiments using exposure times of 96 h.
It is known that fluoroquinolone antibiotics are not completely devoid of cytotoxicity in mammalian cells, but due to their rather low potency the side effects of anti-infective treatment are usually tolerable. Although the cytotoxicity of ofloxacin in vitro has been reported to be somewhat lower than that of the more commonly used ciprofloxacin, IC50 values in the 10−4 molar range in bladder and lung cancer cell lines can be inferred from literature data.58−60 Still, the cytotoxicity of the organometallic ruthenium complex 1 with ofloxacin in CH1 ovarian cancer cells (IC50 = 18 μM) is remarkable and is not paralleled by the two analogues with other quinolone ligands. Which of the structural features account for the observed differences in cytotoxicity remains unclear and requires more extensive structure−activity relationship studies. In analogy to their mechanism of action in bacteria, inhibition of topoisomerase II with the consequence of DNA cleavage is considered the primary cellular effect of ofloxacin and other fluoroquinolones, with prokaryotic topoisomerase being more sensitive by orders of magnitude,(61) and ofloxacin was shown to synergize with the topoisomerase II inhibitor doxorubicin in bladder cancer cells in vitro.(59) However, very little topoisomerase II inhibition was observed in an electrophoretic assay with plasmid DNA, no matter whether ofloxacin or complex 1 was applied, whereas 1 yielded DNA interactions in addition to those observed with ofloxacin alone, most likely as a result of ruthenium binding to DNA.(39)
Conclusions
In this paper the synthesis of organometallic Ru(cym) complexes with quinolone ligands is reported. In addition to ofloxacin, the antibacterials nalidixic acid and cinoxacin were selected as ligands to prepare 1−3. The structures were established by X-ray diffraction analysis and spectroscopic methods. Furthermore, they were characterized with regard to drug-like properties, such as stability in aqueous solution and reaction-with biological target molecules, as well as their tumor-inhibiting potential in a cancer cell line panel. NMR and CZE-ICP-MS studies revealed that the compounds undergo a quick activation step by releasing the chlorido ligand and replacing it with a labile water molecule that allows the compounds to readily interact with target molecules, such as blood proteins and DNA. Compounds 2 and 3 are more stable in aqueous solution than 1; however, all react with similar kinetics with HSA. The binding to 5′-GMP occurs via its nucleophilic N7, but a preassociation via the negatively charged phosphate backbone of DNA cannot be excluded. In an anticancer assay in vitro, only the ofloxacin derivative 1 was active in relevant concentrations in two cell lines.
Further work will be directed toward the development of new complexes with ligands of the quinolone family in order to elucidate the influence of the structure and substitution pattern of the ligands on the anticancer activity of the compound class. Additional studies will aim to enlighten the potential of such compounds, comprising the influence on cancer cell adhesion, migration, and invasion and the use in alternative anticancer treatment approaches, such as electrochemotherapy.62−65
Acknowledgments
We are grateful for financial support from a bilateral Slovenian-Austrian project of the Slovenian Research Agency (ARRS; J1-0200-0103-008) and for a junior researcher grant for J.K. from the ARRS. We acknowledge the support of the Austrian Science Foundation (FWF; I496-B11), the Mahlke-Obermann Foundation, the Hochschuljubiläumsstiftung Vienna, and the Austrian Exchange Service (ÖAD). The project was also supported by COST D39, in particular by a short-term scientific mission (STSM D39-6067) for J.K. We thank Franc Perdih and Andrej Pevec for help and advice in crystal structure solution and refinement and Matthias Skocic for preliminary cytotoxicity tests.
Supporting Information Available
Figures, tables, and CIF files giving X-ray diffraction data for compounds 2 and 3, 1H NMR spectra, the CZE-ICP-MS operational parameters, and HSA binding data. This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Material
References
- Heffeter P.; Jungwirth U.; Jakupec M.; Hartinger C.; Galanski M.; Elbling L.; Micksche M.; Keppler B.; Berger W. Drug Res. Upd. 2008, 11, 1–16. [DOI] [PubMed] [Google Scholar]
- Rademaker-Lakhai J. M.; Van Den Bongard D.; Pluim D.; Beijnen J. H.; Schellens J. H. M. Clin. Cancer Res. 2004, 10, 3717–3727. [DOI] [PubMed] [Google Scholar]
- Depenbrock H.; Schmelcher S.; Peter R.; Keppler B. K.; Weirich G.; Block T.; Rastetter J.; Hanauske A. R. Eur. J. Cancer 1997, 33, 2404–2410. [DOI] [PubMed] [Google Scholar]
- Kapitza S.; Pongratz M.; Jakupec M. A.; Heffeter P.; Berger W.; Lackinger L.; Keppler B. K.; Marian B. J. Cancer Res. Clin. Oncol. 2005, 131, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartinger C. G.; Zorbas-Seifried S.; Jakupec M. A.; Kynast B.; Zorbas H.; Keppler B. K. J. Inorg. Biochem. 2006, 100, 891–904. [DOI] [PubMed] [Google Scholar]
- Hartinger C. G.; Jakupec M. A.; Zorbas-Seifried S.; Groessl M.; Egger A.; Berger W.; Zorbas H.; Dyson P. J.; Keppler B. K. Chem. Biodiversity 2008, 5, 2140–2155. [DOI] [PubMed] [Google Scholar]
- Hartinger C. G.; Dyson P. J. Chem. Soc. Rev. 2009, 38, 391–401. [DOI] [PubMed] [Google Scholar]
- Allardyce C. S.; Dyson P. J.; Ellis D. J.; Heath S. L. Chem. Commun. 2001, 1396–1397. [Google Scholar]
- Scolaro C.; Bergamo A.; Brescacin L.; Delfino R.; Cocchietto M.; Laurenczy G.; Geldbach T. J.; Sava G.; Dyson P. J. J. Med. Chem. 2005, 48, 4161–4171. [DOI] [PubMed] [Google Scholar]
- Dyson P. J. Chimia 2007, 61, 698–703. [Google Scholar]
- Chatterjee S.; Kundu S.; Bhattacharyya A.; Hartinger C. G.; Dyson P. J. J. Biol. Inorg. Chem. 2008, 13, 1149–1155. [DOI] [PubMed] [Google Scholar]
- Morris R. E.; Aird R. E.; Murdoch Pdel S.; Chen H.; Cummings J.; Hughes N. D.; Parsons S.; Parkin A.; Boyd G.; Jodrell D. I.; Sadler P. J. J. Med. Chem. 2001, 44, 3616–21. [DOI] [PubMed] [Google Scholar]
- Chen H.; Parkinson J. A.; Parsons S.; Coxall R. A.; Gould R. O.; Sadler P. J. J. Am. Chem. Soc. 2002, 124, 3064–3082. [DOI] [PubMed] [Google Scholar]
- Chen H.; Parkinson J. A.; Morris R. E.; Sadler P. J. J. Am. Chem. Soc. 2003, 125, 173–186. [DOI] [PubMed] [Google Scholar]
- Mendoza-Ferri M. G.; Hartinger C. G.; Eichinger R. E.; Stolyarova N.; Jakupec M. A.; Nazarov A. A.; Severin K.; Keppler B. K. Organometallics 2008, 27, 2405–2407. [Google Scholar]
- Kandioller W.; Hartinger C. G.; Nazarov A. A.; Bartel C.; Skocic M.; Jakupec M. A.; Arion V. B.; Keppler B. K. Chem. Eur. J. 2009, 15, 12283–12291. [DOI] [PubMed] [Google Scholar]
- Kandioller W.; Hartinger C. G.; Nazarov A. A.; Kuznetsov M. L.; John R. O.; Bartel C.; Jakupec M. A.; Arion V. B.; Keppler B. K. Organometallics 2009, 28, 4249–4251. [Google Scholar]
- Mendoza-Ferri M. G.; Hartinger C. G.; Nazarov A. A.; Eichinger R. E.; Jakupec M. A.; Severin K.; Keppler B. K. Organometallics 2009, 28, 6260–6265. [Google Scholar]
- Nováková O.; Nazarov A. A.; Hartinger C. G.; Keppler B. K.; Brabec V. Biochem. Pharmacol. 2009, 77, 364–374. [DOI] [PubMed] [Google Scholar]
- Hanif M.; Henke H.; Meier S. M.; Martic S.; Labib M.; Kandioller W.; Jakupec M. A.; Arion V. B.; Kraatz H. B.; Keppler B. K.; Hartinger C. G. Inorg. Chem. 2010, 49, 7953–7963. [DOI] [PubMed] [Google Scholar]
- Hanif M.; Schaaf P.; Kandioller W.; Hejl M.; Jakupec M. A.; Roller A.; Keppler B. K.; Hartinger C. G. Aust. J. Chem. 2010, 63, 1521–1528. [Google Scholar]
- Kasser J. H.; Kandioller W.; Hartinger C. G.; Nazarov A. A.; Arion V. B.; Dyson P. J.; Keppler B. K. J. Organomet. Chem. 2010, 695, 875–881. [Google Scholar]
- Andriole V. T., Ed. The Quinolones., 3rd ed.; Academic Press: San Diego, CA, 2000; p 517. [Google Scholar]
- Hooper D., Rubinstein E., Eds. Quinolone Antimicrobial Agents., 3rd ed.; ASM Press: Washington, DC, 2003; p 485. [Google Scholar]
- Ronald A. R., Low D. E., Eds. Fluoroquinolone Antibiotics; Birkhäuser: Basel, Switzerland, 2003; p 250. [Google Scholar]
- Kwok Y.; Sun D.; Clement J. J.; Hurley L. H. Anti-Cancer Drug Des. 1999, 14, 443–50. [PubMed] [Google Scholar]
- Yu H.; Kwok Y.; Hurley L. H.; Kerwin S. M. Biochemistry 2000, 39, 10236–46. [DOI] [PubMed] [Google Scholar]
- Vilfan I. D.; Drevensek P.; Turel I.; Poklar Ulrih N. Biochim. Biophys. Acta, Gene Struct. Expression 2003, 1628, 111–122. [DOI] [PubMed] [Google Scholar]
- Drevensek P.; Turel I.; Poklar Ulrih N. J. Inorg. Biochem. 2003, 96, 407–415. [DOI] [PubMed] [Google Scholar]
- Turel I. Coord. Chem. Rev. 2002, 232, 27–47. [Google Scholar]
- Drevensek P.; Kosmrlj J.; Giester G.; Skauge T.; Sletten E.; Sepcic K.; Turel I. J. Inorg. Biochem. 2006, 100, 1755–1763. [DOI] [PubMed] [Google Scholar]
- Drevensek P.; Ulrih N. P.; Majerle A.; Turel I. J. Inorg. Biochem. 2006, 100, 1705–1713. [DOI] [PubMed] [Google Scholar]
- Skyrianou K. C.; Perdih F.; Turel I.; Kessissoglou D. P.; Psomas G. J. Inorg. Biochem. 2010, 104, 161–170. [DOI] [PubMed] [Google Scholar]
- Mitscher L. A. Chem. Rev. 2005, 105, 559–592. [DOI] [PubMed] [Google Scholar]
- Wohlkonig A.; Chan P. F.; Fosberry A. P.; Homes P.; Huang J.; Kranz M.; Leydon V. R.; Miles T. J.; Pearson N. D.; Perera R. L.; Shillings A. J.; Gwynn M. N.; Bax B. D. Nat. Struct. Mol. Biol. 2010, 17, 1152–1153. [DOI] [PubMed] [Google Scholar]
- Vock C. A.; Ang W. H.; Scolaro C.; Phillips A. D.; Lagopoulos L.; Juillerat-Jeanneret L.; Sava G.; Scopelliti R.; Dyson P. J. J. Med. Chem. 2007, 50, 2166–2175. [DOI] [PubMed] [Google Scholar]
- Ang W. H.; De Luca A.; Chapuis-Bernasconi C.; Juillerat-Jeanneret L.; Lo Bello M.; Dyson P. J. ChemMedChem 2007, 2, 1799–1806. [DOI] [PubMed] [Google Scholar]
- Mulcahy S.; Meggers E., Organometallics as Structural Scaffolds for Enzyme Inhibitor Design. In Medicinal Organometallic Chemistry; Jaouen G., Metzler-Nolte N., Eds.; Springer: Berlin/Heidelberg, 2010; Vol. 32, pp 141−153. [Google Scholar]
- Turel I.; Kljun J.; Perdih F.; Morozova E.; Bakulev V.; Kasyanenko N.; Byl J. A. W.; Osheroff N. Inorg. Chem. 2010, 49, 10750–10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altomare A.; Burla M. C.; Camalli M.; Cascarano G. L.; Giacovazzo C.; Guagliardi A.; Moliterni A. G. G.; Polidori G.; Spagna R. J. Appl. Crystallogr. 1999, 32, 115–119. [Google Scholar]
- Sheldrick G. M. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008, A64, 112–122. [DOI] [PubMed] [Google Scholar]
- Macrae C. F.; Edgington P. R.; McCabe P.; Pidcock E.; Shields G. P.; Taylor R.; Towler M.; van de Streek J. J. Appl. Crystallogr. 2006, 39, 453–457. [Google Scholar]
- Farrugia L. J. J. Appl. Crystallogr. 1997, 30, 565. [Google Scholar]
- Debreczeni J. É.; Bullock A. N.; Atilla G. E.; Williams D. S.; Bregman H.; Knapp S.; Meggers E. Angew. Chem., Int. Ed. Engl. 2006, 45, 1580–1585. [DOI] [PubMed] [Google Scholar]
- Nguyen A.; Vessieres A.; Hillard E. A.; Top S.; Pigeon P.; Jaouen G. Chimia 2007, 61, 716–724. [Google Scholar]
- Ott I.; Kircher B.; Bagowski C. P.; Vlecken D. H. W.; Ott E. B.; Will J.; Bensdorf K.; Sheldrick W. S.; Gust R. Angew. Chem., Int. Ed. Engl. 2009, 48, 1160–1163. [DOI] [PubMed] [Google Scholar]
- Van der Sluis P.; Spek A. L. Acta Crystallogr., Sect. A: Found. Crystallogr. 1990, A46, 194–201. [Google Scholar]
- Peacock A. F. A.; Sadler P. J. Chem. Asian J. 2008, 3, 1890–1899. [DOI] [PubMed] [Google Scholar]
- Peacock A. F. A.; Melchart M.; Deeth R. J.; Habtemariam A.; Parsons S.; Sadler P. J. Chem. Eur. J. 2007, 13, 2601–2613. [DOI] [PubMed] [Google Scholar]
- Timerbaev A. R.; Hartinger C. G.; Aleksenko S. S.; Keppler B. K. Chem. Rev. 2006, 106, 2224–2248. [DOI] [PubMed] [Google Scholar]
- Groessl M.; Reisner E.; Hartinger C. G.; Eichinger R.; Semenova O.; Timerbaev A. R.; Jakupec M. A.; Arion V. B.; Keppler B. K. J. Med. Chem. 2007, 50, 2185–2193. [DOI] [PubMed] [Google Scholar]
- Groessl M.; Hartinger C. G.; Polec-Pawlak K.; Jarosz M.; Keppler B. K. Electrophoresis 2008, 29, 2224–2232. [DOI] [PubMed] [Google Scholar]
- Groessl M.; Bytzek A.; Hartinger C. G. Electrophoresis 2009, 30, 2720–2727. [DOI] [PubMed] [Google Scholar]
- Dorcier A.; Hartinger C. G.; Scopelliti R.; Fish R. H.; Keppler B. K.; Dyson P. J. J. Inorg. Biochem. 2008, 102, 1066–1076. [DOI] [PubMed] [Google Scholar]
- Zorbas-Seifried S.; Hartinger C. G.; Meelich K.; Galanski M.; Keppler B. K.; Zorbas H. Biochemistry 2006, 45, 14817–14825. [DOI] [PubMed] [Google Scholar]
- Sava G.; Capozzi I.; Clerici K.; Gagliardi G.; Alessio E.; Mestroni G. Clin. Exp. Metastasis 1998, 16, 371–379. [DOI] [PubMed] [Google Scholar]
- Sava G.; Gagliardi R.; Cocchietto M.; Clerici K.; Capozzi I.; Marrella M.; Alessio E.; Mestroni G.; Milanino R. Pathol. Oncol. Res. 1998, 4, 30–36. [DOI] [PubMed] [Google Scholar]
- Seay T. M.; Peretsman S. J.; Dixon P. S. J. Urol. 1996, 155, 757–762. [PubMed] [Google Scholar]
- Kamat A. M.; DeHaven J. I.; Lamm D. L. Urology 1999, 54, 56–61. [DOI] [PubMed] [Google Scholar]
- Chen C.-Y.; Chen Q.-Z.; Wang X.-F.; Liu M.-S.; Chen Y.-F. Transition Met. Chem. 2009, 34, 757–763. [Google Scholar]
- Yamashita Y.; Ashizawa T.; Morimoto M.; Hosomi J.; Nakano H. Cancer Res. 1992, 52, 2818–22. [PubMed] [Google Scholar]
- Bicek A.; Turel I.; Kanduser M.; Miklavcic D. Bioelectrochemistry 2007, 71, 113–117. [DOI] [PubMed] [Google Scholar]
- Sersa G.; Miklavcic D.; Cemazar M.; Rudolf Z.; Pucihar G.; Snoj M. Eur. J. Surg. Oncol. 2008, 34, 232–40. [DOI] [PubMed] [Google Scholar]
- Hudej R.; Turel I.; Kanduser M.; Scancar J.; Kranjc S.; Sersa G.; Miklavcic D.; Jakupec M. A.; Keppler B. K.; Cemazar M. Anticancer Res. 2010, 30, 2055–2063. [PubMed] [Google Scholar]
- Kljun J.; Petricek S.; Zigon D.; Hudej R.; Miklavcic D.; Turel I.. Bioinorg. Chem. Appl. 2010, Article ID 183097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.